Zona Pellucida Protein 2 (ZP2) Is Expressed in Colon Cancer and Promotes Cell Proliferation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Tumor Tissue Samples
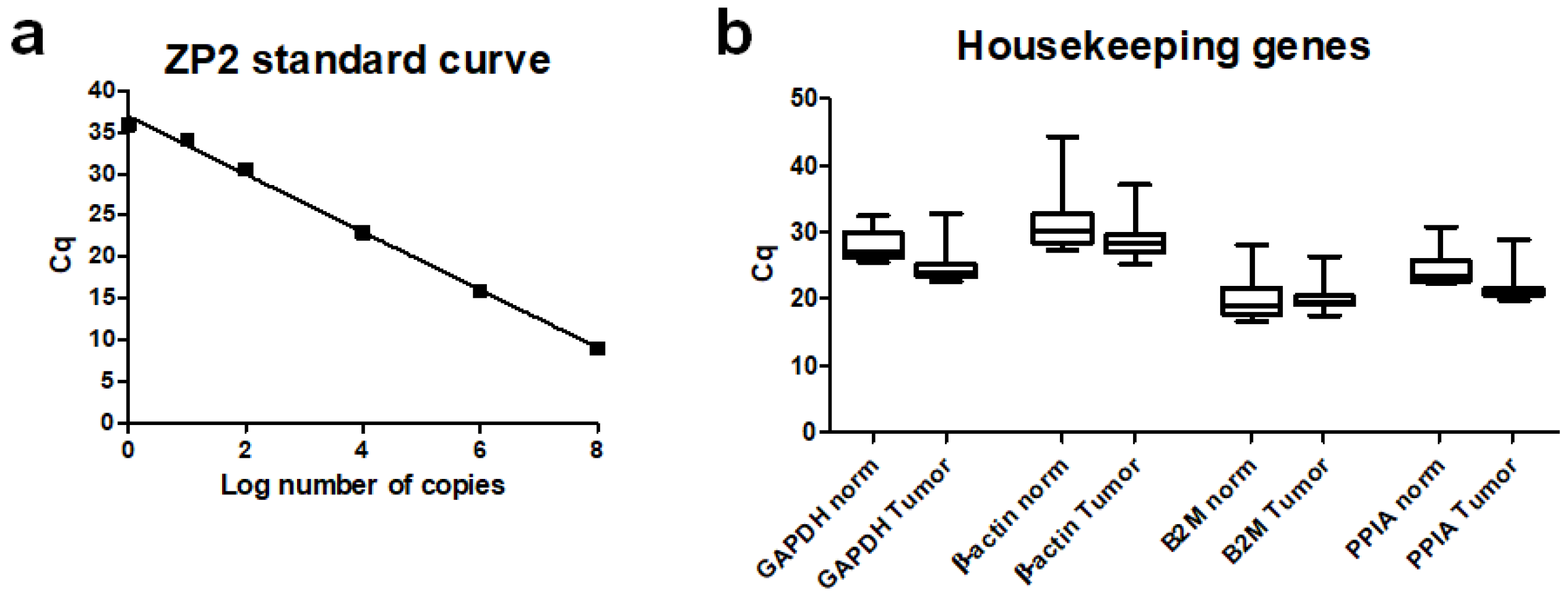
2.3. Quantitative Reverse Transcription-PCR (qPCR)
2.4. Quantification of ZP2 Molecules
2.5. Small Interfering RNA (siRNA) Experiments
2.6. Protein Extraction and Western Blot
2.7. Immunohistochemistry
2.8. Cell Proliferation Assay
2.9. Statistics
3. Results
3.1. ZP2 Is Expressed in Subsets of Human Tumor Cell Lines
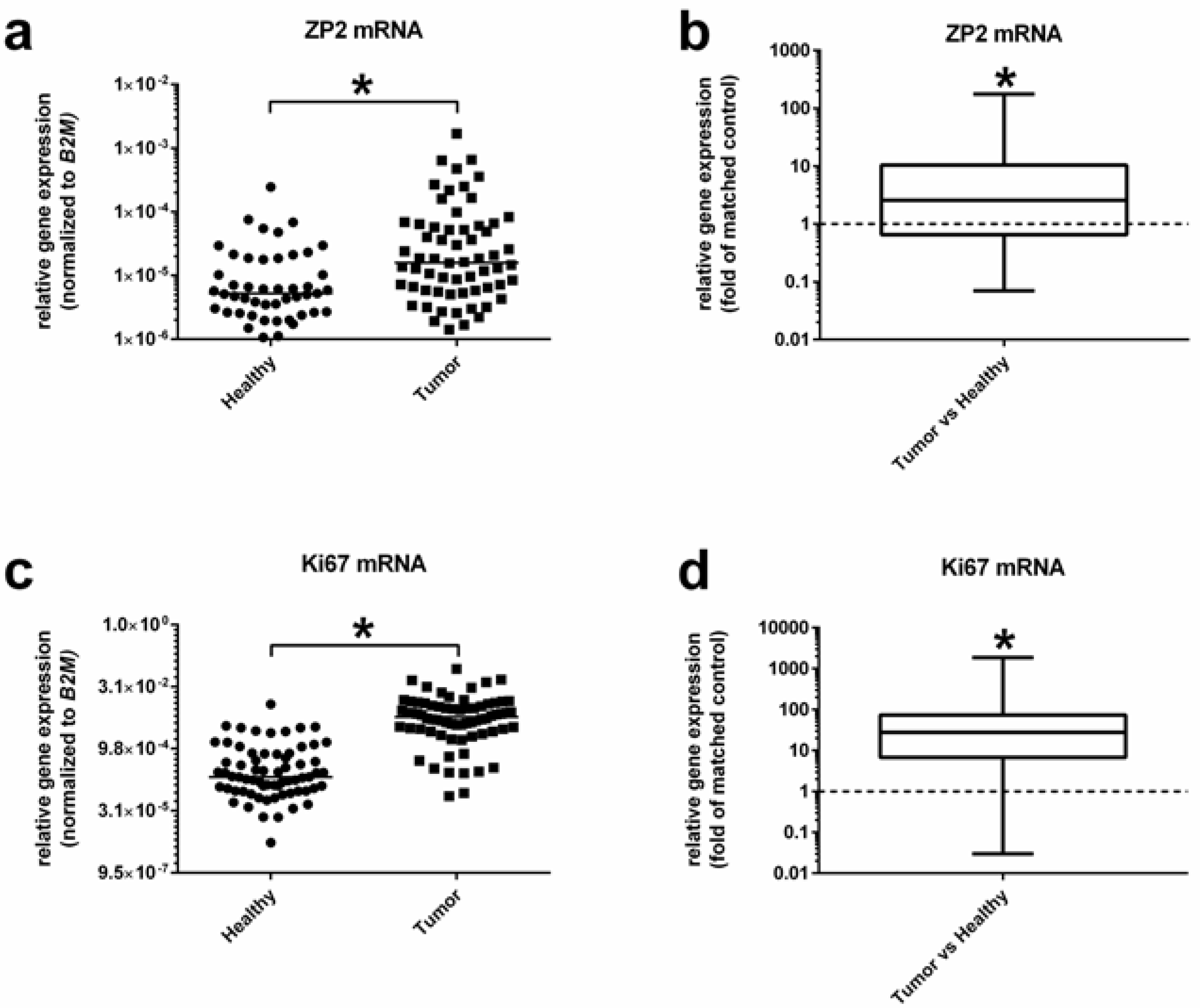
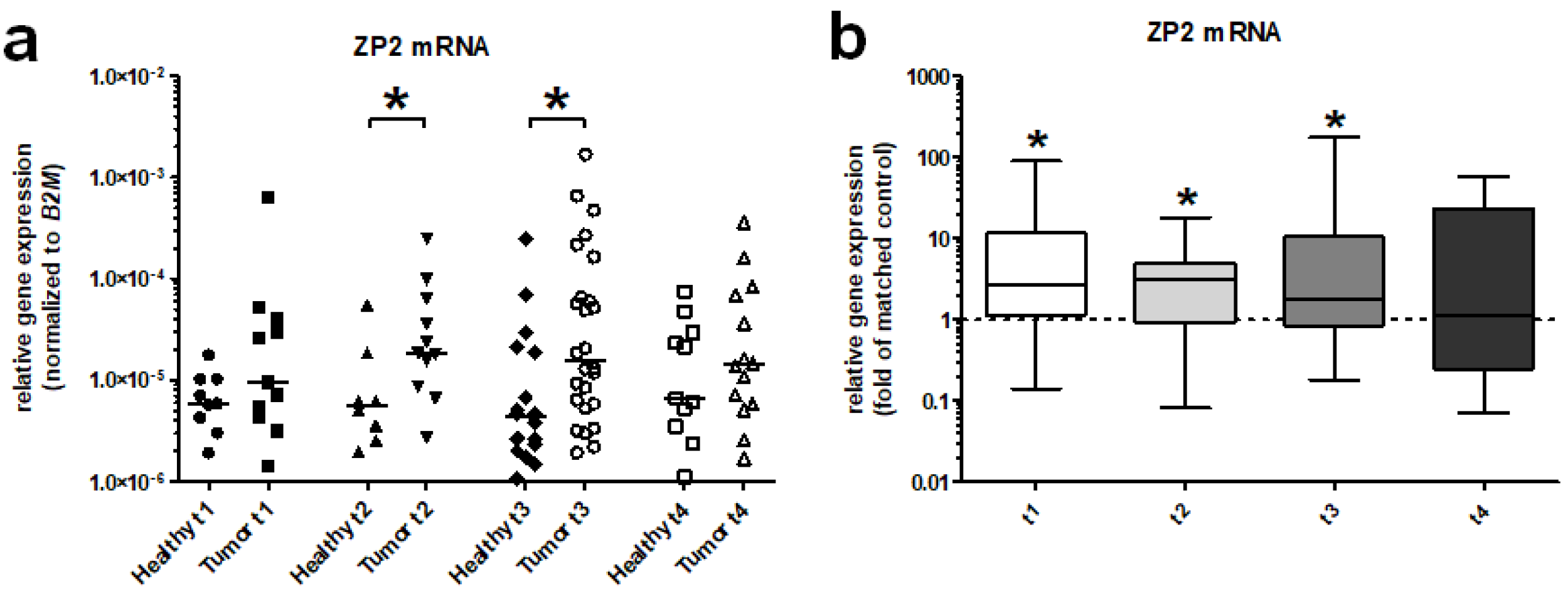
3.2. ZP2 Is Expressed in Subsets of Primary Colon Cancer Tissues in a Low Abundant Manner
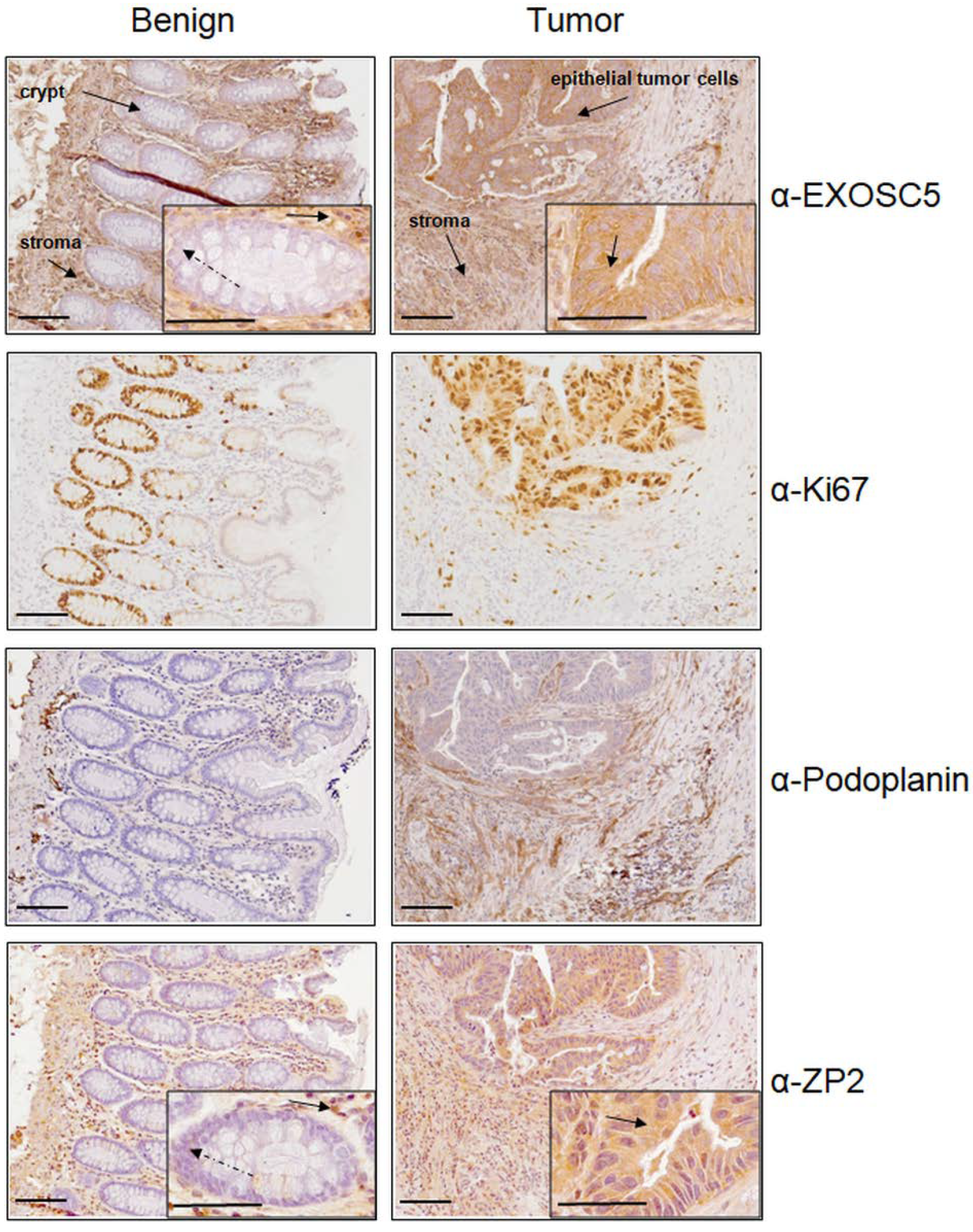
3.3. Immunohistochemical Analysis of ZP2 in Colon Cancer Tissue
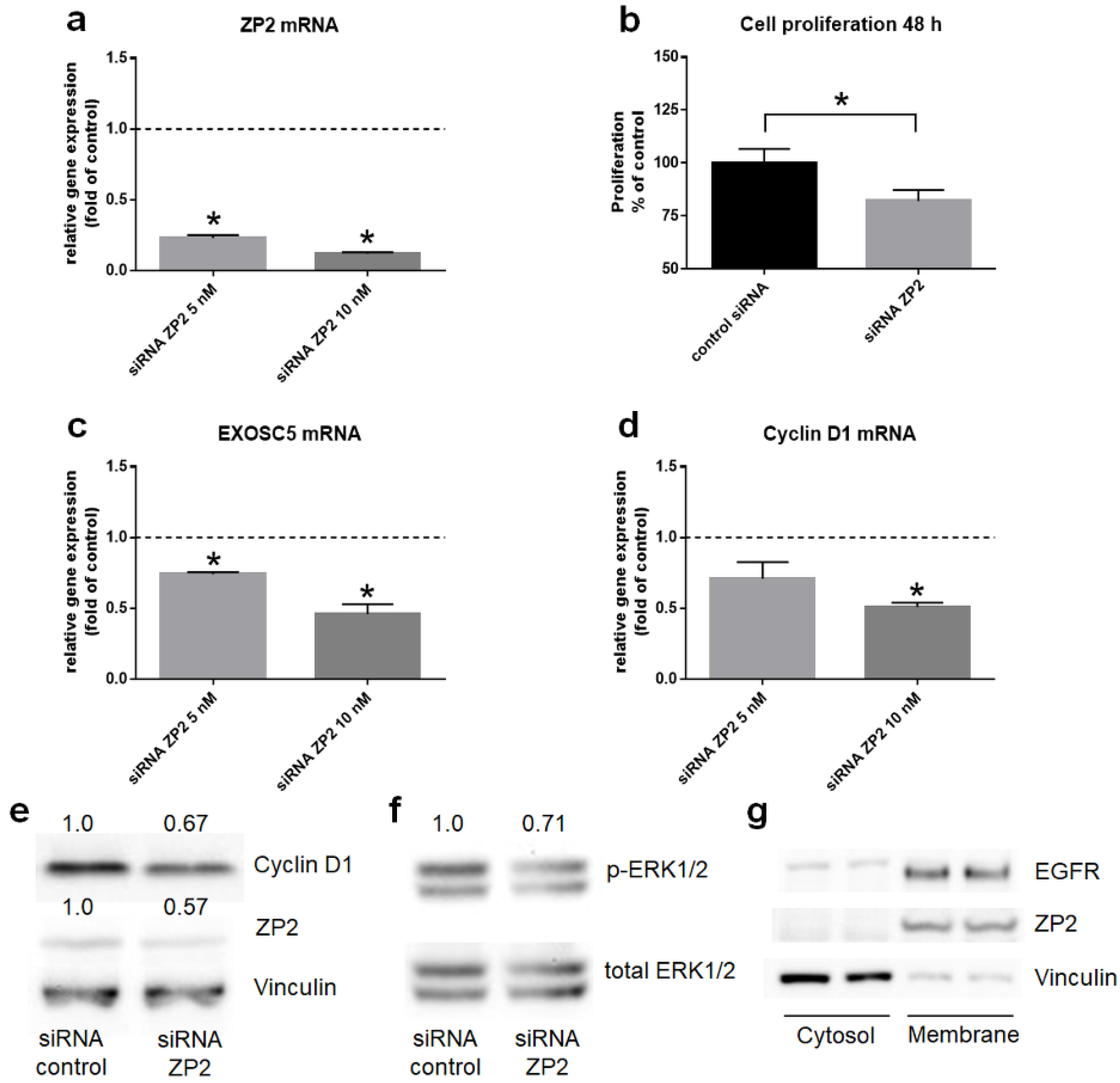
3.4. ZP2 Promotes Colon Cancer Cell Proliferation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loeb, L.A. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat. Rev. Cancer 2011, 11, 450–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar-Maia, A.; Alajem, A.; Meshorer, E.; Ramalho-Santos, M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011, 12, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Stergachis, A.B.; Neph, S.; Reynolds, A.; Humbert, R.; Miller, B.; Paige, S.L.; Vernot, B.; Cheng, J.B.; Thurman, R.E.; Sandstrom, R.; et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 2013, 154, 888–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaern, M.; Elston, T.C.; Blake, W.J.; Collins, J.J. Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 2005, 6, 451–464. [Google Scholar] [CrossRef]
- Landan, G.; Cohen, N.M.; Mukamel, Z.; Bar, A.; Molchadsky, A.; Brosh, R.; Horn-Saban, S.; Zalcenstein, D.A.; Goldfinger, N.; Zundelevich, A.; et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat. Genet. 2012, 44, 1207–1214. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraus, D.; Reckenbeil, J.; Perner, S.; Winter, J.; Probstmeier, R. Expression pattern of matrix metalloproteinase 20 (MMP20) in human tumors. Anticancer Res. 2016, 36, 2713–2718. [Google Scholar]
- Gupta, S.K.; Bhandari, B.; Shrestha, A.; Biswal, B.K.; Palaniappan, C.; Malhotra, S.S.; Gupta, N. Mammalian zona pellucida glycoproteins: Structure and function during fertilization. Cell Tissue Res. 2012, 349, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K. Role of zona pellucida glycoproteins during fertilization in humans. J. Reprod. Immunol. 2015, 108, 90–97. [Google Scholar] [CrossRef]
- Jimenez-Movilla, M.; Dean, J. ZP2 and ZP3 cytoplasmic tails prevent premature interactions and ensure incorporation into the zona pellucida. J. Cell Sci. 2011, 124, 940–950. [Google Scholar] [CrossRef] [Green Version]
- Costa, J.; Pereira, R.; Oliveira, J.; Alves, Â.; Marques-Magalhães, Â.; Frutuoso, A.; Leal, C.; Barros, N.; Fernandes, R.; Queiroz Almeida, D.; et al. Structural and molecular analysis of the cancer prostate cell line PC3: Oocyte zona pellucida glycoproteins. Tissue Cell 2018, 55, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Kraus, D.; Probstmeier, R.; Winter, J. Tumor-Associated Antigens for Diagnosis and Therapy. Germany Patent DE 10 2014 002 256 A1, 20 February 2014. [Google Scholar]
- Kraus, D.; Probstmeier, R.; Winter, J. Tumor-Associated Antigens for Diagnosis and Therapy. Germany Patent DE 10 2014 011 258 A1, 30 July 2014. [Google Scholar]
- Buschmann, T.; Fotiadis, N.-K.; Fuchs, M.; Heim, S.; Lehnherr-Ilina, T.; Lamer, S.; Meuer, J.; Rothmann-Cosic, K.; Seibert, V.; Tortola Perez, S.; et al. Differentially Expressed Tumour-Specific Polypeptides for Use in the Diagnosis and Treatment of Cancer. Canada Patent CA2539490A1, 20 September 2004. Available online: https://patents.google.com/patent/CA2539490A1/un (accessed on 24 March 2005).
- Rahman, N.A.; Bennink, H.J.; Chrusciel, M.; Sharp, V.; Zimmerman, Y.; Dina, R.; Li, X.; Ellonen, A.; Rivero-Müller, A.; Dilworth, S.; et al. A novel treatment strategy for ovarian cancer based on immunization against zona pellucida protein (ZP) 3. FASEB J. 2012, 26, 324–333. [Google Scholar] [CrossRef]
- Watanabe, M.; Iizumi, Y.; Sukeno, M.; Iizuka-Ohashi, M.; Sowa, Y.; Sakai, T. The pleiotropic regulation of cyclin D1 by newly identified sesaminol-binding protein ANT2. Oncogenesis 2017, 6, e311. [Google Scholar] [CrossRef] [Green Version]
- Meloche, S.; Pouysségur, J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 2007, 26, 3227–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, J.; Kraus, D.; Reckenbeil, J.; Probstmeier, R. Oncogenic relevant defensins: Expression pattern and proliferation characteristics of human tumor cell lines. Tumour Biol. 2016, 37, 7959–7966. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef] [PubMed]
- Parr, C.; Jiang, W.G. Quantitative analysis of lymphangiogenic markers in human colorectal cancer. Int. J. Oncol. 2003, 23, 533–539. [Google Scholar] [CrossRef]
- Pan, H.; Pan, J.; Song, S.; Ji, L.; Lv, H.; Yang, Z. EXOSC5 as a novel prognostic marker promotes proliferation of colorectal cancer via activating the ERK and Akt pathways. Front. Oncol. 2019, 9, 643. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004, 15, 155–166. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feinberg, A. DNA methylation in cancer: Three decades of discovery. Genome. Med. 2014, 6, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.I.; Morgan, B.; Trautwein, J.; et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Marjani, S.L.; Hu, Z.; Weissman, S.M.; Pan, X.; Wu, S. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res. 2016, 76, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amant, F.; Verheecke, M.; Wlodarska, I.; Dehaspe, L.; Brady, P.; Brison, N.; Van Den Bogaert, K.; Dierickx, D.; Vandecaveye, V.; Tousseyn, T.; et al. Presymptomatic Identification of Cancers in Pregnant Women During Noninvasive Prenatal Testing. JAMA Oncol. 2015, 1, 814–819. [Google Scholar] [CrossRef] [Green Version]
- Sobecki, M.; Mrouj, K.; Colinge, J.; Gerbe, F.; Jay, P.; Krasinska, L.; Dulic, V.; Fisher, D. Cell-Cycle Regulation Accounts for Variability in Ki-67 Expression Levels. Cancer Res. 2017, 77, 2722–2734. [Google Scholar] [CrossRef] [Green Version]
- Mulyawan, I.M. Role of Ki67 protein in colorectal cancer. Int. J. Res. Med. Sci. 2019, 7, 644–648. [Google Scholar] [CrossRef]
- Choi, S.Y.; Sung, R.; Lee, S.J.; Lee, T.G.; Kim, N.; Yoon, S.M.; Lee, E.J.; Chae, H.B.; Youn, S.J.; Park, S.M. Podoplanin, α-smooth muscle actin or S100A4 expressing cancer-associated fibroblasts are associated with different prognosis in colorectal cancers. J. Korean Med. Sci. 2013, 28, 1293–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avella, M.A.; Xiong, B.; Dean, J. The molecular basis of gamete recognition in mice and human. Mol. Hum. Reprod. 2013, 19, 279–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoppe, T.; Kraus, D.; Novak, N.; Probstmeier, R.; Frentzen, M.; Wenghoefer, M.; Jepsen, S.; Winter, J. Oral pathogens change proliferation properties of oral tumor cells by affecting gene expression of human defensins. Tumour Biol. 2016, 37, 13789–13798. [Google Scholar] [CrossRef] [PubMed]


| Cell line | Origin | Ct1 | Ct2 | Ct3 | Ct4 | % |
|---|---|---|---|---|---|---|
| A-172 | Glioblastoma | nd | nd | nd | nd | 0 |
| A549 | Non-small cell lung carcinoma | 34.4 | nd | nd | nd | 25 |
| A64-CLS | Submaxillary gland adenoma | nd | nd | nd | nd | 0 |
| B-CPAP | Papillary thyroid carcinoma | nd | nd | nd | nd | 0 |
| BHY | Oral squamous cell carcinoma | nd | nd | nd | nd | 0 |
| CAL-62 | Thyroid anaplastic carcinoma | 34.5 | nd | nd | nd | 25 |
| COLO 320HSR | Colon carcinoma | 34.3 | 33.9 | nd | 35.0 | 75 |
| FTC-133 | Follicular thyroid carcinoma | nd | nd | nd | nd | 0 |
| H1184 | Small cell lung carcinoma | nd | nd | nd | nd | 0 |
| H146 | Small lung cell carcinoma | 35.0 | nd | nd | nd | 25 |
| HeLa | Cervix adenocarcinoma | nd | nd | nd | nd | 0 |
| HL-60 | Leukemia (promyelocytic) | nd | nd | nd | nd | 0 |
| HN | Oral squamous cell carcinoma | 34.9 | nd | nd | nd | 25 |
| HT-29 | Colon carcinoma | 31.0 | 31.9 | 31.2 | 31.8 | 100 |
| LNCaP | Prostate carcinoma | nd | nd | nd | nd | 0 |
| LoVo | Colon carcinoma | nd | nd | 34.5 | nd | 25 |
| MCF-7 | Breast carcinoma | nd | nd | nd | nd | 0 |
| MG-63 | Osteosarcoma | 34.6 | nd | nd | nd | 25 |
| PC-3 | Prostate adenocarcinoma | 35.0 | nd | nd | nd | 25 |
| SaOs | Osteosarcoma | nd | nd | nd | nd | 0 |
| SKN-MC | Neuroblastoma | nd | nd | nd | nd | 0 |
| SW-403 | Colon carcinoma | 31.4 | 31.9 | 31.7 | 31.9 | 100 |
| SW-480 | Colon carcinoma | 33.3 | 34.5 | nd | 35.0 | 75 |
| SW-948 | Colon carcinoma | nd | nd | 34.9 | 34.7 | 50 |
| THP-1 | Leukemia | nd | nd | nd | nd | 0 |
| U251-MG | Glioblastoma | 35.0 | 34.7 | nd | nd | 50 |
| U373-MG | Glioblastoma (astrocytoma) | nd | nd | nd | nd | 0 |
| U87-MG | Glioblastoma | 34.6 | nd | nd | nd | 25 |
| Tissue Samples | Healthy | Tumor | ∑ |
|---|---|---|---|
| positive | 48 | 61 | 109 |
| negative | 21 | 8 | 29 |
| ∑ | 69 | 69 | 138 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kraus, D.; Glassmann, A.; Golletz, C.; Kristiansen, G.; Winter, J.; Probstmeier, R. Zona Pellucida Protein 2 (ZP2) Is Expressed in Colon Cancer and Promotes Cell Proliferation. Cancers 2021, 13, 1759. https://doi.org/10.3390/cancers13081759
Kraus D, Glassmann A, Golletz C, Kristiansen G, Winter J, Probstmeier R. Zona Pellucida Protein 2 (ZP2) Is Expressed in Colon Cancer and Promotes Cell Proliferation. Cancers. 2021; 13(8):1759. https://doi.org/10.3390/cancers13081759
Chicago/Turabian StyleKraus, Dominik, Alexander Glassmann, Carsten Golletz, Glen Kristiansen, Jochen Winter, and Rainer Probstmeier. 2021. "Zona Pellucida Protein 2 (ZP2) Is Expressed in Colon Cancer and Promotes Cell Proliferation" Cancers 13, no. 8: 1759. https://doi.org/10.3390/cancers13081759
APA StyleKraus, D., Glassmann, A., Golletz, C., Kristiansen, G., Winter, J., & Probstmeier, R. (2021). Zona Pellucida Protein 2 (ZP2) Is Expressed in Colon Cancer and Promotes Cell Proliferation. Cancers, 13(8), 1759. https://doi.org/10.3390/cancers13081759







