A Systematic Review of the Use of Circulating Cell-Free DNA Dynamics to Monitor Response to Treatment in Metastatic Breast Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Selection of Studies
2.3. Data Extraction
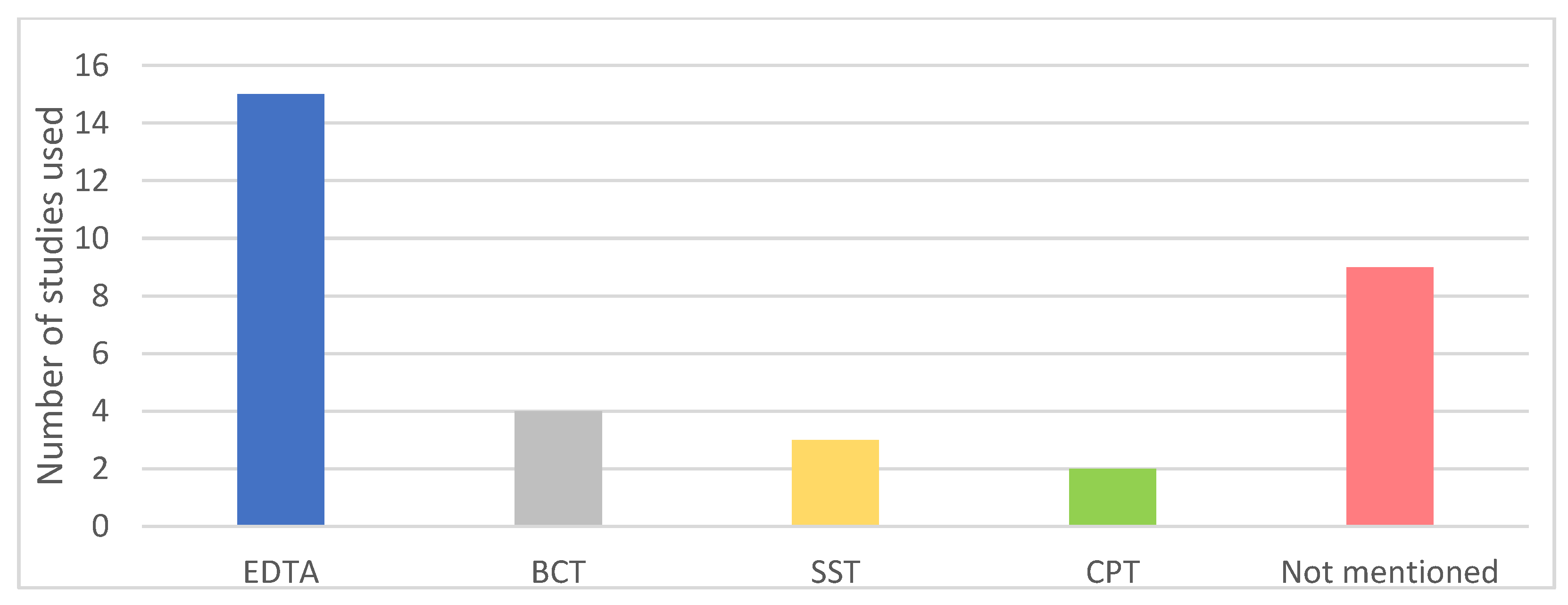
2.4. Methodological Quality
3. Results
3.1. Literature Search and Selection
3.2. Characteristics of the Included Studies
| Detection | First Author | Year | Method | Target(s) | Baseline | % | Longitudinal | % | Correlative | Measurement | Determined by | Risk of Bias (QUIPS) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | J.A. Garcia-Saenz [16] | 2017 | dPCR | PIK3CA | 8/32 | 25.0% | 8/8 * | 100.0% | Partial | Continuous | RECIST, PTM | Low |
| W. Jacot [17] | 2019 | dPCR | PIK3CA | 10/36 | 27.8% | 10/36 | 27.8% | Yes | Endpoint | RECIST | Low | |
| A.R. Kodahl [18] | 2018 | dPCR | PIK3CA | 20/60 | 33.3% | 4/6 * | 66.7% | Yes | Continuous | RECIST | Moderate | |
| X. Li [19] | 2020 | NGS | ESR1 | 9/45 | 20.0% | 5/5 * | 100.0% | Partial | Continuous | RECIST, PTM | Moderate | |
| P. Wang [20] | 2015 | dPCR | ESR1 | 7/29 | 24.1% | 4/4 * | 100.0% | Yes | Continuous | PTM | Moderate | |
| S.R. Vitale [21] | 2018 | dPCR | ESR1 | 3/67 | 4.5% | 4/17 | 23.5% | Yes | Continuous | Not defined | Moderate | |
| C. Paoletti [22] | 2018 | dPCR | ESR1 | 14/45 | 31.1% | 17/45 | 37.8% | Yes | Endpoint | RECIST, PTM | Low | |
| D. Sefrioui [23] | 2015 | dPCR | ESR1 | 4/7 | 57.1% | 4/7 | 57.1% | Yes | Continuous | Not defined | Low | |
| E. Jeannot [24] | 2020 | dPCR | ESR1 | 17/59 | 28.8% | 15/15 * | 100.0% | Yes | Endpoint | RECIST | Low | |
| F. Clatot + [15] | 2020 | dPCR | ESR1 | 22/70 | 31.4% | 22/22 * | 100.0% | Yes | Continuous | RECIST | Low | |
| T. Takeshita [25] | 2016 | dPCR | ESR1 | 12/42 | 28.6% | 12/42 | 28.6% | Yes | Both | RECIST, CA 15-3, CEA | Low | |
| T. Takeshita [26] | 2017 | dPCR | PIK3CA | 17/69 | 24.6% | 21/52 | 40.4% | No | Both | RECIST | Low | |
| ““ | ESR1 | 20/69 | 29.0% | 24/52 | 46.2% | Yes | Both | ““ | ||||
| J.M. Spoerke [27] | 2016 | dPCR | PIK3CA | 62/156 | 39.7% | 41/60 | 68.3% | Yes | Both | RECIST | Low | |
| ““ | ESR1 | 57/153 | 37.3% | 42/60 | 70.0% | No | Both | ““ | ||||
| B. O’Leary [5] | 2018 | dPCR | PIK3CA | 100/455 | 22.0% | 65/65 * | 100.0% | Yes | Endpoint | RECIST | Low | |
| ““ | ESR1 | 114/445 | 25.6% | 73/73 * | 100.0% | No | ““ | ““ | ||||
| J.S. Frenel [28] | 2015 | dPCR | PIK3CA | 1/7 | 14.3% | 1/7 | 14.3% | No | Endpoint | RECIST | Low | |
| ““ | TP53 | 5/7 | 71.4% | 2/7 | 28.6% | Yes | ““ | ““ | ||||
| S.J. Dawson [4] | 2013 | TAm-Seq | PIK3CA and/or TP53 | 24/52 | 46.2% | 25/30 | 83.3% | Yes | Continuous | RECIST | Low | |
| ““ | PIK3CA | 9/30 | 30.0% | 11/30 | 36.7% | n.d. | ||||||
| ““ | TP53 | 15/30 | 50.0% | 17/30 | 56.7% | n.d. | ||||||
| S. Hrebien [29] | 2019 | dPCR | PIK3CA, GATA3, ESR1 and/or TP53 | 38/58 | 78.1% | 35/35 * | 100.0% | Yes | Endpoint | n.a. | Low | |
| PIK3CA | 30/58 | 51.7% | 32/35 | 91.4% | n.d. | |||||||
| TP53 | 4/58 | 6.9% | 4/31 | 12.9% | n.d. | |||||||
| F. Ma + [14] | 2016 | NGS | PIK3CA, mTOR, PTEN and/or TP53 | 9/18 | 50.0% | 11/18 | 61.1% | Yes | Endpoint | RECIST | Low | |
| ““ | PIK3CA | 6/18 | 33.3% | 8/18 | 44.4% | n.d. | ||||||
| ““ | TP53 | 3/18 | 16.7% | 7/18 | 39.9% | n.d. | ||||||
| F. Ma [13] | 2019 | NGS | 193 gene panel | 37/37 | 100.0% | 21/21 | 100.0% | Yes | Endpoint | RECIST | Low | |
| ““ | PIK3CA | 13/37 | 35.1% | 10/21 | 47.6% | n.d. | ||||||
| ““ | TP53 | 20/37 | 54.1% | 13/21 | 61.9% | n.d. | ||||||
| S.W. Lok [30] | 2018 | dPCR | PIK3CA, ESR1, GATA3 and/or MAP3K1 | 28/33 | 84.8% | 28/33 | 84.8% | n.d. | Low | |||
| ““ | PIK3CA | 14/33 | 42.4% | 14/33 | 42.4% | n.d. | ||||||
| ““ | ESR1 | 10/33 | 30.3% | 10/33 | 30.3% | Yes | Both | RECIST | ||||
| ““ | GATA3 | 5/33 | 15.2% | 5/33 | 15.2% | n.d. | ||||||
| ““ | MAP3K1 | 4/33 | 12.1% | 4/33 | 12.1% | n.d. | ||||||
| C.X. Ma [31] | 2017 | NGS | HER2 | 9/381 | 2.4% | 11/11 * | 100.0% | Yes | Continuous | RECIST | Low | |
| ““ | PIK3CA | n.a. | 5/11 | 45.5% | n.d. | |||||||
| ““ | ESR1 | n.a. | 2/11 | 18.1% | n.d. | |||||||
| ““ | TP53 | n.a. | 6/11 | 54.5% | n.d. | |||||||
| C. Hufnagl [32] | 2020 | TAm-Seq | 8 gene panel | 4/4 | 100.0% | 4/4 | 100.0% | No | Continuous | RECIST, CA 15-3 | Moderate | |
| K. Page [33] | 2016 | dPCR | 16 gene panel | 21/42 | 50.0% | 9/9 * | 100.0% | Yes | Continuous | RECIST, CA 15-3 | Moderate | |
| ““ | PIK3CA | 12/42 | 28.6% | 6/9 | 66.7% | n.d. | ||||||
| ““ | ESR1 | 6/42 | 14.3% | 2/9 | 22.2% | n.d. | ||||||
| ““ | TP53 | 6/42 | 14.3% | 2/9 | 22.2% | n.d. | ||||||
| R.D. Baird [34] | 2019 | TAm-Seq | 20-gene panel | 12/30 | 40% | 4/4 * | 100% | Yes | Continuous | RECIST | Low | |
| ““ | PIK3CA | 7/30 | 23.3% | 2/4 | 50% | n.d. | ||||||
| ““ | ESR1 | 3/30 | 10% | 1/4 | 25% | n.d. | ||||||
| ““ | TP53 | 5/30 | 16.7% | 2/4 | 50% | n.d. | ||||||
| CNV | Guan [35] | 2020 | NGS | HER2 | 47/105 | 44.8% | 19/26 | 73.1% | Yes | Endpoint | Not defined | Low |
| B.S. Sorensen [36] | 2010 | qPCR | HER2 | 14/28 | 50.0% | 22/22 | 100.0% | Yes | Endpoint | Not defined | Low | |
| F. Ma + [14] | 2016 | NGS | HER2 | 13/17 | 76.5% | 13/17 | 76.5% | Yes | Continuous | RECIST | Low | |
| C. Suppan [6] | 2019 | FastSeq | Line1 | 10/29 | 34.5% | 29/29 | 100.0% | Yes | Continuous | CA 15-3 | Moderate | |
| Methylation | M.J. Fackler [7] | 2014 | qMSP | Cumulative gene index (10 genes) | 52/57 | 91.2% | 13/13 * | 100.0% | Yes | Continuous | RECIST | Low |
| 29/29 * | 100.0% | Yes | Endpoint | |||||||||
| K. Visvanathan [37] | 2016 | qMSP | Cumulative gene index (6/10 genes) | 129/129 | 100.0% | 129/129 | 100.0% | Yes | Endpoint | RECIST | Low | |
| M. Zurita [38] | 2010 | qMSP | 14-3-3-σ | 34/34 | 100.0% | 34/34 | 100.0% | Yes | Endpoint | RECIST | Low | |
| S. Kristiansen [39] | 2015 | qMSP | RASSF1A | n.a. | n.a. | 29/29 | 100.0% | Yes | Continuous | PTM | Moderate | |
| qMSP | LINE-1 | n.a. | n.a. | n.a | n.a | No | Continuous | PTM | ||||
| X.L. Liu [40] | 2020 | WGBS | Whole genome | 16/16 | 100.0% | 16/16 | 100.0% | Yes | Endpoint | Not defined | Moderate | |
| Size/Conc. | Z. Ye [41] | 2019 | qPCR | Alu115 and Alu81 | 117/117 | 100.0% | 22/22 | 100.0% | Yes | Endpoint | RECIST | Low |
| F. Clatot + [15] | 2020 | dPCR | cfDNA conc. | 103/103 | 100.0% | 70/70 | 100.0% | No | Continuous | RECIST | Low |
3.3. Risk of Bias
3.4. Mutation-Based ctDNA Detection
3.5. Copy Number Based ctDNA Detection
3.6. DNA Methylation-Based ctDNA Detection
3.7. cfDNA Abundance
4. Discussion
4.1. Analytical Validity
4.2. Clinical Validity
4.3. Clinical Utility
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W.; et al. Circulating Tumor Cells versus Imaging—Predicting Overall Survival in Metastatic Breast Cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef] [Green Version]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, S.-J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Leary, B.; Hrebien, S.; Morden, J.P.; Beaney, M.; Fribbens, C.; Huang, X.; Liu, Y.; Bartlett, C.H.; Koehler, M.; Cristofanilli, M.; et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suppan, C.; Brcic, I.; Tiran, V.; Mueller, H.D.; Posch, F.; Auer, M.; Ercan, E.; Ulz, P.; Cote, R.J.; Datar, R.H.; et al. Untargeted Assessment of Tumor Fractions in Plasma for Monitoring and Prognostication from Metastatic Breast Cancer Patients Undergoing Systemic Treatment. Cancers 2019, 11, 1171. [Google Scholar] [CrossRef] [Green Version]
- Fackler, M.J.; Bujanda, Z.L.; Umbricht, C.; Teo, W.W.; Cho, S.; Zhang, Z.; Visvanathan, K.; Jeter, S.; Argani, P.; Wang, C.; et al. Novel Methylated Biomarkers and a Robust Assay to Detect Circulating Tumor DNA in Metastatic Breast Cancer. Cancer Res. 2014, 74, 2160–2170. [Google Scholar] [CrossRef] [Green Version]
- Hrebien, S.; O’Leary, B.; Beaney, M.; Schiavon, G.; Fribbens, C.; Bhambra, A.; Johnson, R.; García-Murillas, I.; Turner, N. Reproducibility of Digital PCR Assays for Circulating Tumor DNA Analysis in Advanced Breast Cancer. PLoS ONE 2016, 11, e0165023. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.A.; Chung, J.H.; Kennedy, M.; Hughes, J.D.; Chennagiri, N.; Lieber, D.S.; Fendler, B.; Young, L.; Zhao, M.; Coyne, M.; et al. Analytical Validation of a Hybrid Capture–Based Next-Generation Sequencing Clinical Assay for Genomic Profiling of Cell-Free Circulating Tumor DNA. J. Mol. Diagn. 2018, 20, 686–702. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [Green Version]
- Hayden, J.A.; Van Der Windt, D.A.; Cartwright, J.L.; Côté, P.; Bombardier, C. Assessing Bias in Studies of Prognostic Factors. Ann. Intern. Med. 2013, 158, 280–286. [Google Scholar] [CrossRef]
- van Dessel, L.F.; Beije, N.; Helmijr, J.C.A.; Vitale, S.R.; Kraan, J.; Look, M.P.; de Wit, R.; Sleijfer, S.; Jansen, M.P.H.M.; Martens, J.M.N.; et al. Application of circulating tumor DNA in prospective clinical oncology trials–standardization of preanalytical conditions. Mol. Oncol. 2017, 11, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.; Guan, Y.; Yi, Z.; Chang, L.; Li, Q.; Chen, S.; Zhu, W.; Guan, X.; Li, C.; Qian, H.; et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int. J. Cancer 2020, 146, 1359–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.; Zhu, W.; Guan, Y.; Yang, L.; Xia, X.; Chen, S.; Li, Q.; Guan, X.; Yi, Z.; Qian, H.; et al. ctDNA dynamics: A novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 2016, 7, 66020–66031. [Google Scholar] [CrossRef] [PubMed]
- Clatot, F.; Perdrix, A.; Beaussire, L.; LeQuesne, J.; Lévy, C.; Emile, G.; Bubenheim, M.; Lacaille, S.; Calbrix, C.; Augusto, L.; et al. Risk of early progression according to circulating ESR1 mutation, CA-15.3 and cfDNA increases under first-line anti-aromatase treatment in metastatic breast cancer. Breast Cancer Res. 2020, 22, 1–12. [Google Scholar] [CrossRef]
- García-Saenz, J.A.; Ayllón, P.; Laig, M.; Acosta-Eyzaguirre, D.; García-Esquinas, M.; Montes, M.; Sanz, J.; Barquín, M.; Moreno, F.; Garcia-Barberan, V.; et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 2017, 17, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacot, W.; Dalenc, F.; Lopez-Crapez, E.; Chaltiel, L.; Durigova, A.; Gros, N.; Lozano, N.; Lacaze, J.-L.; Pouderoux, S.; Gladieff, L.; et al. PIK3CA mutations early persistence in cell-free tumor DNA as a negative prognostic factor in metastatic breast cancer patients treated with hormonal therapy. Breast Cancer Res. Treat. 2019, 177, 659–667. [Google Scholar] [CrossRef]
- Kodahl, A.R.; Ehmsen, S.; Pallisgaard, N.; Jylling, A.M.B.; Jensen, J.D.; Laenkholm, A.-V.; Knoop, A.S.; Ditzel, H.J. Correlation between circulating cell-free PIK 3 CA tumor DNA levels and treatment response in patients with PIK 3 CA -mutated metastatic breast cancer. Mol. Oncol. 2018, 12, 925–935. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Lu, J.; Zhang, L.; Luo, Y.; Zhao, Z.; Li, M. Clinical Implications of Monitoring ESR1 Mutations by Circulating Tumor DNA in Estrogen Receptor Positive Metastatic Breast Cancer: A Pilot Study. Transl. Oncol. 2020, 13, 321–328. [Google Scholar] [CrossRef]
- Wang, P.; Bahreini, A.; Gyanchandani, R.; Lucas, P.C.; Hartmaier, R.J.; Watters, R.J.; Jonnalagadda, A.R.; Bittar, H.E.T.; Berg, A.; Hamilton, R.L.; et al. Sensitive Detection of Mono- and Polyclonal ESR1 Mutations in Primary Tumors, Metastatic Lesions, and Cell-Free DNA of Breast Cancer Patients. Clin. Cancer Res. 2016, 22, 1130–1137. [Google Scholar] [CrossRef] [Green Version]
- Vitale, S.R.; Sieuwerts, A.M.; Beije, N.; Kraan, J.; Angus, L.; Mostert, B.; Reijm, E.A.; Van, N.M.; Van Marion, R.; Dirix, L.Y.; et al. An Optimized Workflow to Evaluate Estrogen Receptor Gene Mutations in Small Amounts of Cell-Free DNA. J. Mol. Diagn. 2019, 21, 123–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paoletti, C.; Schiavon, G.; Dolce, E.M.; Darga, E.P.; Carr, T.H.; Geradts, J.; Hoch, M.; Klinowska, T.; Lindemann, J.; Marshall, G.; et al. Circulating Biomarkers and Resistance to Endocrine Therapy in Metastatic Breast Cancers: Correlative Results from AZD9496 Oral SERD Phase I Trial. Clin. Cancer Res. 2018, 24, 5860–5872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sefrioui, D.; Perdrix, A.; Sarafan-Vasseur, N.; Dolfus, C.; Dujon, A.; Picquenot, J.M.; Delacour, J.; Cornic, M.; Bohers, E.; Leheurteur, M.; et al. Monitoring ESR 1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int. J. Cancer 2015, 137, 2513–2519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeannot, E.; Darrigues, L.; Michel, M.; Stern, M.-H.; Pierga, J.-Y.; Rampanou, A.; Melaabi, S.; Benoist, C.; Bièche, I.; Vincent-Salomon, A.; et al. A single droplet digital PCR for ESR1 activating mutations detection in plasma. Oncogene 2020, 39, 2987–2995. [Google Scholar] [CrossRef]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Inao, T.; Sueta, A.; Fujiwara, S.; Omoto, Y.; Iwase, H. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget 2016, 7, 32504–32518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeshita, T.; Yamamoto, Y.; Yamamoto-Ibusuki, M.; Tomiguchi, M.; Sueta, A.; Murakami, K.; Omoto, Y.; Iwase, H. Analysis of ESR1 and PIK3CA mutations in plasma cell-free DNA from ER-positive breast cancer patients. Oncotarget 2017, 8, 52142–52155. [Google Scholar] [CrossRef] [Green Version]
- Spoerke, J.M.; Gendreau, S.; Walter, K.; Qiu, J.; Wilson, T.R.; Savage, H.; Aimi, J.; Derynck, M.K.; Chen, M.; Chan, I.T.; et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Frenel, J.S.; Carreira, S.; Goodall, J.; Roda, D.; Perez-Lopez, R.; Tunariu, N.; Riisnaes, R.; Miranda, S.; Figueiredo, I.; Nava-Rodrigues, D.; et al. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clin. Cancer Res. 2015, 21, 4586–4596. [Google Scholar] [CrossRef] [Green Version]
- Hrebien, S.; Citi, V.; Garcia-Murillas, I.; Cutts, R.; Fenwick, K.; Kozarewa, I.; McEwen, R.; Ratnayake, J.; Maudsley, R.; Carr, T.; et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019, 30, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Lok, S.W.; Whittle, J.R.; Vaillant, F.; Teh, C.E.; Lo, L.L.; Policheni, A.N.; Bergin, A.R.T.; Desai, J.; Ftouni, S.; Gandolfo, L.C.; et al. A phase Ib dose-escalation and expansion study of the BCL2 inhibitor venetoclax combined with tamoxifen in ER and BCL2–positive metastatic breast cancer. Cancer Discov. 2019, 9, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.X.; Bose, R.; Gao, F.; Freedman, R.A.; Telli, M.L.; Kimmick, G.; Winer, E.; Naughton, M.; Goetz, M.P.; Russell, C.; et al. Neratinib Efficacy and Circulating Tumor DNA Detection of HER2 Mutations in HER2 Nonamplified Metastatic Breast Cancer. Clin. Cancer Res. 2017, 23, 5687–5695. [Google Scholar] [CrossRef] [Green Version]
- Hufnagl, C.; Leisch, M.; Weiss, L.; Melchardt, T.; Moik, M.; Asslaber, D.; Roland, G.; Steininger, P.; Meissnitzer, T.; Neureiter, D.; et al. Evaluation of circulating cell-free DNA as a molecular monitoring tool in patients with metastatic cancer. Oncol. Lett. 2019, 19, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Page, K.; Guttery, D.S.; Fernandez-Garcia, D.; Hills, A.; Hastings, R.K.; Luo, J.; Goddard, K.; Shahin, V.; Woodley-Barker, L.; Rosales, B.M.; et al. Next Generation Sequencing of Circulating Cell-Free DNA for Evaluating Mutations and Gene Amplification in Metastatic Breast Cancer. Clin. Chem. 2017, 63, 532–541. [Google Scholar] [CrossRef] [Green Version]
- Baird, R.D.; Van Rossum, A.G.; Oliveira, M.; Beelen, K.; Gao, M.; Schrier, M.; Mandjes, I.A.; Garcia-Corbacho, J.; Vallier, A.-L.; Dougall, G.; et al. POSEIDON Trial Phase 1b Results: Safety, Efficacy and Circulating Tumor DNA Response of the Beta Isoform-Sparing PI3K Inhibitor Taselisib (GDC-0032) Combined with Tamoxifen in Hormone Receptor Positive Metastatic Breast Cancer Patients. Clin. Cancer Res. 2019, 25, 6598–6605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, X.; Liu, B.; Niu, Y.; Dong, X.; Zhu, X.; Li, C.; Li, L.; Yi, Z.; Sun, X.; Chen, H.; et al. Longitudinal HER2 amplification tracked in circulating tumor DNA for therapeutic effect monitoring and prognostic evaluation in patients with breast cancer. Breast 2020, 49, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, B.S.; Mortensen, L.S.; Andersen, J.; Nexo, E. Circulating HER2 DNA after trastuzumab treatment predicts survival and response in breast cancer. Anticancer Res. 2010, 30, 2463–2468. [Google Scholar] [PubMed]
- Visvanathan, K.; Fackler, M.S.; Zhang, Z.; Lopez-Bujanda, Z.A.; Jeter, S.C.; Sokoll, L.J.; Garrett-Mayer, E.; Cope, L.M.; Umbricht, C.B.; Euhus, D.M.; et al. Monitoring of Serum DNA Methylation as an Early Independent Marker of Response and Survival in Metastatic Breast Cancer: TBCRC 005 Prospective Biomarker Study. J. Clin. Oncol. 2017, 35, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Zurita, M.; Lara, P.C.; Del Moral, R.; Torres, B.; Linares-Fernández, J.L.; Arrabal, S.R.; Martínez-Galán, J.; Oliver, F.J.; De Almodóvar, J.M.R. Hypermethylated 14-3-3-σ and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer 2010, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristiansen, S.; Jørgensen, L.M.; Hansen, M.H.; Nielsen, D.; Sölétormos, G. Concordance of Hypermethylated DNA and the Tumor Markers CA 15-3, CEA, and TPA in Serum during Monitoring of Patients with Advanced Breast Cancer. BioMed Res. Int. 2015, 2015, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-R.; Zhang, R.-Y.; Gong, H.; Rugo, H.S.; Chen, L.-B.; Fu, Y.; Che, J.-W.; Tie, J.; Shao, B.; Wan, F.-L.; et al. Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033819896331. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Wang, C.; Wan, S.; Mu, Z.; Zhang, Z.; Abu-Khalaf, M.M.; Fellin, F.M.; Silver, D.P.; Neupane, M.; Jaslow, R.J.; et al. Association of clinical outcomes in metastatic breast cancer patients with circulating tumour cell and circulating cell-free DNA. Eur. J. Cancer 2019, 106, 133–143. [Google Scholar] [CrossRef]
- Razavi, P.; Chang, M.T.; Xu, G.; Bandlamudi, C.; Ross, D.S.; Vasan, N.; Cai, Y.; Bielski, C.M.; Donoghue, M.T.A.; Jonsson, P.; et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Angus, L.; Smid, M.; Wilting, S.M.; Van Riet, J.; Van Hoeck, A.; Nguyen, L.; Nik-Zainal, S.; Steenbruggen, T.G.; Tjan-Heijnen, V.C.G.; Labots, M.; et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat. Genet. 2019, 51, 1450–1458. [Google Scholar] [CrossRef]
- McMahon, K.W.; Karunasena, E.; Ahuja, N. The Roles of DNA Methylation in the Stages of Cancer. Cancer J. 2017, 23, 257–261. [Google Scholar] [CrossRef]
- Lampignano, R.; Neumann, M.H.D.; Weber, S.; Kloten, V.; Herdean, A.; Voss, T.; Groelz, D.; Babayan, A.; Tibbesma, M.; Schlumpberger, M.; et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2019, 66, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Bos, M.K.; Nasserinejad, K.; Jansen, M.P.H.M.; Angus, L.; Atmodimedjo, P.N.; De Jonge, E.; Dinjens, W.N.M.; Van Schaik, R.H.N.; Del Re, M.; Dubbink, H.J.; et al. Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol. Oncol. 2021, 15, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belic, J.; Koch, M.; Ulz, P.; Auer, M.; Gerhalter, T.; Mohan, S.; Fischereder, K.; Petru, E.; Bauernhofer, T.; Geigl, J.B.; et al. Rapid Identification of Plasma DNA Samples with Increased ctDNA Levels by a Modified FAST-SeqS Approach. Clin. Chem. 2015, 61, 838–849. [Google Scholar] [CrossRef] [Green Version]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Juric, D.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.; Iwata, H.; Conte, P.; Mayer, I.; Kaufman, B.; et al. Abstract GS3-08: Alpelisib + fulvestrant for advanced breast cancer: Subgroup analyses from the phase III SOLAR-1 trial. Gen. Sess. Abstr. 2019, 79, GS3-08. [Google Scholar] [CrossRef]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.D.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef]
- Zivanovic, B.A.; Weng, C.F.; Silva, M.J.; Yeung, M.; Lo, L.; Ftouni, S.; Litchfield, C.; Ko, Y.-A.; Kuykhoven, K.; Van Geelen, C.; et al. Circulating tumour DNA in metastatic breast cancer to guide clinical trial enrolment and precision oncology: A cohort study. PLoS Med. 2020, 17, e1003363. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef] [PubMed]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating Tumor Cells and Response to Chemotherapy in Metastatic Breast Cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef] [PubMed]
- Tellez-Gabriel, M.; Knutsen, E.; Perander, M. Current Status of Circulating Tumor Cells, Circulating Tumor DNA, and Exosomes in Breast Cancer Liquid Biopsies. Int. J. Mol. Sci. 2020, 21, 9457. [Google Scholar] [CrossRef]


| Assay | Advantages | Drawbacks |
|---|---|---|
| Mutations |
|
|
| DNA Methylation |
|
|
| Copy number variations |
|
|
| cfDNA concentration |
|
|
| ctDNA Application | Study Name/Author | Status | Patients | Intervention | Control | Outcome |
|---|---|---|---|---|---|---|
| Acting on resistance mutations | PADA-1 (NCT03079011) | Ongoing | 800 | Palbociclib treated patient with rising ESR1 mutation levels will receive additional fulvestrant | Patients will continue with palbociclib. A subset will be crossed over | t.b.a. |
| INTERACT Study (NCT04256941) | Ongoing | 124 | AI and CDK4/6i treated patients with ESR1 mutation after 12 months will switch to fulvestrant | Patients will continue with AI | t.b.a. | |
| Targeting actionable mutations | PlasmaMATCH [52] | Published | 1150 | Patients with detected mutations will enter a specific treatment cohort: (1) ESR1, fulvestrant; (2) HER2, neratinib; (3) AKT (and ER+), capivasertib plus fulvestrant; (4) AKT pathway activation, capivasertib monotherapy | None | 357 patients (34%) with targetable mutation and 136 patients (13%) included in a treatment cohort |
| Zivanovic et al. [53] | Published | 234 | Treatment based on detected actionable mutations | None | 104 patients (44%) with actionable mutations, clinical management was changed in 40 patients (17%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jongbloed, E.M.; Deger, T.; Sleijfer, S.; Martens, J.W.M.; Jager, A.; Wilting, S.M. A Systematic Review of the Use of Circulating Cell-Free DNA Dynamics to Monitor Response to Treatment in Metastatic Breast Cancer Patients. Cancers 2021, 13, 1811. https://doi.org/10.3390/cancers13081811
Jongbloed EM, Deger T, Sleijfer S, Martens JWM, Jager A, Wilting SM. A Systematic Review of the Use of Circulating Cell-Free DNA Dynamics to Monitor Response to Treatment in Metastatic Breast Cancer Patients. Cancers. 2021; 13(8):1811. https://doi.org/10.3390/cancers13081811
Chicago/Turabian StyleJongbloed, Elisabeth M., Teoman Deger, Stefan Sleijfer, John W. M. Martens, Agnes Jager, and Saskia M. Wilting. 2021. "A Systematic Review of the Use of Circulating Cell-Free DNA Dynamics to Monitor Response to Treatment in Metastatic Breast Cancer Patients" Cancers 13, no. 8: 1811. https://doi.org/10.3390/cancers13081811
APA StyleJongbloed, E. M., Deger, T., Sleijfer, S., Martens, J. W. M., Jager, A., & Wilting, S. M. (2021). A Systematic Review of the Use of Circulating Cell-Free DNA Dynamics to Monitor Response to Treatment in Metastatic Breast Cancer Patients. Cancers, 13(8), 1811. https://doi.org/10.3390/cancers13081811






