Simultaneous Identification of EGFR,KRAS,ERBB2, and TP53 Mutations in Patients with Non-Small Cell Lung Cancer by Machine Learning-Derived Three-Dimensional Radiomics
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
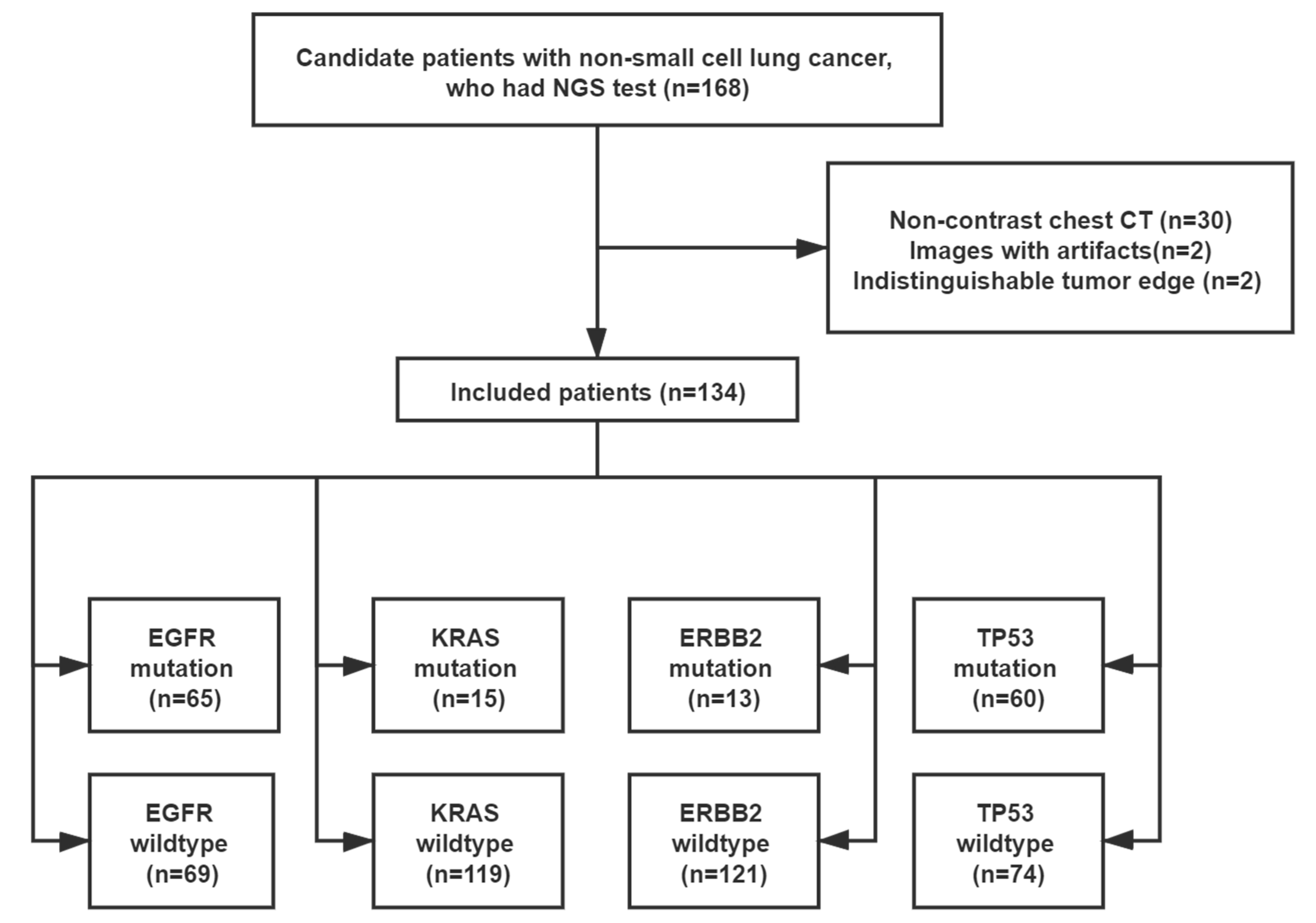
2.1. Study Population
2.2. NGS
2.3. CT Image Acquisition
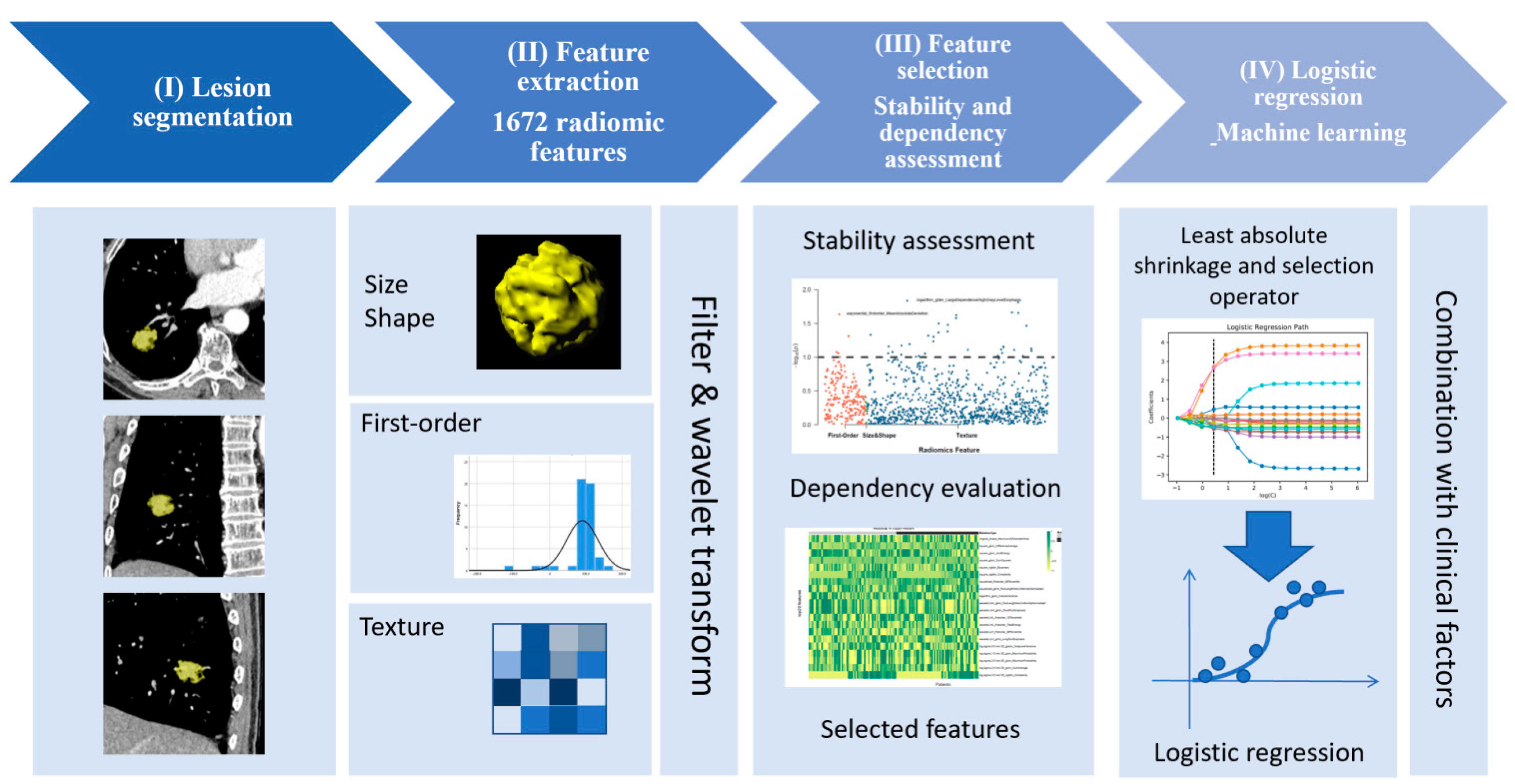
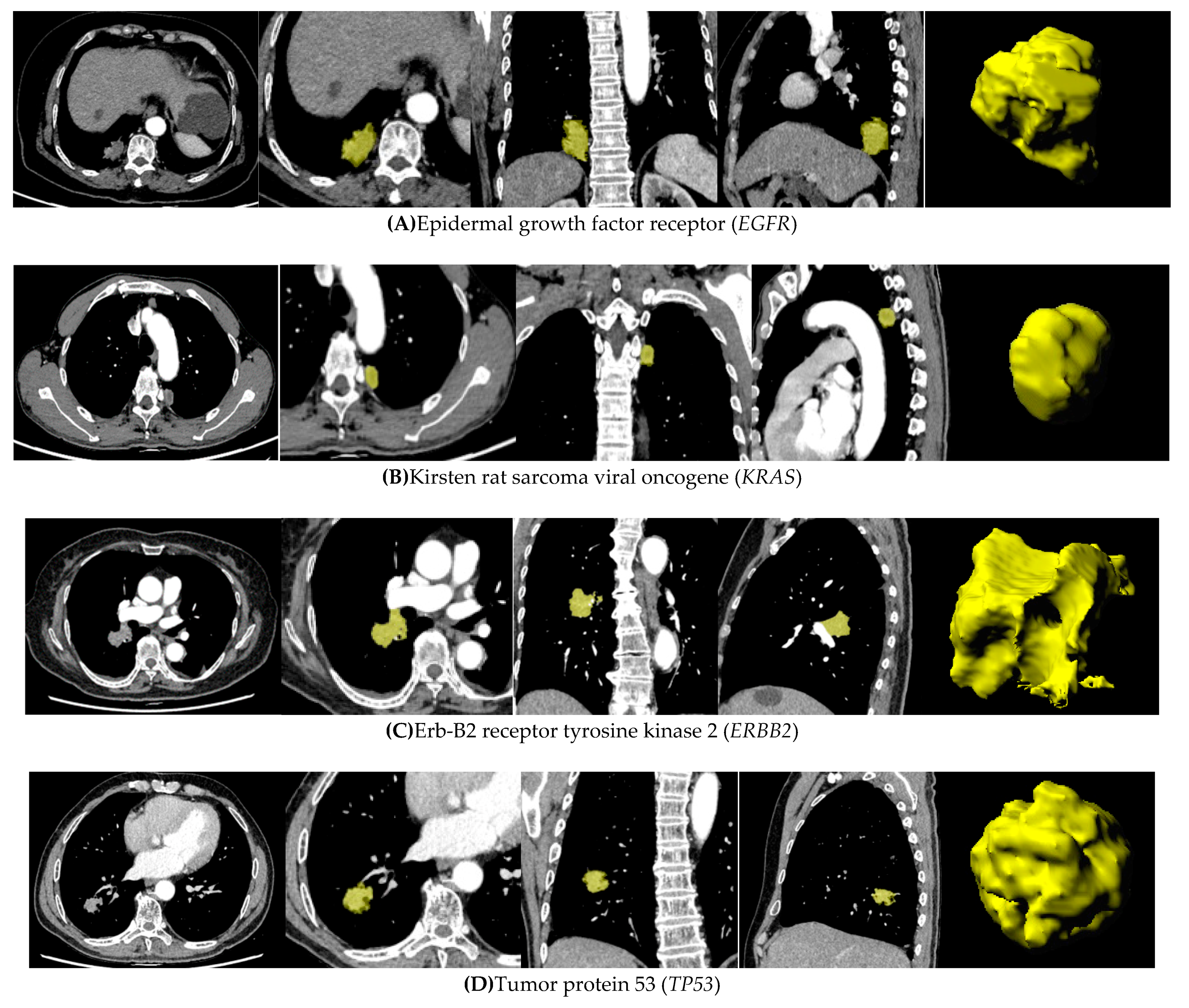
2.4. Lesion Segmentation and Extraction of Radiomic Features
2.5. Selection of Radiomic Features
2.6. Model Development
2.7. Statistical Analysis
3. Results
3.1. Demographics
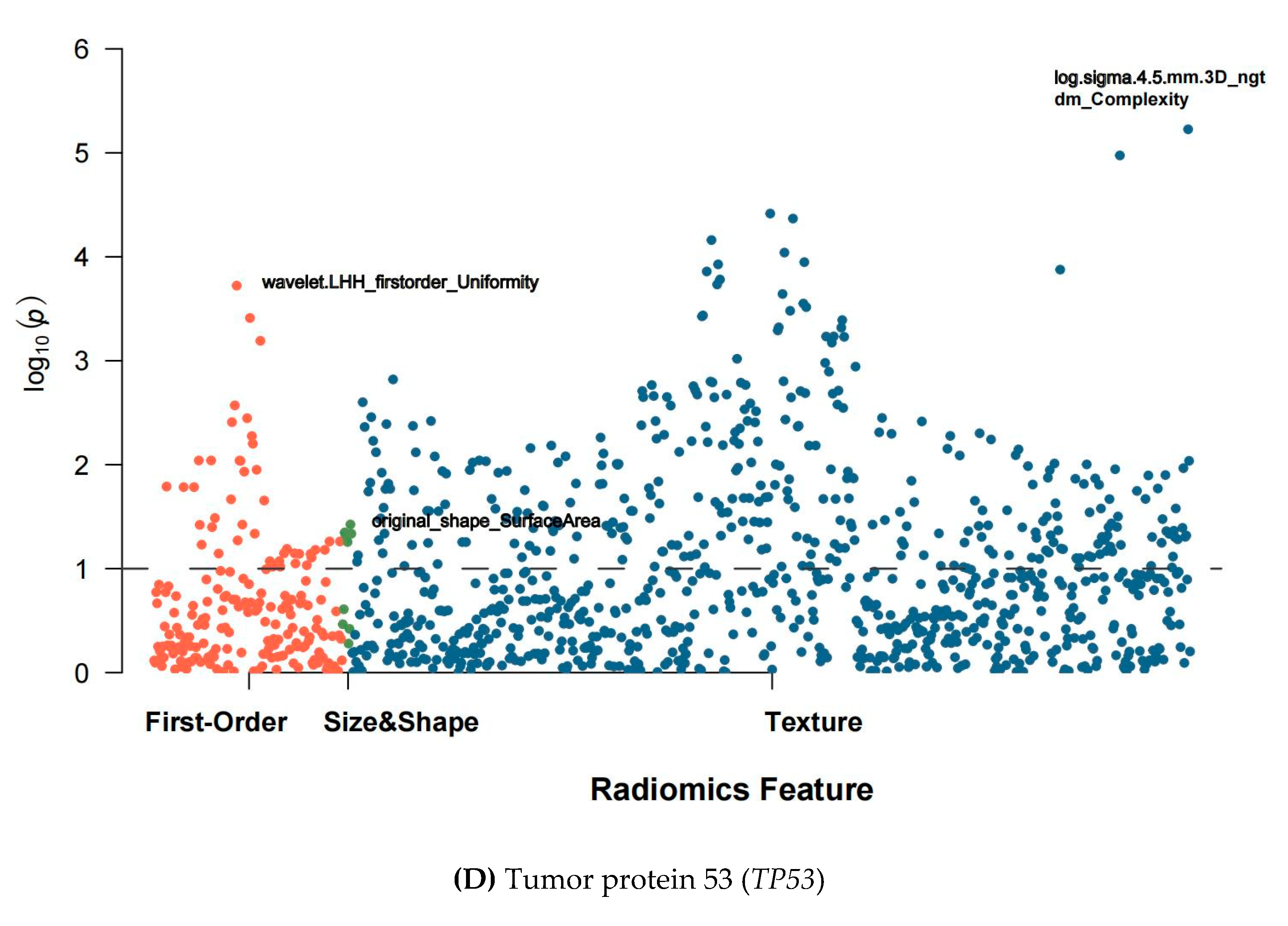
3.2. Extraction and Selection of Radiomic Features
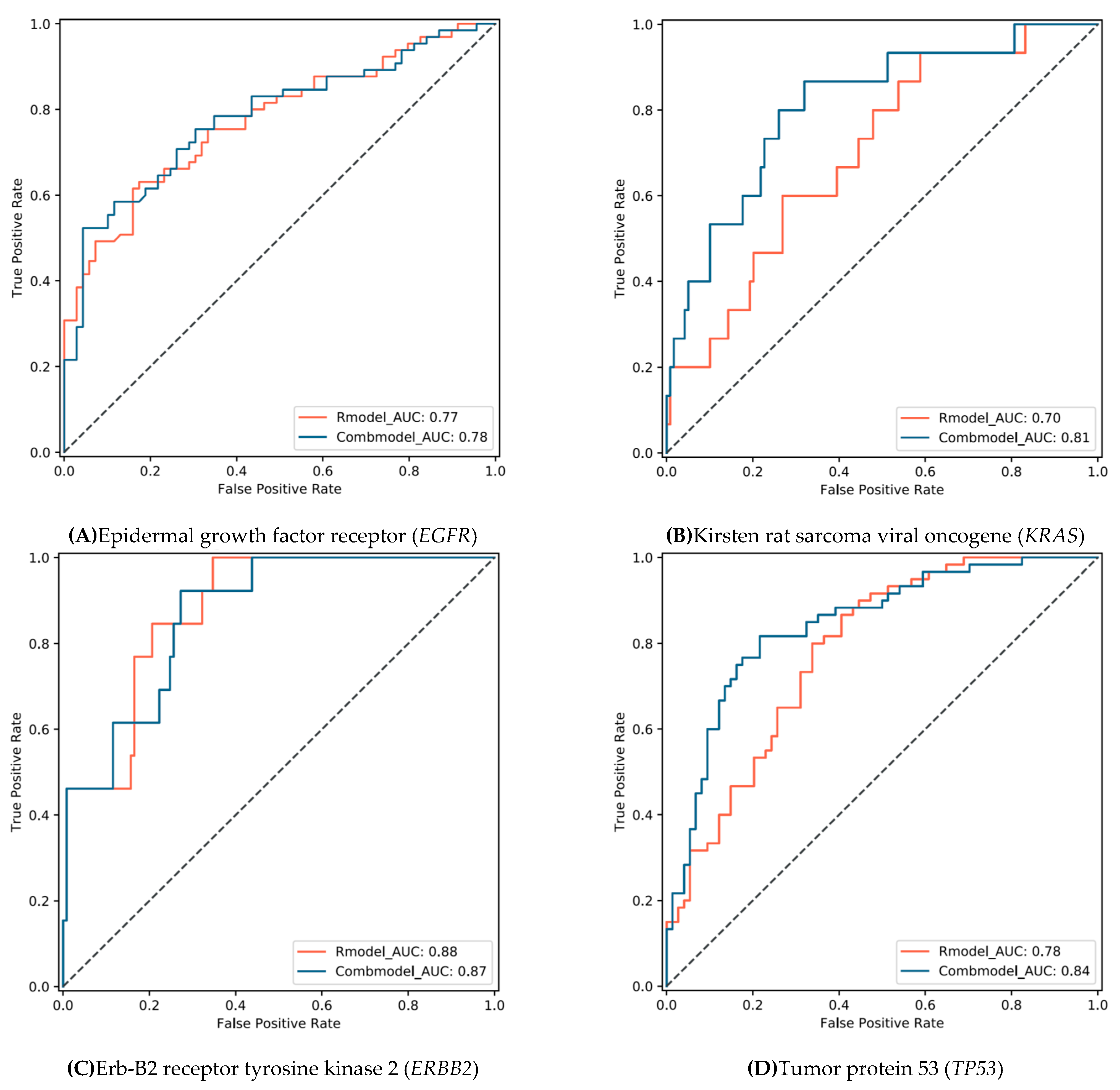
3.3. Model Performance
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nat. Cell Biol. 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Zhang, Y.-L.; Yuan, J.-Q.; Wang, K.-F.; Fu, X.-H.; Han, X.-R.; Threapleton, D.; Yang, Z.-Y.; Mao, C.; Tang, J.-L. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016, 7, 78985–78993. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Yang, H.; Teo, A.S.M.; Amer, L.B.; Sherbaf, F.G.; Tan, C.Q.; Alvarez, J.J.S.; Lu, B.; Lim, J.Q.; Takano, A.; et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020, 52, 177–186. [Google Scholar] [CrossRef]
- Roberts, P.J.; Stinchcombe, T.E.; Der, C.J.; Socinski, M.A. Personalized Medicine in Non–Small-Cell Lung Cancer: Is KRAS a Useful Marker in Selecting Patients for Epidermal Growth Factor Receptor–Targeted Therapy? J. Clin. Oncol. 2010, 28, 4769–4777. [Google Scholar] [CrossRef]
- Steuer, C.E.; Behera, M.; Berry, L.; Kim, S.; Rossi, M.; Sica, G.; Owonikoko, T.K.; Johnson, B.E.; Kris, M.G.; Bunn, P.A. Role of race in oncogenic driver prevalence and outcomes in lung adenocarcinoma: Results from the Lung Cancer Mutation Consor-tium. Cancer 2016, 122, 766–772. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, H.; Kawasaki, N.; Taguchi, M.; Kabasawa, K. Association of HER-2 overexpression with prognosis in nonsmall cell lung carcinoma: A metaanalysis. Cancer 2005, 103, 1865–1873. [Google Scholar] [CrossRef]
- Mar, N.; Vredenburgh, J.J.; Wasser, J.S. Targeting HER2 in the treatment of non-small cell lung cancer. Lung Cancer 2015, 87, 220–225. [Google Scholar] [CrossRef]
- Wang, X.; Kong, C.; Xu, W.; Yang, S.; Shi, D.; Zhang, J.; Du, M.; Wang, S.; Bai, Y.; Zhang, T.; et al. Decoding tumor mutation burden and driver mutations in early stage lung adenocarcinoma using CT-based radiomics signature. Thorac. Cancer 2019, 10, 1904–1912. [Google Scholar] [CrossRef] [Green Version]
- VanderLaan, P.A.; Rangachari, D.; Mockus, S.M.; Spotlow, V.; Reddi, H.V.; Malcolm, J.; Huberman, M.S.; Joseph, L.J.; Kobayashi, S.S.; Costa, D.B. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017, 106, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Castellanos, E.; Feld, E.; Horn, L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR -Mutated Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 612–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. New Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- D’Haene, N.; Le Mercier, M.; De Nève, N.; Blanchard, O.; Delaunoy, M.; El Housni, H.; Dessars, B.; Heimann, P.; Remmelink, M.; Demetter, P.; et al. Clinical Validation of Targeted Next Generation Sequencing for Colon and Lung Cancers. PLOS ONE 2015, 10, e0138245. [Google Scholar] [CrossRef]
- Frampton, G.M.; Fichtenholtz, A.; A Otto, G.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef]
- Fernandes, M.G.O.; Jacob, M.; Martins, N.; Moura, C.S.; Guimarães, S.; Reis, J.P.; Justino, A.; Pina, M.J.; Cirnes, L.; Sousa, C.; et al. Targeted Gene Next-Generation Sequencing Panel in Patients with Advanced Lung Adenocarcinoma: Paving the Way for Clinical Implementation. Cancers 2019, 11, 1229. [Google Scholar] [CrossRef] [Green Version]
- Van Haele, M.; Borght, S.V.; Ceulemans, A.; Wieërs, M.; Metsu, S.; Sagaert, X.; Weynand, B. Rapid clinical mutational testing of KRAS, BRAF and EGFR: a prospective comparative analysis of the Idylla technique with high-throughput next-generation sequencing. J. Clin. Pathol. 2019, 73, 35–41. [Google Scholar] [CrossRef]
- Chan, B.A.; Hughes, B.G. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl. Lung Cancer Res. 2015, 4, 36–54. [Google Scholar]
- Thawani, R.; McLane, M.; Beig, N.; Ghose, S.; Prasanna, P.; Velcheti, V.; Madabhushi, A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018, 115, 34–41. [Google Scholar] [CrossRef]
- Velazquez, E.R.; Parmar, C.; Liu, Y.; Coroller, T.P.; Cruz, G.; Stringfield, O.; Ye, Z.; Makrigiorgos, M.; Fennessy, F.; Mak, R.H.; et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Xu, Z.; Ge, Y.; Jiang, B.; Groen, H.; Vliegenthart, R.; Xie, X. 3D radiomics predicts EGFR mutation, exon-19 deletion and exon-21 L858R mutation in lung adenocarcinoma. Transl. Lung Cancer Res. 2020, 9, 1212–1224. [Google Scholar] [CrossRef]
- Jia, T.-Y.; Xiong, J.-F.; Li, X.-Y.; Yu, W.; Xu, Z.-Y.; Cai, X.-W.; Ma, J.-C.; Ren, Y.-C.; Larsson, R.; Zhang, J.; et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur. Radiol. 2019, 29, 4742–4750. [Google Scholar] [CrossRef]
- Pinheiro, G.; Pereira, T.; Dias, C.; Freitas, C.; Hespanhol, V.; Costa, J.L.; Cunha, A.; Oliveira, H.P. Identifying relationships between imaging phenotypes and lung cancer-related mutation status: EGFR and KRAS. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, Y.; An, T.; Gao, H.; Wang, K.; Zhou, Q.; Hu, Y.; Song, Y.; Ding, C.; Peng, F.; et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir. Med. 2018, 6, 681–690. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiol. 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fave, X.; Cook, M.; Frederick, A.; Zhang, L.; Yang, J.; Fried, D.; Stingo, F.; Court, L. Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput. Med Imaging Graph. 2015, 44, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kim, J.; Balagurunathan, Y.; Li, Q.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin. Lung Cancer 2016, 17, 441–448.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagher-Ebadian, H.; Janic, B.; Liu, C.; Pantelic, M.; Hearshen, D.; Elshaikh, M.; Movsas, B.; Chetty, I.J.; Wen, N. Detection of Dominant Intra-prostatic Lesions in Patients With Prostate Cancer Using an Artificial Neural Network and MR Multi-modal Radiomics Analysis. Front. Oncol. 2019, 9, 1313. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Iearn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Xie, D.; Wang, T.-T.; Huang, S.-J.; Deng, J.-J.; Ren, Y.-J.; Yang, Y.; Wu, J.-Q.; Zhang, L.; Fei, K.; Sun, X.-W.; et al. Radiomics nomogram for prediction disease-free survival and adjuvant chemotherapy benefits in patients with resected stage I lung adenocarcinoma. Transl. Lung Cancer Res. 2020, 9, 1112–1123. [Google Scholar] [CrossRef]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nat. Cell Biol. 2013, 503, 548–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patricelli, M.P.; Janes, M.R.; Li, L.-S.; Hansen, R.; Peters, U.; Kessler, L.V.; Chen, Y.; Kucharski, J.M.; Feng, J.; Ely, T.; et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 2016, 6, 316–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schram, A.M.; O’Reilly, E.M.; Somwar, R.; Benayed, R.; Shameem, S.; Chauhan, T.; Torrisi, J.; Ford, J.; Maussang, D.; Wasserman, E.; et al. Abstract PR02: Clinical proof of concept for MCLA-128, a bispecific HER2/3 antibody therapy, inNRG1fusion-positive cancers. In Monoclonal Antibodies; American Association for Cancer Research (AACR): Philadelphia, PA, USA, 2019; Volume 18, p. PR02. [Google Scholar]
- Canale, M.; Petracci, E.; Delmonte, A.; Chiadini, E.; Ulivi, P. Impact of TP53 Mutations on Outcome in EGFR-Mutated Pa-tients Treated with First-Line Tyrosine Kinase Inhibitors. Clin. Cancer Res. 2017, 23, 2195–2202. [Google Scholar] [CrossRef] [Green Version]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-R.; Schultheis, A.M.; Yu, H.; Mandelker, D.; Ladanyi, M.; Büttner, R. Precision medicine in non-small cell lung cancer: Current applications and future directions. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, B.; Liu, X.; Song, J.; Fang, M.; Hu, C.; Dong, D.; Li, W.; Tian, J. Quantitative Biomarkers for Prediction of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Lung Cancer. Transl. Oncol. 2018, 11, 94–101. [Google Scholar] [CrossRef]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef]


| True Label | Radiomic Features | Combined Model (Radiomic Features and Clinical Factors) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wildtype | Mutation | Sensitivity | Specificity | Accuracy | AUC | Wildtype | Mutation | Sensitivity | Specificity | Accuracy | AUC | ||
| EGFR | Wildtype (n = 69) | 57 | 12 | 0.63(0.50–0.74) | 0.83(0.71–0.90) | 0.73(0.65–0.80) | 0.77(0.70–0.85) | 66 | 3 | 0.52(0.40–0.65) | 0.96(0.87–0.99) | 0.75(0.66–0.82) | 0.78(0.70–0.86) |
| Mutation (n = 65) | 24 | 41 | 31 | 34 | |||||||||
| KRAS | Wildtype (n = 119) | 49 | 70 | 0.93(0.66–0.99) | 0.41(0.32–0.51) | 0.47(0.38–0.56) | 0.70(0.57–0.83) | 81 | 38 | 0.87(0.58–0.97) | 0.68(0.59–0.76) | 0.70(0.62- 0.78) | 0.81(0.69–0.93) |
| Mutation (n = 15) | 1 | 14 | 2 | 13 | |||||||||
| ERBB2 | Wildtype (n = 121) | 42 | 79 | 1.00(0.72–1.00) | 0.65(0.56–0.74) | 0.69(0.60–0.76) | 0.88(0.80–0.96) | 88 | 33 | 0.92(0.62–0.99) | 0.73(0.64–0.80) | 0.75(0.66–0.82) | 0.87(0.78–0.95) |
| Mutation (n = 13) | 0 | 13 | 1 | 12 | |||||||||
| TP53 | Wildtype (n = 74) | 49 | 25 | 0.80(0.67–0.89) | 0.66(0.54–0.77) | 0.72(0.64- 0.80) | 0.78(0.71–0.86) | 58 | 16 | 0.82(0.69–0.90) | 0.78(0.67–0.87) | 0.80(0.72–0.87) | 0.84(0.78–0.91) |
| Mutation (n = 60) | 12 | 48 | 11 | 49 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Xu, Z.; Liu, G.; Jiang, B.; de Bock, G.H.; Groen, H.J.M.; Vliegenthart, R.; Xie, X. Simultaneous Identification of EGFR,KRAS,ERBB2, and TP53 Mutations in Patients with Non-Small Cell Lung Cancer by Machine Learning-Derived Three-Dimensional Radiomics. Cancers 2021, 13, 1814. https://doi.org/10.3390/cancers13081814
Zhang T, Xu Z, Liu G, Jiang B, de Bock GH, Groen HJM, Vliegenthart R, Xie X. Simultaneous Identification of EGFR,KRAS,ERBB2, and TP53 Mutations in Patients with Non-Small Cell Lung Cancer by Machine Learning-Derived Three-Dimensional Radiomics. Cancers. 2021; 13(8):1814. https://doi.org/10.3390/cancers13081814
Chicago/Turabian StyleZhang, Tiening, Zhihan Xu, Guixue Liu, Beibei Jiang, Geertruida H. de Bock, Harry J. M. Groen, Rozemarijn Vliegenthart, and Xueqian Xie. 2021. "Simultaneous Identification of EGFR,KRAS,ERBB2, and TP53 Mutations in Patients with Non-Small Cell Lung Cancer by Machine Learning-Derived Three-Dimensional Radiomics" Cancers 13, no. 8: 1814. https://doi.org/10.3390/cancers13081814






