Next Generation Sequencing Technology in the Clinic and Its Challenges
Abstract
Simple Summary
Abstract
1. Introduction
2. Sequencing Technologies
3. Extend of Sequencing
4. Targeted Drug Therapies
Additional Annotation Tool—Drug Databases
5. Precautions of Data Output from Sequencing
5.1. Type of Biological Specimen
5.1.1. FF and FFPE Tissue
5.1.2. Liquid Biopsies
5.2. Homopolymers, Repetitive Regions and Pseudogenes
5.3. Bioinformatics
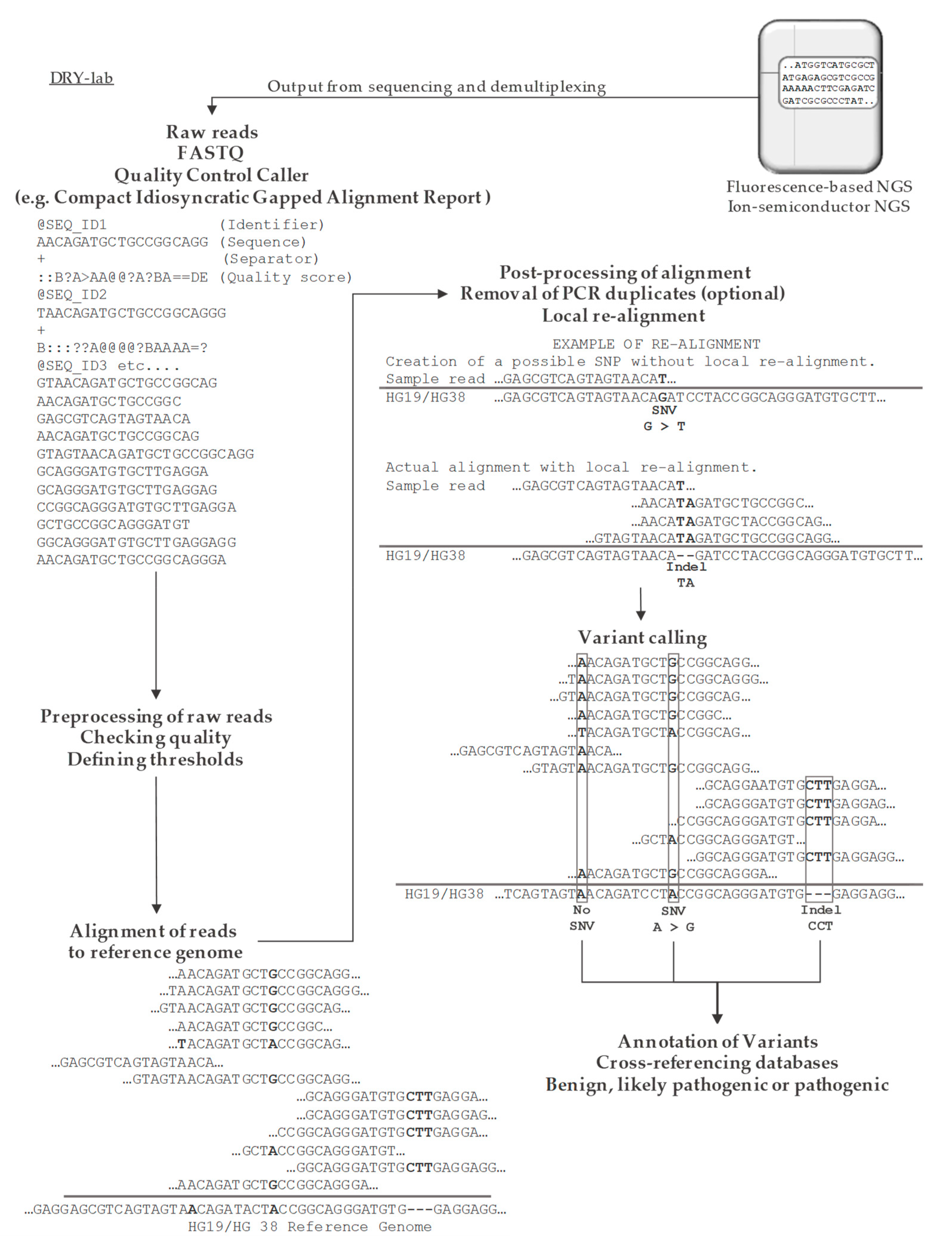
5.3.1. Alignment
| Alignment Tools | Model | Latest Version | Ref. |
|---|---|---|---|
| Bowtie2 | FM-index | v2.4.2 | [2] |
| BWA-MEM | FM-index | v0.7.17 | [3] |
| CUSHAW3 | FM-index | v3.0.3 | [72] |
| GSNAP | Hash-table | N/A | [73] |
| ISAAC | FM-index | v4 | [74] |
| MOSAIK | Hash-table | v2.6.0 | [75] |
| Novoalign | Hash-table | v4.03.01 | http://www.novocraft.com/ * |
| SOAPv2 | FM-index | v2.20 | [76] |
5.3.2. Variant Calling for SNV and Small Indels
5.3.3. Variant Calling for CNV and SV
6. Clinical Demand
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Sato, T.; Cibulskis, K.; Getz, G.; Stewart, C.; Lichtenstein, L. Calling Somatic SNVs and Indels with Mutect2. bioRxiv 2019, 861054. [Google Scholar] [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, M.; Subramanian, U.; Devarajan, B. Performance assessment of variant calling pipelines using human whole exome sequencing and simulated data. BMC Bioinform. 2019, 20, 342. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Han, S.; Wang, Z.; Gelernter, J.; Yang, B.Z. Variant Callers for Next-Generation Sequencing Data: A Comparison Study. PLoS ONE 2013, 8, e75619. [Google Scholar] [CrossRef]
- Maxam, A.M.; Gilbert, W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 1977, 74, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Olson, M.V. The human genome project. Proc. Natl. Acad. Sci. USA 1993, 90, 4338–4344. [Google Scholar] [CrossRef]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Xiao, T.; Zhou, W. The third generation sequencing: The advanced approach to genetic diseases. Transl. Pediatr. 2020, 9, 163–173. [Google Scholar] [CrossRef]
- Pertea, M.; Shumate, A.; Pertea, G.; Varabyou, A.; Breitwieser, F.P.; Chang, Y.; Madugundu, A.K.; Pandey, A.; Salzberg, L.S. CHESS: A new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 2018, 19, 208. [Google Scholar] [CrossRef]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- McFarland, C.D.; Mirny, L.A.; Korolev, K.S. Tug-of-war between driver and passenger mutations in cancer and other adaptive processes. Proc. Natl. Acad. Sci. USA 2014, 111, 15138–15143. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Dietlein, F.; Weghorn, D.; Taylor-Weiner, A.; Richters, A.; Reardon, B.; Liu, D.; Lander, E.S.; Van Allen, E.M.; Sunyaev, S.R. Identification of cancer driver genes based on nucleotide context. Nat. Genet. 2020, 52, 208–218. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 173, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, A.; Mustonen, V.; Reva, B.; Ritchie, G.R.S.; Creixell, P.; Karchin, R.; Vazquez, M.; Fink, J.L.; Kassahn, K.S.; Pearson, J.V.; et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat. Methods 2013, 10, 723–729. [Google Scholar] [CrossRef]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-Generation Sequencing and Emerging Technologies. Semin. Thromb. Hemost. 2019, 45, 661–673. [Google Scholar] [CrossRef]
- Shin, S.H.; Bode, A.M.; Dong, Z. Precision medicine: The foundation of future cancer therapeutics. Npj Precis. Oncol. 2017, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.A.; Lisowska, K.M. Advances in ovarian cancer therapy. Cancer Chemother. Pharmacol. 2018, 81, 17–38. [Google Scholar] [CrossRef]
- Caulfield, S.E.; Davis, C.C.; Byers, K.F. Olaparib: A Novel Therapy for Metastatic Breast Cancer in Patients With a BRCA1/2 Mutation. J. Adv. Pract. Oncol. 2019, 10, 167–174. [Google Scholar] [CrossRef] [PubMed]
- DeMatteo, R.P.; Ballman, K.V.; Antonescu, C.R.; Make, R.G.; Pisters, P.W.T.; Demetri, G.D.; Blackstein, M.E.; Blanke, C.D.; Von Mehren, M.; Brennan, M.F.; et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: A randomised, double-blind, placebo-controlled trial. Lancet 2009, 373, 1097–1104. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Dinu, D.; Dobre, M.; Panaitescu, E.; Bîrlă, R.; Iosif, C.; Hoara, P.; Caragui, A.; Boeriu, M.; Constantinoiu, S.; Ardeleanu, C. Prognostic significance of KRAS gene mutations in colorectal cancer—Preliminary study. J. Med. Life 2014, 7, 581–587. [Google Scholar]
- Maus, M.K.H.; Grimminger, P.P.; Mack, P.C.; Astrow, S.H.; Stephens, C.; Zeger, G.; Hsiang, J.; Brabender, J.; Friedrich, M.; Alakus, H.; et al. KRAS mutations in non-small-cell lung cancer and colorectal cancer: Implications for EGFR-targeted therapies. Lung Cancer 2014, 83, 163–167. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, 777–783. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, 941–947. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, 980–985. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, 1062–1067. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Bewicke-Copley, F.; Arjun Kumar, E.; Palladino, G.; Korfi, K.; Wang, J. Applications and analysis of targeted genomic sequencing in cancer studies. Comput. Struct. Biotechnol. J. 2019, 17, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Miller, C.A.; Griffith, O.L.; Krysiak, K.; Skidmore, Z.L.; Ramu, A.; Walker, J.R.; Dang, H.X.; Trani, L.; Larson, D.E.; et al. Optimizing Cancer Genome Sequencing and Analysis. Cell Syst. 2015, 1, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Pontén, F.; Moberg, C.; Söderkvist, P.; Uhlén, M.; Pontén, J.; Sitbon, G.; Lundeberg, J. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am. J. Pathol. 1999, 155, 1467–1471. [Google Scholar] [CrossRef]
- Kim, S.; Park, C.; Ji, Y.; Kim, D.G.; Bae, H.; Vrancken, M.; Kim, D.; Kim, K. Deamination Effects in Formalin-Fixed, Paraffin-Embedded Tissue Samples in the Era of Precision Medicine. J. Mol. Diagn. 2017, 19, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.H.; Li, J.; Gong, H.F.; Yu, G.Y.; Liu, P.; Hao, L.Q.; Liu, L.J.; Bai, C.G.; Zhang, W. Comparison of Fresh Frozen Tissue With Formalin-Fixed Paraffin-Embedded Tissue for Mutation Analysis Using a Multi-Gene Panel in Patients with Colorectal Cancer. Front. Oncol. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kerick, M.; Isau, M.; Timmermann, B.; Sültmann, H.; Herwig, R.; Krobitsch, S.; Schaefer, G.; Verdorfer, I.; Bartsch, G.; Klocker, H.; et al. Targeted high throughput sequencing in clinical cancer Settings: Formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med. Genom. 2011, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Feeney, M.; Lazarus, H.; Nadler, L.M.; Colvin, R.B.; Knapp, R.C. Reactivity of a Monoclonal Antibody with Human Ovarian Carcinoma. J. Clin. Investig. 1981, 68, 1331–1337. [Google Scholar] [CrossRef]
- Loktionov, A. Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins. World J. Gastrointest. Oncol. 2020, 12, 124–128. [Google Scholar] [CrossRef]
- Mamdani, H.; Ahmed, S.; Armstrong, S.; Mok, T.; Jalal, S.I. Blood-based tumor biomarkers in lung cancer for detection and treatment. Transl. Lung Cancer Res. 2017, 6, 648–660. [Google Scholar] [CrossRef]
- Oloomi, M.; Moazzezy, N.; Bouzari, S. Comparing blood versus tissue-based biomarkers expression in breast cancer patients. Heliyon 2020, 6, 1–7. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2020, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, 1–14. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Dawson, S.J. Circulating tumor cells and circulating tumor DNA for precision medicine: Dream or reality? Ann. Oncol. 2014, 25, 2304–2313. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing–Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef] [PubMed]
- Xu, C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J. 2018, 16, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2012, 13, 36–46. [Google Scholar] [CrossRef]
- Lee, C.Y.; Yen, H.Y.; Zhong, A.W.; Gao, H. Resolving misalignment interference for NGS-based clinical diagnostics. Hum. Genet. 2020, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R.F. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 2011, 11, 792–798. [Google Scholar] [CrossRef]
- Han, L.; Yuan, Y.; Zheng, S.; Yang, Y.; Li, J.; Edgerton, M.E.; Diao, L.; Xu, Y.; Verhaak, R.G.W.; Liang, H. The Pan-Cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Puget, N.; Gad, S.; Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Lenoir, G.M.; Mazoyer, S. Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am. J. Hum. Genet. 2002, 70, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Oliveira, J.; Sousa, M. Bioinformatics and Computational Tools for Next-Generation Sequencing Analysis in Clinical Genetics. J. Clin. Med. 2020, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Meynert, A.M.; Ansari, M.; FitzPatrick, D.R.; Taylor, M.S. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinform. 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cornish, A.; Guda, C. A Comparison of Variant Calling Pipelines Using Genome in a Bottle as a Reference. BioMed Res. Int. 2015, 456479, 1–11. [Google Scholar] [CrossRef]
- Guo, Y.; Dai, Y.; Yu, H.; Zhao, S.; Samuels, D.C.; Shyr, Y. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics 2017, 109, 83–90. [Google Scholar] [CrossRef]
- Pan, B.; Kusko, R.; Xiao, W.; Zheng, Y.; Liu, Z.; Xiao, C.; Sakkiah, S.; Guo, W.; Gong, P.; Zhang, C.; et al. Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinform. 2019, 20, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Guda, K.; Willis, J.; Veigl, M.; Wang, Z.; Markowitz, S.; Adams, M.D.; Sun, S. How do alignment programs perform on sequencing data with varying qualities and from repetitive regions? BioData Min. 2012, 5, 1–12. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Zhong, H.; Meng, Y.; Du, H. Systematic comparison of germline variant calling pipelines cross multiple next-generation sequencers. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yuan, Y.; Chen, X.; Chen, J.; Lin, S.; Li, X.; Du, H. Systematic comparison of somatic variant calling performance among different sequencing depth and mutation frequency. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.B.; Lee, I.; Li, H.; Won, D.; Hernandez-Ferrer, C.; Negron, J.A.; Kong, S.W. Comparative analysis of whole-genome sequencing pipelines to minimize false negative findings. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kim, E.; Lee, I.; Marcotte, E.M. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Meng, J.; Chen, Y.P.P. A database of simulated tumor genomes towards accurate detection of somatic small variants in cancer. PLoS ONE 2018, 13, e202982. [Google Scholar] [CrossRef]
- Supernat, A.; Vidarsson, O.V.; Steen, V.M.; Stokowy, T. Comparison of three variant callers for human whole genome sequencing. Sci. Rep. 2018, 8, 1–6. [Google Scholar] [CrossRef]
- Xu, H.; DiCarlo, J.; Satya, R.V.; Peng, Q.; Wang, Y. Comparison of somatic mutation calling methods in amplicon and whole exome sequence data. BMC Genom. 2014, 15, 1–10. [Google Scholar] [CrossRef]
- Mielczarek, M.; Szyda, J. Review of alignment and SNP calling algorithms for next-generation sequencing data. J. Appl. Genet. 2016, 57, 71–79. [Google Scholar] [CrossRef]
- Kuhnle, A.; Mun, T.; Boucher, C.; Gagie, T.; Langmead, B.; Manzini, G. Efficient Construction of a Complete Index for Pan-Genomics Read Alignment. J. Comput. Biol. 2020, 27, 500–513. [Google Scholar] [CrossRef]
- Lindner, R.; Friedel, C.C. A Comprehensive Evaluation of Alignment Algorithms in the Context of RNA-Seq. PLoS ONE 2012, 7, e52403. [Google Scholar] [CrossRef]
- Zhang, H.; Chan, Y.; Fan, K.; Schmidt, B.; Liu, W. Fast and efficient short read mapping based on a succinct hash index. BMC Bioinform. 2018, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Popp, B.; Schmidt, B. CUSHAW3: Sensitive and accurate base-space and color-space short-read alignment with hybrid seeding. PLoS ONE 2014, 9, e86869. [Google Scholar] [CrossRef]
- Wu, T.D.; Nuca, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef]
- Raczy, C.; Petrovski, R.; Saunders, C.T.; Chorny, I.; Kruglyak, S.; Margulies, E.H.; Chuang, H.; Källberg, M.; Kumar, S.A.; Liao, A.; et al. Isaac: Ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013, 29, 2041–2043. [Google Scholar] [CrossRef]
- Lee, W.P.; Stromberg, M.P.; Ward, A.; Stewart, C.; Garrison, E.P.; Marth, G.T. MOSAIK: A hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS ONE 2014, 9, e90581. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.; Yiu, S.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Challis, D.; Yu, J.; Evani, U.S.; Jackson, A.R.; Paithankar, S.; Coarfa, C.; Milosavljevic, A.; Gibbs, R.A.; Yu, F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinform. 2012, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lupat, R.; Amarasinghe, K.C.; Thompson, E.R.; Doyle, M.A.; Ryland, G.L.; Tothill, R.W.; Halgamuge, S.K.; Campbell, I.G.; Gorringe, K.L. CONTRA: Copy number analysis for targeted resequencing. Bioinformatics 2012, 28, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Amarasinghe, K.C.; Li, J.; Halgamuge, S.K. Correction to CoNVEX: Copy number variation estimation in exome sequencing data using HMM. BMC Bioinform. 2013, 14, 1–9. [Google Scholar] [CrossRef]
- Poplin, R.; Chang, P.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; Newburger, D.; Dijamco, J.; Nguyen, N.; Afshar, P.T.; et al. A universal snp and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Sathirapongsasuti, J.F.; Lee, H.; Horst, B.A.J.; Brunner, G.; Cochran, A.J.; Binder, S.; Quackenbush, J.; Nelson, S.F. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 2011, 27, 2648–2654. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2009, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Bertels, K.; Al Ars, Z. Efficient Acceleration of the Pair-HMMs Forward Algorithm for GATK HaplotypeCaller on Graphics Processing Units. Evol. Bioinform. 2018, 14, 1–12. [Google Scholar] [CrossRef]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014, 15, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Nijkamp, J.F.; Van Den Broek, M.A.; Geertman, J.M.A.; Reinders, M.J.T.; Daran, J.M.G.; De Ridder, D. De novo detection of copy number variation by co-assembly. Bioinformatics 2012, 28, 3195–3202. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef]
- Rimmer, A.; Phan, H.; Mathieson, I.; Iqbal, Z.; Twigg, S.R.F.; Wilkie, A.O.M.; McVean, G.; Lunter, G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014, 46, 912–918. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- O’Fallon, B.D.; Wooderchak-Donahue, W.; Crockett, D.K. A support vector machine for identification of single-nucleotide polymorphisms from next-generation sequencing data. Bioinformatics 2013, 29, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.E.; Harris, C.C.; Chen, K.; Koboldt, D.C.; Abbott, T.E.; Dooling, D.J.; Ley, T.J.; Mardis, E.R.; Wilson, R.K.; Ding, L. Somaticsniper: Identification of somatic point mutations in whole genome sequencing data. Bioinformatics 2012, 28, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Layer, R.M.; Faust, G.G.; Lindberg, M.R.; Rose, D.B.; Garrison, E.P.; Marth, G.T.; Quinlan, A.R.; Hall, I.M. SpeedSeq: Ultra-fast personal genome analysis and interpretation. Nat. Methods 2015, 12, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Saunders, C.T.; Wong, W.S.W.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.; Keane, T.M.; Stalker, J.; Adams, D.J. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef]
- Gillet-Markowska, A.; Richard, H.; Fischer, G.; Lafontaine, I. Ulysses: Accurate detection of low-frequency structural variations in large insert-size sequencing libraries. Bioinformatics 2015, 31, 801–808. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Li, Y.; Roberts, N.D.; Wala, J.A.; Shapira, O.; Schumacher, S.E.; Kumar, K.; Khurana, E.; Waszak, S.; Korbel, J.O.; Haber, J.E.; et al. Patterns of somatic structural variation in human cancer genomes. Nature 2020, 578, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Marotta, M.; Chen, X.; Inoshita, A.; Stephens, R.; Budd, G.T.; Crowe, J.P.; Lyons, J.; Kondratova, A.; Tubbs, R.; Tanaka, H. A common copy-number breakpoint of ERBB2 amplification in breast cancer colocalizes with a complex block of segmental duplications. Breast Cancer Res. 2012, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, Z.N.; Fojo, T.; Blagosklonny, M.V. Complementation of two mutant p53: Implications for loss of heterozygosity in cancer. FEBS Lett. 2005, 579, 2231–2235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alaei-Mahabadi, B.; Bhadury, J.; Karlsson, J.W.; Nilsson, J.A.; Larsson, E. Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc. Natl. Acad. Sci. USA 2016, 113, 13768–13773. [Google Scholar] [CrossRef]
- Alkan, C.; Coe, B.P.; Eichler, E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C. Best practices for variant calling in clinical sequencing. Genome Med. 2020, 12, 1–13. [Google Scholar] [CrossRef]
- Le Scouarnec, S.; Gribble, S.M. Characterising chromosome rearrangements: Recent technical advances in molecular cytogenetics. Heredity 2012, 108, 75–85. [Google Scholar] [CrossRef]
- Escaramís, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief. Funct. Genom. 2015, 14, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Pirooznia, M.; Goes, F.; Zandi, P.P. Whole-genome CNV analysis: Advances in computational approaches. Front. Genet. 2015, 6, 1–9. [Google Scholar] [CrossRef]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three NGS sequencing platforms. BMC Genom. 2012, 13, 1–13. [Google Scholar] [CrossRef]
- Chowell, D.; Napier, J.; Gupta, R.; Anderson, K.S.; Maley, C.C.; Wilson Sayres, M.A. Modeling the subclonal evolution of cancer cell populations. Cancer Res. 2018, 78, 830–839. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, E.F.; Riegman, P.H.; Grizzle, W.E.; Watson, P.H. Factors that drive the increasing use of FFPE tissue in basic and translational cancer research. Biotech. Histochem. 2018, 93, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Reumers, J.; Rijk, P.D.; Zhao, H.; Liekens, A.; Smeets, D.; Cleary, J.; Loo, P.V.; Bossche, M.V.D.; Catthoor, K.; Sabbe, B.; et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat. Biotechnol. 2012, 30, 61–68. [Google Scholar] [CrossRef] [PubMed]
| Platform | Immobilization | Amplification | Sequencing Technology | Limitations & Error Rate | Read Length (bp *) | Run Time (h **) | Output (Gb ***) |
|---|---|---|---|---|---|---|---|
| 1st generation technologies | |||||||
| Sanger | N/A | PCR with dNTPs and ddNTPs | Irreversible chain termination | 0.001% | ≤900 | ~4 | ≤0.002 |
| Maxam-Gilbert | N/A | N/A | Chemical termination of 32P labeled ssDNA | 0.001% | ≤900 | N/A | ≤0.002 |
| 2nd generation technologies | |||||||
| HiSeq2000 | Flow cell | Bridge amplification | Cyclic reversible dye chain termination | GC-rich regions 0.2% | ≤125 | 7–144 | ≤1600 |
| MiSeq | Flow cell | Bridge amplification | Cyclic reversible dye chain termination | GC-rich regions 0.2% | ≤300 | 4–55 | ≤15 |
| Ion Torrent PGM | Bead emulsion | Emulsion PCR | Synthesis depended H+ detection | Homo-polymers 1% Indel | ≤400 | 2–7.5 | ≤2 |
| Ion Torrent S5XL | Bead emulsion | Emulsion PCR | Synthesis depended H+ detection | Homo-polymers 1% Indel | ≤600 | 2.5–4 | ≤25 |
| 3rd generation technologies | |||||||
| ONT MinION | Processive enzyme | N/A | Monitoring the current of a nucleotide in ssDNA | 5–20% | 10,000–30,000 | Real time | ≤25 |
| PacBio SMRT | DNA attachment to the bottom of each Zero Mode Waveguide | N/A | Detection of incorporation of fluorescent nucleotides during real time synthesis | 10–15% | 10,000–30,000 | Real time | ≤4 |
| Gene | Aberration | Targeting Drug | Cancer Type |
|---|---|---|---|
| BRCA-1/2 | Loss-of-function | PARP-inhibitor | Breast cancer, Ovarian cancer, Prostate cancer |
| ERBB2/HER2 | Amplification | Dimerization-inhibitor of HER2-HER3 | Breast cancer |
| PIK3CA | Gain-of-function | PIK3 kinase-inhibitor | Breast cancer |
| BCL2 | Gain-of-function (17 bp deletion) | Blocker of Bcl-2 | Chronic lymphocytic leukemia |
| RAS | Wild type | EGFR-inhibitor | Colorectal cancer |
| c-KIT | Gain-of-function (exon 9, 11, 13, and 17) | Tyrosine kinase-inhibitors | Gastrointestinal Stromal Tumor |
| EGFR | Gain-of-function (exon 19 deletion and/or L858R) | Tyrosine kinase-inhibitors | Lung cancer, Brain cancer |
| BRAF | Gain-of-function (V600E/K) | Kinase-inhibitor | Melanoma |
| CDK12 | Loss-of-function | PARP-inhibitor | Prostate cancer |
| Variant Calling Tools | Variant Detection | Latest Version | Ref. |
|---|---|---|---|
| Atlas2 suite | SNV, indels | v1.4.1 | [77] |
| CONTRA | CNV, SV | v2.0.8 | [78] |
| CNVnator | CNV, SV | v0.4 | [79] |
| CoNVEX | CNV, SV | N/A | [80] |
| DeepVariant | SNV, indels | v1.0 | [81] |
| DELLY | CNV, SV | v0.8.7 | [82] |
| ExomeCNV | CNV, SV | v1.4 | [83] |
| FreeBayes | SNV, indels | v1.3.4 | [84] |
| GATK Haplotype Caller (GATK-HC) | SNV, indels | v4.1.9.0 | [85,86] |
| GlfSingle | SNV, indels | N/A | N/A |
| ISAAC Variant Caller (IVC) | SNV, indels | V2.0.13 | [74] |
| LUMPY | CNV, SV | v0.3.1 | [87] |
| Magnolya | CNV, SV | v0.15 | [88] |
| Mutect | SNV, indels | v1.1.5 | [89] |
| Mutect2 | SNV, indels | v4.1.9.0 | [4] |
| Pindel | CNV, SV | N/A | [90] |
| Platypus | SNV, indels, SV | N/A | [91] |
| SAMtools | SNV, indels | v1.11 | [92] |
| SNPSVM | SNV | N/A | [93] |
| SomaticSniper | SNV | v1.0.5.0 | [94] |
| SpeedSeq | SNV, indels | v0.1.2 | [95] |
| Strelka | SNV, indels | N/A | [96] |
| Strelka2 | SNV, indels | v2.9.10 | [5] |
| SVMerge | CNV, SV | v1.2 | [97] |
| Torrent Variant Caller (TVC) | SNV, indels, SV | v5.12.0 | N/A |
| Ulysses | CNV, SV | v1.0 | [98] |
| Varscan2 | SNV, Indel | v2.4.4 | [99] |
| Research Paper | Subjected Sample | Reference Genome | Alignment Tool | Variant Calling Tool |
|---|---|---|---|---|
| Chen et al. 2019 [61] | NA12878 | WES WGS | BWA-MEM | GATK-HC SAMtools Strelka2 VarScan2 |
| Chen et al. 2020 [62] | WES | BWA-MEM | Mutect2 Strelka2 | |
| Cornish et al. 2015 [57] | WES | Bowtie2 BWA_MEM CUSHAW3 MOSAIK Novoalign | SAMtools SNPSVM | |
| Hwang et al. 2015 [64] | WES WGS | Bowtie2 BWA-MEM Novoalign | FreeBayes GATK-HC SAMtools TVC | |
| Hwang et al. 2019 [63] | WES WGS | Bowtie2 BWA-MEM GSNAP ISAAC Novoalign SOAP2 | Atlas2 FreeBayes GATK-HC glfSingle IVC Platypus SAMtools suite VarScan2 | |
| Kumaran et al. 2019 [6] | WES | Bowtie2 BWA-MEM MOSAIK Novoalign SOAP2 | GATK-HC DeepVariant FreeBayes SAMtools | |
| Meng et al. 2018 [65] | TES WES WGS | BWA-MEM | DeepVariant Lancet Strelka2 VarScan2 | |
| Pan et al. 2019 [59] | WGS | Bowtie2 BWA-MEM ISAAC Novoalign | FreeBayes GATK-HC IVC SAMtools | |
| Supernat et al. 2018 [66] | WES WGS | BWA-MEM | DeepVariant GATK-HC SpeedSeq | |
| Xu et al. 2014 [67] | WES | BWA-MEM | Mutect SomaticSniper Strelka VarScan2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vestergaard, L.K.; Oliveira, D.N.P.; Høgdall, C.K.; Høgdall, E.V. Next Generation Sequencing Technology in the Clinic and Its Challenges. Cancers 2021, 13, 1751. https://doi.org/10.3390/cancers13081751
Vestergaard LK, Oliveira DNP, Høgdall CK, Høgdall EV. Next Generation Sequencing Technology in the Clinic and Its Challenges. Cancers. 2021; 13(8):1751. https://doi.org/10.3390/cancers13081751
Chicago/Turabian StyleVestergaard, Lau K., Douglas N. P. Oliveira, Claus K. Høgdall, and Estrid V. Høgdall. 2021. "Next Generation Sequencing Technology in the Clinic and Its Challenges" Cancers 13, no. 8: 1751. https://doi.org/10.3390/cancers13081751
APA StyleVestergaard, L. K., Oliveira, D. N. P., Høgdall, C. K., & Høgdall, E. V. (2021). Next Generation Sequencing Technology in the Clinic and Its Challenges. Cancers, 13(8), 1751. https://doi.org/10.3390/cancers13081751







