Is Carboxypeptidase B1 a Prognostic Marker for Ductal Carcinoma In Situ?
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Breast Tissue Samples Selection
2.2. Tissue Microarray (TMA) Preparation
2.3. Cell Lines
2.4. RNA Extraction and Quantitative Real-Time PCR (qPCR) Analysis
2.5. Quantitative PCR Was Performed Using SyBr Green Technology as Described Previously
2.6. Analysis of Public BC Datasets
2.7. Lentiviral Production and Infection
2.8. Proliferation
2.9. Migration
2.10. Spheroid Assay
2.11. Flag Immunoprecipitation (IP)
2.12. Western Blotting
2.13. Mass Spectrometry
2.14. IHC Analysis
2.15. Statistical Analysis
3. Results
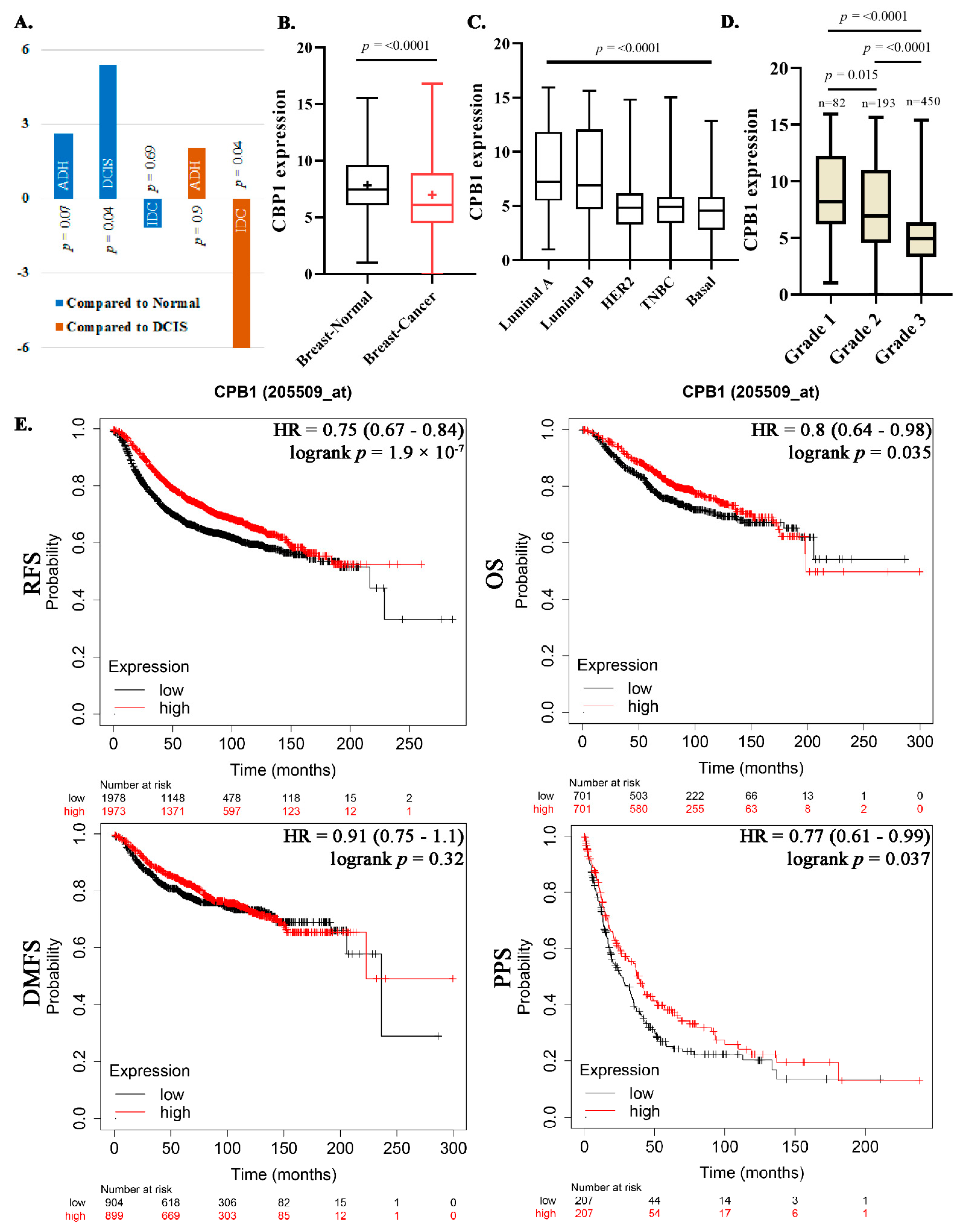
3.1. HTA Analysis Identified CPB1 as Specifically Expressed in DCIS
3.2. Association of CPB1 with Better Survival Outcomes
3.3. Validation of CPB1 Expression in DCIS
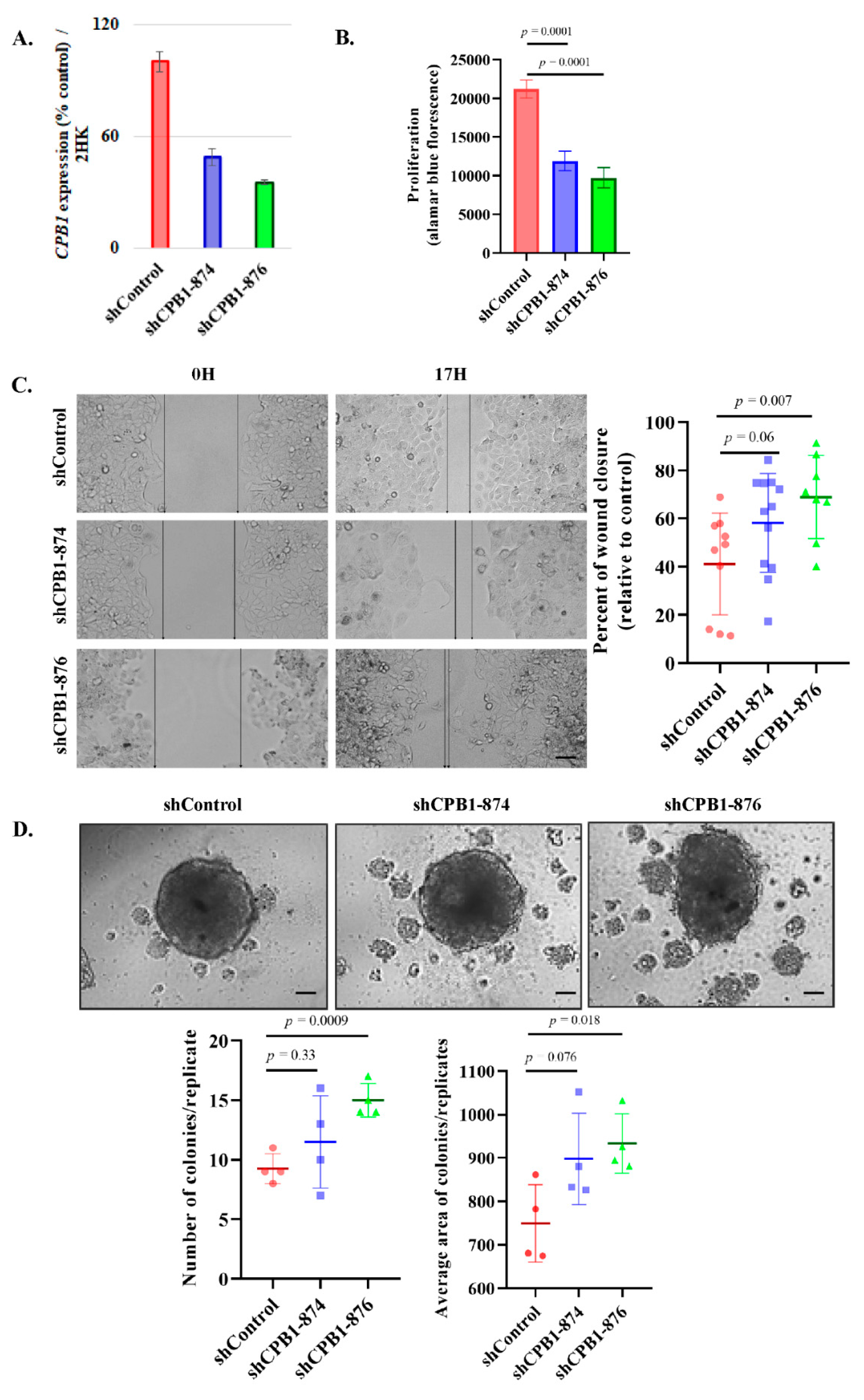
3.4. Knockdown of CPB1 Leads to an Increase in the Invasive Properties of the DCIS Cell Line
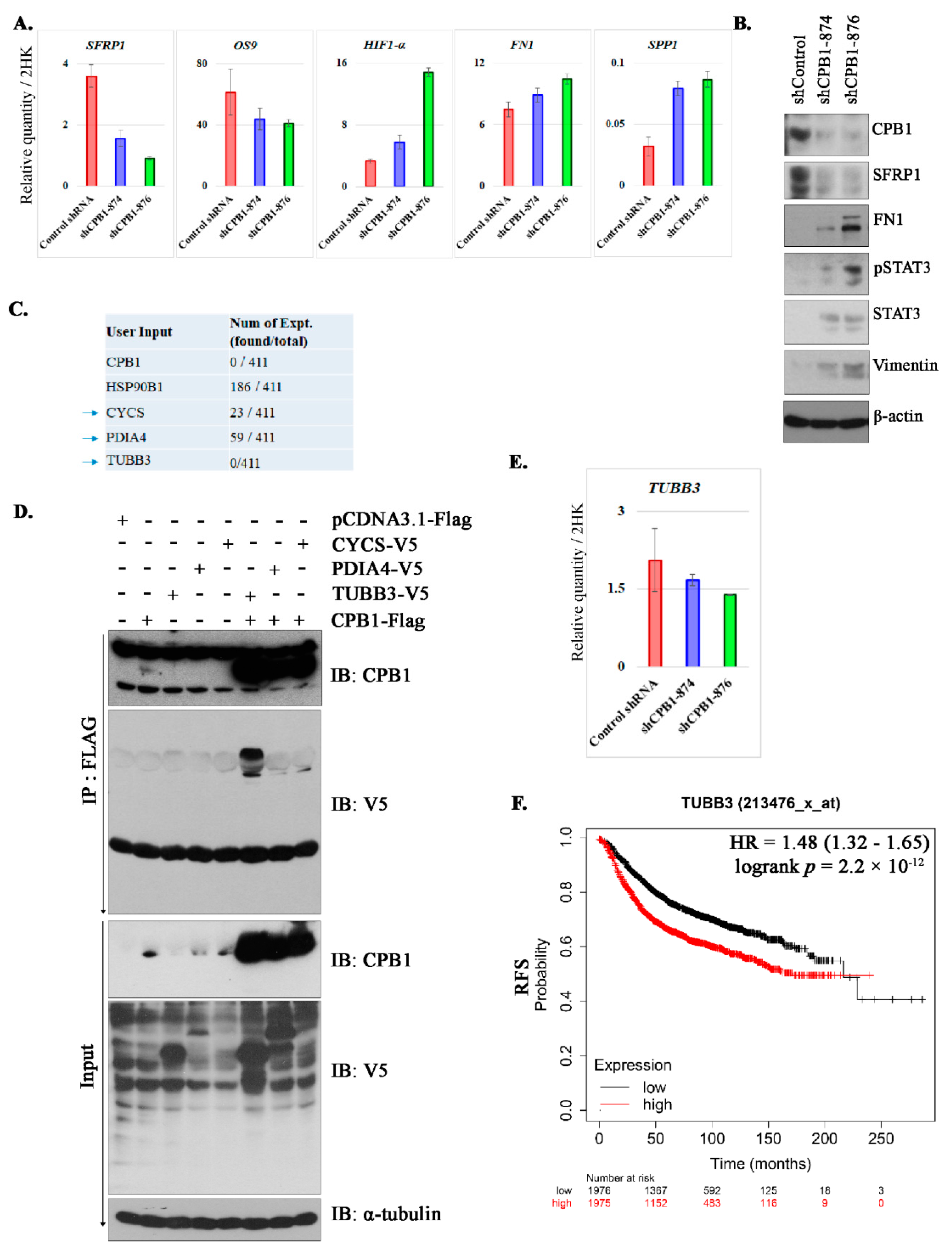
3.5. Changes in Signaling after Knockdown of CPB1 in MCF10DCIS.com Cell Line
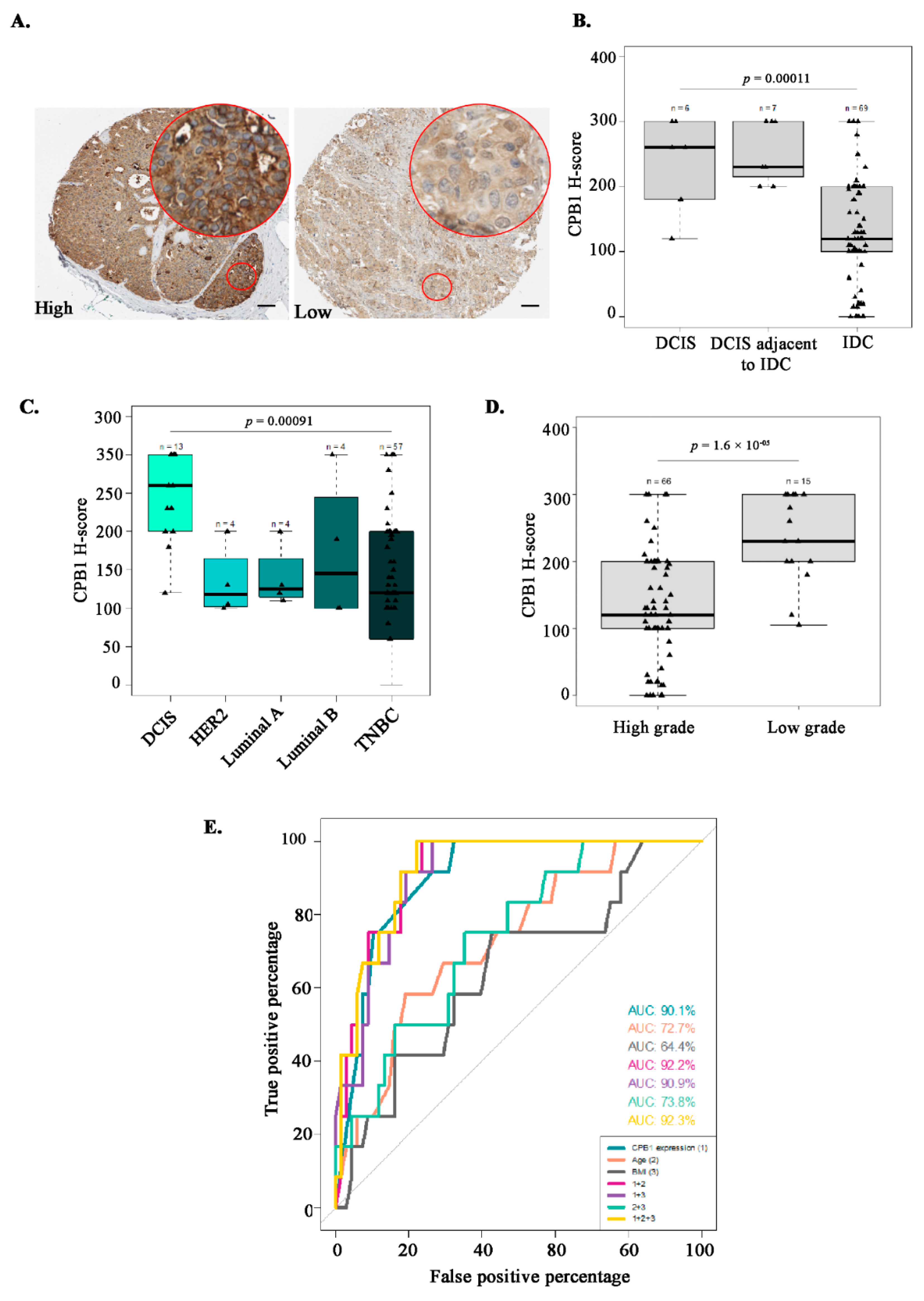
3.6. Characterization of the Study Population Based on CPB1 Expression by IHC
3.7. Association Studies Confirm CPB1 as a Predictive Marker of DCIS
4. Discussion
5. Conclusions
- It is important to validate our finding in a larger cohort comprising ADH, ADH adjacent to DCIS, DCIS, DCIS adjacent to IDC and IDC. This will be of great importance in the clinical setting as this could truly aid clinicians in both making priority decisions and follow-up of DCIS patients.
- It would be interesting to study whether cellular reprogramming during the involution process somehow triggers CPB1 expression leading to DCIS or tumorigenic changes due to DCIS leads to upregulation of CPB1.
- It would be worth investigating further the relationship of CPB1 expression with microcalcifications to understand if it is merely due to the presence of microcalcifications in DCIS or if CPB1 does have any role in microcalcifications formation.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kothari, C.; Diorio, C.; Durocher, F. Gene Signatures of Breast Cancer Development and the Potential for Novel Targeted Treatments. Pharmacogenomics 2020, 21, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Luzzi, V.; Holtschlag, V.; Watson, M.A. Expression Profiling of Ductal Carcinoma in Situ by Laser Capture Microdissection and High-Density Oligonucleotide Arrays. Am. J. Pathol. 2001, 158, 2005–2010. [Google Scholar] [CrossRef] [Green Version]
- Porter, D.A.; Krop, I.E.; Nasser, S.; Sgroi, D.; Kaelin, C.M.; Marks, J.R.; Riggins, G.; Polyak, K. A SAGE (Serial Analysis of Gene Expression) View of Breast Tumor Progression. Cancer Res. 2001, 61, 5697. [Google Scholar]
- Wulfkuhle, J.D.; Sgroi, D.C.; Krutzsch, H.; McLean, K.; McGarvey, K.; Knowlton, M.; Chen, S.; Shu, H.; Sahin, A.; Kurek, R.; et al. Proteomics of Human Breast Ductal Carcinoma in Situ. Cancer Res. 2002, 62, 6740–6749. [Google Scholar] [PubMed]
- Kothari, C.; Ouellette, G.; Labrie, Y.; Jacob, S.; Diorio, C.; Durocher, F. Identification of a Gene Signature for Different Stages of Breast Cancer Development That Could Be Used for Early Diagnosis and Specific Therapy. Oncotarget 2018, 9, 37407–37420. [Google Scholar] [CrossRef] [Green Version]
- Makki, J. Diversity of Breast Carcinoma: Histological Subtypes and Clinical Relevance. Clin. Med. Insights Pathol. 2015, 8, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burstein, H.J.; Polyak, K.; Wong, J.S.; Lester, S.C.; Kaelin, C.M. Ductal Carcinoma in Situ of the Breast. N. Engl. J. Med. 2004, 350, 1430–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshaies, I.; Provencher, L.; Jacob, S.; Côté, G.; Robert, J.; Desbiens, C.; Poirier, B.; Hogue, J.-C.; Vachon, E.; Diorio, C. Factors Associated with Upgrading to Malignancy at Surgery of Atypical Ductal Hyperplasia Diagnosed on Core Biopsy. Breast Edinb. Scotl. 2011, 20, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Hogue, J.-C.; Morais, L.; Provencher, L.; Desbiens, C.; Poirier, B.; Poirier, É.; Jacob, S.; Diorio, C. Characteristics Associated with Upgrading to Invasiveness after Surgery of a DCIS Diagnosed Using Percutaneous Biopsy. Anticancer Res. 2014, 34, 1183–1191. [Google Scholar]
- Nicosia, L.; di Giulio, G.; Bozzini, A.C.; Fanizza, M.; Ballati, F.; Rotili, A.; Lazzeroni, M.; Latronico, A.; Abbate, F.; Renne, G.; et al. Complete Removal of the Lesion as a Guidance in the Management of Patients with Breast Ductal Carcinoma In Situ. Cancers 2021, 13, 868. [Google Scholar] [CrossRef]
- Collins, L.C.; Aroner, S.A.; Connolly, J.L.; Colditz, G.A.; Schnitt, S.J.; Tamimi, R.M. Breast Cancer Risk by Extent and Type of Atypical Hyperplasia: An Update from the Nurses’ Health Studies. Cancer 2016, 122, 515–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, P.J.; Wolman, S.R.; Tait, L.; Heppner, G.H.; Miller, F.R. MCF10AT: A Model for the Evolution of Cancer from Proliferative Breast Disease. Am. J. Pathol. 1996, 148, 313–319. [Google Scholar] [PubMed]
- Santner, S.J.; Dawson, P.J.; Tait, L.; Soule, H.D.; Eliason, J.; Mohamed, A.N.; Wolman, S.R.; Heppner, G.H.; Miller, F.R. Malignant MCF10CA1 Cell Lines Derived from Premalignant Human Breast Epithelial MCF10AT Cells. Breast Cancer Res. Treat. 2001, 65, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Pouliot, M.-C.; Kothari, C.; Joly-Beauparlant, C.; Labrie, Y.; Ouellette, G.; Simard, J.; Droit, A.; Durocher, F. Transcriptional Signature of Lymphoblastoid Cell Lines of BRCA1, BRCA2 and Non-BRCA1/2 High Risk Breast Cancer Families. Oncotarget 2017, 8, 78691–78712. [Google Scholar] [CrossRef] [Green Version]
- Luu-The, V.; Paquet, N.; Calvo, E.; Cumps, J. Improved Real-Time RT-PCR Method for High-Throughput Measurements Using Second Derivative Calculation and Double Correction. BioTechniques 2005, 38, 287–293. [Google Scholar] [CrossRef]
- Narod, S.A.; Iqbal, J.; Giannakeas, V.; Sopik, V.; Sun, P. Breast Cancer Mortality After a Diagnosis of Ductal Carcinoma In Situ. JAMA Oncol. 2015, 1, 888–896. [Google Scholar] [CrossRef]
- Giannakeas, V.; Sopik, V.; Narod, S.A. Association of a Diagnosis of Ductal Carcinoma In Situ With Death From Breast Cancer. JAMA Netw. Open 2020, 3, e2017124. [Google Scholar] [CrossRef] [PubMed]
- Welch, H.G.; Black, W.C. Using Autopsy Series to Estimate the Disease “Reservoir” for Ductal Carcinoma in Situ of the Breast: How Much More Breast Cancer Can We Find? Ann. Intern. Med. 1997, 127, 1023–1028. [Google Scholar] [CrossRef]
- Betsill, W.L.J.; Rosen, P.P.; Lieberman, P.H.; Robbins, G.F. Intraductal Carcinoma. Long-Term Follow-up after Treatment by Biopsy Alone. JAMA 1978, 239, 1863–1867. [Google Scholar] [CrossRef]
- Rosen, P.P.; Braun, D.W.J.; Kinne, D.E. The Clinical Significance of Pre-Invasive Breast Carcinoma. Cancer 1980, 46, 919–925. [Google Scholar] [CrossRef]
- Page, D.L.; Dupont, W.D.; Rogers, L.W.; Landenberger, M. Intraductal Carcinoma of the Breast: Follow-up after Biopsy Only. Cancer 1982, 49, 751–758. [Google Scholar] [CrossRef]
- Cuzick, J.; Sestak, I.; Pinder, S.E.; Ellis, I.O.; Forsyth, S.; Bundred, N.J.; Forbes, J.F.; Bishop, H.; Fentiman, I.S.; George, W.D. Effect of Tamoxifen and Radiotherapy in Women with Locally Excised Ductal Carcinoma in Situ: Long-Term Results from the UK/ANZ DCIS Trial. Lancet Oncol. 2011, 12, 21–29. [Google Scholar] [CrossRef]
- Donker, M.; Litière, S.; Werutsky, G.; Julien, J.-P.; Fentiman, I.S.; Agresti, R.; Rouanet, P.; de Lara, C.T.; Bartelink, H.; Duez, N.; et al. Breast-Conserving Treatment with or without Radiotherapy in Ductal Carcinoma In Situ: 15-Year Recurrence Rates and Outcome after a Recurrence, from the EORTC 10853 Randomized Phase III Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 4054–4059. [Google Scholar] [CrossRef] [PubMed]
- Wapnir, I.L.; Dignam, J.J.; Fisher, B.; Mamounas, E.P.; Anderson, S.J.; Julian, T.B.; Land, S.R.; Margolese, R.G.; Swain, S.M.; Costantino, J.P.; et al. Long-Term Outcomes of Invasive Ipsilateral Breast Tumor Recurrences after Lumpectomy in NSABP B-17 and B-24 Randomized Clinical Trials for DCIS. J. Natl. Cancer Inst. 2011, 103, 478–488. [Google Scholar] [CrossRef]
- Wärnberg, F.; Garmo, H.; Emdin, S.; Hedberg, V.; Adwall, L.; Sandelin, K.; Ringberg, A.; Karlsson, P.; Arnesson, L.-G.; Anderson, H.; et al. Effect of Radiotherapy after Breast-Conserving Surgery for Ductal Carcinoma in Situ: 20 Years Follow-up in the Randomized SweDCIS Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 3613–3618. [Google Scholar] [CrossRef]
- Collins, L.C.; Tamimi, R.M.; Baer, H.J.; Connolly, J.L.; Colditz, G.A.; Schnitt, S.J. Outcome of Patients with Ductal Carcinoma in Situ Untreated after Diagnostic Biopsy: Results from the Nurses’ Health Study. Cancer 2005, 103, 1778–1784. [Google Scholar] [CrossRef]
- Pinder, S.E.; Ellis, I.O. The Diagnosis and Management of Pre-Invasive Breast Disease: Ductal Carcinoma in Situ (DCIS) and Atypical Ductal Hyperplasia (ADH)--Current Definitions and Classification. Breast Cancer Res. 2003, 5, 254–257. [Google Scholar] [CrossRef] [Green Version]
- Vincent-Salomon, A.; Lucchesi, C.; Gruel, N.; Raynal, V.; Pierron, G.; Goudefroye, R.; Reyal, F.; Radvanyi, F.; Salmon, R.; Thiery, J.-P.; et al. Integrated Genomic and Transcriptomic Analysis of Ductal Carcinoma In Situ of the Breast. Clin. Cancer Res. 2008, 14, 1956. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, L.; Wilkerson, P.M.; Lambros, M.B.; Campion-Flora, A.; Rodrigues, D.N.; Gauthier, A.; Cabral, C.; Pawar, V.; Mackay, A.; A’Hern, R.; et al. Genomic and Mutational Profiling of Ductal Carcinomas in Situ and Matched Adjacent Invasive Breast Cancers Reveals Intra-Tumour Genetic Heterogeneity and Clonal Selection. J. Pathol. 2012, 227, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Napier, B.A.; Brubaker, S.W.; Sweeney, T.E.; Monette, P.; Rothmeier, G.H.; Gertsvolf, N.A.; Puschnik, A.; Carette, J.E.; Khatri, P.; Monack, D.M. Complement Pathway Amplifies Caspase-11-Dependent Cell Death and Endotoxin-Induced Sepsis Severity. J. Exp. Med. 2016, 213, 2365–2382. [Google Scholar] [CrossRef] [Green Version]
- Swaisgood, C.M.; Schmitt, D.; Eaton, D.; Plow, E.F. In Vivo Regulation of Plasminogen Function by Plasma Carboxypeptidase B. J. Clin. Investig. 2002, 110, 1275–1282. [Google Scholar] [CrossRef]
- Gouri, A.; Dekaken, A.; El Bairi, K.; Aissaoui, A.; Laabed, N.; Chefrour, M.; Ciccolini, J.; Milano, G.; Benharkat, S. Plasminogen Activator System and Breast Cancer: Potential Role in Therapy Decision Making and Precision Medicine. Biomark. Insights 2016, 11, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Ferroni, P.; Roselli, M.; Portarena, I.; Formica, V.; Riondino, S.; LA Farina, F.; Costarelli, L.; Melino, A.; Massimiani, G.; Cavaliere, F.; et al. Plasma Plasminogen Activator Inhibitor-1 (PAI-1) Levels in Breast Cancer - Relationship with Clinical Outcome. Anticancer Res. 2014, 34, 1153–1161. [Google Scholar] [PubMed]
- Grøndahl-Hansen, J.; Christensen, I.J.; Briand, P.; Pappot, H.; Mouridsen, H.T.; Blichert-Toft, M.; Danø, K.; Brünner, N. Plasminogen Activator Inhibitor Type 1 in Cytosolic Tumor Extracts Predicts Prognosis in Low-Risk Breast Cancer Patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 1997, 3, 233–239. [Google Scholar]
- Bouchal, P.; Dvořáková, M.; Roumeliotis, T.; Bortlíček, Z.; Ihnatová, I.; Procházková, I.; Ho, J.T.C.; Maryáš, J.; Imrichová, H.; Budinská, E.; et al. Combined Proteomics and Transcriptomics Identifies Carboxypeptidase B1 and Nuclear Factor ΚB (NF-ΚB) Associated Proteins as Putative Biomarkers of Metastasis in Low Grade Breast Cancer. Mol. Cell. Proteom. 2015, 14, 1814–1830. [Google Scholar] [CrossRef] [Green Version]
- Radisky, D.C.; Hartmann, L.C. Mammary Involution and Breast Cancer Risk: Transgenic Models and Clinical Studies. J. Mammary Gland Biol. Neoplasia 2009, 14, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Clemenceau, A.; Diorio, C.; Durocher, F. Role of Secreted Frizzled-Related Protein 1 in Early Mammary Gland Tumorigenesis and Its Regulation in Breast Microenvironment. Cells 2020, 9, 208. [Google Scholar] [CrossRef] [Green Version]
- Figueroa, J.D.; Pfeiffer, R.M.; Brinton, L.A.; Palakal, M.M.; Degnim, A.C.; Radisky, D.; Hartmann, L.C.; Frost, M.H.; Stallings Mann, M.L.; Papathomas, D.; et al. Standardized Measures of Lobular Involution and Subsequent Breast Cancer Risk among Women with Benign Breast Disease: A Nested Case-Control Study. Breast Cancer Res. Treat. 2016, 159, 163–172. [Google Scholar] [CrossRef]
- Kothari, C.; Diorio, C.; Durocher, F. The Importance of Breast Adipose Tissue in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 5760. [Google Scholar] [CrossRef]
- Hanna, M.; Dumas, I.; Orain, M.; Jacob, S.; Têtu, B.; Sanschagrin, F.; Bureau, A.; Poirier, B.; Diorio, C. Association between Local Inflammation and Breast Tissue Age-Related Lobular Involution among Premenopausal and Postmenopausal Breast Cancer Patients. PLoS ONE 2017, 12, e0183579. [Google Scholar] [CrossRef]
- Wilkinson, L.; Thomas, V.; Sharma, N. Microcalcification on Mammography: Approaches to Interpretation and Biopsy. Br. J. Radiol. 2017, 90, 20160594. [Google Scholar] [CrossRef] [Green Version]
- Shanahan, C.M. Inflammation Ushers in Calcification. Circulation 2007, 116, 2782–2785. [Google Scholar] [CrossRef] [PubMed]
- Gauger, K.J.; Chenausky, K.L.; Murray, M.E.; Schneider, S.S. SFRP1 Reduction Results in an Increased Sensitivity to TGF-β Signaling. BMC Cancer 2011, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Wendt, M.K.; Allington, T.M.; Schiemann, W.P. Mechanisms of the Epithelial-Mesenchymal Transition by TGF-Beta. Future Oncol. Lond. Engl. 2009, 5, 1145–1168. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Wang, X.; Ran, L.; Song, J.; Luo, R.; Wang, Y. Hypoxia-Inducible Factor 1α Regulates the Transforming Growth Factor Β1/SMAD Family Member 3 Pathway to Promote Breast Cancer Progression. J. Breast Cancer 2018, 21, 259–266. [Google Scholar] [CrossRef]
- Dunn, L.K.; Mohammad, K.S.; Fournier, P.G.J.; McKenna, C.R.; Davis, H.W.; Niewolna, M.; Peng, X.H.; Chirgwin, J.M.; Guise, T.A. Hypoxia and TGF-Beta Drive Breast Cancer Bone Metastases through Parallel Signaling Pathways in Tumor Cells and the Bone Microenvironment. PLoS ONE 2009, 4, e6896. [Google Scholar] [CrossRef] [Green Version]
- Nami, B.; Wang, Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Galmarini, C.M.; Treilleux, I.; Cardoso, F.; Bernard-Marty, C.; Durbecq, V.; Gancberg, D.; Bissery, M.-C.; Paesmans, M.; Larsimont, D.; Piccart, M.J.; et al. Class III Beta-Tubulin Isotype Predicts Response in Advanced Breast Cancer Patients Randomly Treated Either with Single-Agent Doxorubicin or Docetaxel. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 4511–4516. [Google Scholar] [CrossRef] [Green Version]
- Aoki, D.; Oda, Y.; Hattori, S.; Taguchi, K.; Ohishi, Y.; Basaki, Y.; Oie, S.; Suzuki, N.; Kono, S.; Tsuneyoshi, M.; et al. Overexpression of Class III Beta-Tubulin Predicts Good Response to Taxane-Based Chemotherapy in Ovarian Clear Cell Adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 1473–1480. [Google Scholar] [CrossRef] [Green Version]
- Tame, M.A.; Manjón, A.G.; Belokhvostova, D.; Raaijmakers, J.A.; Medema, R.H. TUBB3 Overexpression Has a Negligible Effect on the Sensitivity to Taxol in Cultured Cell Lines. Oncotarget 2017, 8, 71536–71547. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Vo, T.; Hajar, A.; Li, S.; Chen, X.; Parissenti, A.M.; Brindley, D.N.; Wang, Z. Multiple Mechanisms Underlying Acquired Resistance to Taxanes in Selected Docetaxel-Resistant MCF-7 Breast Cancer Cells. BMC Cancer 2014, 14, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebok, P.; Öztürk, M.; Heilenkötter, U.; Jaenicke, F.; Müller, V.; Paluchowski, P.; Geist, S.; Wilke, C.; Burandt, E.; Lebeau, A.; et al. High Levels of Class III β-Tubulin Expression Are Associated with Aggressive Tumor Features in Breast Cancer. Oncol. Lett. 2016, 11, 1987–1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieterich, M.; Hartwig, F.; Stubert, J.; Klöcking, S.; Kundt, G.; Stengel, B.; Reimer, T.; Gerber, B. Accompanying DCIS in Breast Cancer Patients with Invasive Ductal Carcinoma Is Predictive of Improved Local Recurrence-Free Survival. Breast 2014, 23, 346–351. [Google Scholar] [CrossRef]
- Kole, A.J.; Park, H.S.; Johnson, S.B.; Kelly, J.R.; Moran, M.S.; Patel, A.A. Overall Survival Is Improved When DCIS Accompanies Invasive Breast Cancer. Sci. Rep. 2019, 9, 9934. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Han, W.; Moon, H.-G.; Park, I.-A.; Ahn, S.K.; Kim, J.; Lee, J.W.; Kim, T.; Kim, M.K.; Noh, D.-Y. Grade of Ductal Carcinoma in Situ Accompanying Infiltrating Ductal Carcinoma as an Independent Prognostic Factor. Clin. Breast Cancer 2013, 13, 385–391. [Google Scholar] [CrossRef]
- Chen, H.; Bai, F.; Wang, M.; Zhang, M.; Zhang, P.; Wu, K. The Prognostic Significance of Co-Existence Ductal Carcinoma in Situ in Invasive Ductal Breast Cancer: A Large Population-Based Study and a Matched Case-Control Analysis. Ann. Transl. Med. 2019, 7. [Google Scholar] [CrossRef]

| Characteristics | All (n = 82) | Low CPB1 Expression * (n = 43) | High CPB1 Expression * (n = 39) | p-Value |
|---|---|---|---|---|
| Clinical | ||||
| Age at mastectomy (years) | 55.8 ± 13.9 | 57.6 ± 14.0 | 53.9 ± 13.6 | 0.22 |
| Age at menarche (years) (NA = 5) | 12.8 ± 1.7 | 13.1 ± 1.6 (NA = 3) | 12.4 ± 1.74 (NA = 2) | 0.08 |
| Menopause status (%) | 51 (62.2) | 32 (74.4) | 19 (48.7) | 0.03 |
| Body mass index (NA = 2) | 27.1 ± 5.7 | 28.6 ± 5.7 (NA = 2) | 25.5 ± 5.30 | 0.01 |
| Histopathological | ||||
| Grade ** (NA = 1) | ||||
| 1 | 1 (1.2) | 0 (0.0) | 1 (2.5) | 0.003 |
| 2 | 14 (17.1) | 2 (4.76) | 12 (30.8) | |
| 3 | 66 (80.5) | 40 (95.23) | 26 (66.7) | |
| Tumor size (mm) | 38.2 ± 27.7 | 44.3 ± 33.7 | 31.6 ± 17.1 | 0.03 |
| Microcalcifications | 22 (26.8) | 9 (20.9) | 13 (33.3) | 0.30 |
| Characteristics | Multivariate | |||||
|---|---|---|---|---|---|---|
| Premenopausal (n = 31) | Postmenopausal (n = 51) | All (n = 82) | ||||
| OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| CPB1 expression | 1.02 (1.00–1.03) | 0.022 | 1.03 (1.01–1.05) | 0.016 | 1.02 (1.01–1.03) | 0.00022 |
| CPB1 expression adjusted for age at mastectomy | 1.02 (1.01–1.04) | 0.016 | 1.05 (1.02–1.14) | 0.049 | 1.02 (1.01–1.03) | 0.00056 |
| CPB1 expression adjusted for age at mastectomy and body mass index | 1.025 (1.01–1.05) | 0.017 | 1.06 (1.02–1.15) | 0.066 | 1.02 (1.01–1.04) | 0.00069 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kothari, C.; Clemenceau, A.; Ouellette, G.; Ennour-Idrissi, K.; Michaud, A.; Diorio, C.; Durocher, F. Is Carboxypeptidase B1 a Prognostic Marker for Ductal Carcinoma In Situ? Cancers 2021, 13, 1726. https://doi.org/10.3390/cancers13071726
Kothari C, Clemenceau A, Ouellette G, Ennour-Idrissi K, Michaud A, Diorio C, Durocher F. Is Carboxypeptidase B1 a Prognostic Marker for Ductal Carcinoma In Situ? Cancers. 2021; 13(7):1726. https://doi.org/10.3390/cancers13071726
Chicago/Turabian StyleKothari, Charu, Alisson Clemenceau, Geneviève Ouellette, Kaoutar Ennour-Idrissi, Annick Michaud, Caroline Diorio, and Francine Durocher. 2021. "Is Carboxypeptidase B1 a Prognostic Marker for Ductal Carcinoma In Situ?" Cancers 13, no. 7: 1726. https://doi.org/10.3390/cancers13071726






