Find the Flame: Predictive Biomarkers for Immunotherapy in Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Searching Details
2.2. Selection Criteria
2.3. Data Extraction
2.4. Statistical Analysis
2.5. Results
3. Tumor-Intrinsic Biomarkers
3.1. Tumor Mutational Burden
3.2. MHC-I and MHC-II
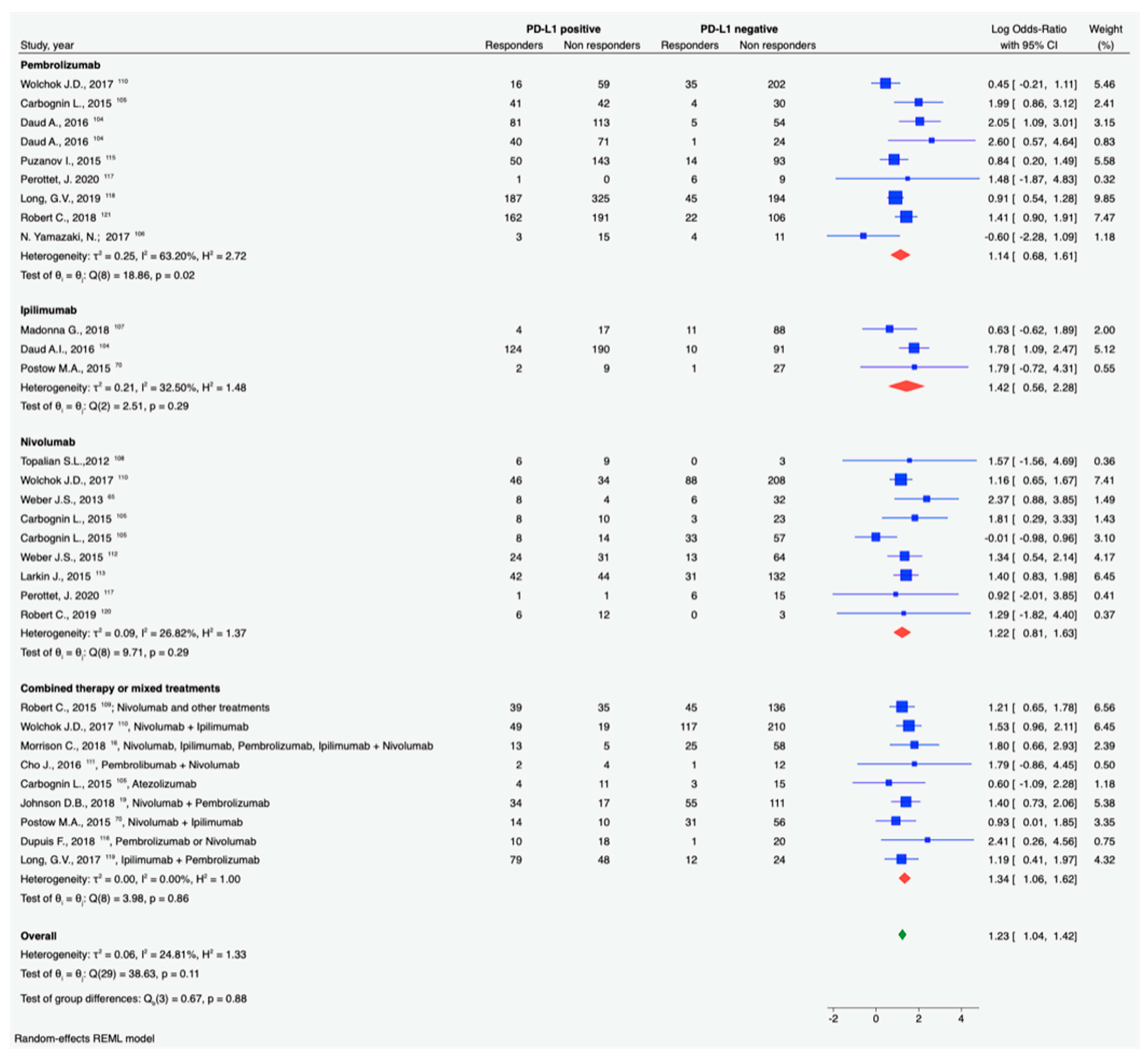
3.3. PD-1/PDL-1
4. Microenvironment Biomarkers
4.1. TILs
4.2. Gene Signatures
5. Systemic Biomarkers
5.1. Circulating Factors
5.2. Circulating Lymphocytes
5.3. Microbiota
5.4. Others
5.5. MDSCs
5.6. ctDNA
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CI | confidence interval |
| ctDNA | circulating tumor DNA |
| ICIs | immune-checkpoint inhibitors |
| IHC | Immunohistochemistry |
| LDH | lactate dehydrogenase |
| MDSC | myeloid derived suppressor cells |
| MHC | major histocompatibility complex |
| MM | metastatic melanoma |
| NGS | next-generation sequencing |
| NLR | neutrophil to lymphocyte ratio |
| ORR | objective response rate |
| RR | response rate |
| SD | standard deviation |
| TCR | T-cell receptor |
| TMB | tumor mutational burden |
References
- Pollack, L.A.; Li, J.; Berkowitz, Z.; Weir, H.K.; Wu, X.-C.; Ajani, U.A.; Ekwueme, D.U.; Li, C.; Pollack, B.P. Melanoma Survival in the United States, 1992 to 2005. J. Am. Acad. Dermatol. 2011, 65, S78–S86. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, P.A.; Vecchio, M.D.; Mandalá, M.; Gogas, H.; Arance, A.M.; Dalle, S.; Cowey, C.L.; Schenker, M.; Grob, J.-J.; Chiarion-Sileni, V.; et al. Adjuvant Nivolumab versus Ipilimumab in Resected Stage IIIB–C and Stage IV Melanoma (CheckMate 238): 4-Year Results from a Multicentre, Double-Blind, Randomised, Controlled, Phase 3 Trial. Lancet Oncol. 2020, 21, 1465–1477. [Google Scholar] [CrossRef]
- Eggermont, A.M.M.; Blank, C.U.; Mandala, M.; Long, G.V.; Atkinson, V.; Dalle, S.; Haydon, A.; Lichinitser, M.; Khattak, A.; Carlino, M.S.; et al. Adjuvant Pembrolizumab versus Placebo in Resected Stage III Melanoma. N. Engl. J. Med. 2018, 378, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Couzin-Frankel, J. Cancer Immunotherapy. Science 2013, 342, 1432–1433. [Google Scholar] [CrossRef]
- Adashek, J.J.; Subbiah, I.M.; Matos, I.; Garralda, E.; Menta, A.K.; Ganeshan, D.M.; Subbiah, V. Hyperprogression and Immunotherapy: Fact, Fiction, or Alternative Fact? Trends Cancer 2020, 6, 181–191. [Google Scholar] [CrossRef]
- Allen, E.M.V.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef]
- Johnson, D.B.; Frampton, G.M.; Rioth, M.J.; Yusko, E.; Xu, Y.; Guo, X.; Ennis, R.C.; Fabrizio, D.; Chalmers, Z.R.; Greenbowe, J.; et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol. Res. 2016, 4, 959–967. [Google Scholar] [CrossRef]
- Weber, J.; Horak, C.; Hodi, F.S.; Chang, H.; Woods, D.; Sanders, C.; Robins, H.; Yusko, E. Baseline Tumor T Cell Receptor (TcR) Sequencing Analysis and Neo Antigen Load Is Associated with Benefit in Melanoma Patients Receiving Sequential Nivolumab and Ipilimumab. Ann. Oncol. 2016, 27, vi359. [Google Scholar] [CrossRef]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martín-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-Tumor Genomic Biomarkers for PD-1 Checkpoint Blockade–Based Immunotherapy. Science 2018, 362. [Google Scholar] [CrossRef]
- Roszik, J.; Haydu, L.E.; Hess, K.R.; Oba, J.; Joon, A.Y.; Siroy, A.E.; Karpinets, T.V.; Stingo, F.C.; Baladandayuthapani, V.; Tetzlaff, M.T.; et al. Novel Algorithmic Approach Predicts Tumor Mutation Load and Correlates with Immunotherapy Clinical Outcomes Using a Defined Gene Mutation Set. BMC Med. 2016, 14, 168. [Google Scholar] [CrossRef] [PubMed]
- Roh, W.; Chen, P.-L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated Molecular Analysis of Tumor Biopsies on Sequential CTLA-4 and PD-1 Blockade Reveals Markers of Response and Resistance. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Morrison, C.; Pabla, S.; Conroy, J.M.; Nesline, M.K.; Glenn, S.T.; Dressman, D.; Papanicolau-Sengos, A.; Burgher, B.; Andreas, J.; Giamo, V.; et al. Predicting Response to Checkpoint Inhibitors in Melanoma beyond PD-L1 and Mutational Burden. J. Immunother. Cancer 2018, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative Molecular and Clinical Modeling of Clinical Outcomes to PD1 Blockade in Patients with Metastatic Melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Estrada, M.V.; Salgado, R.; Sanchez, V.; Doxie, D.B.; Opalenik, S.R.; Vilgelm, A.E.; Feld, E.; Johnson, A.S.; Greenplate, A.R.; et al. Melanoma-Specific MHC-II Expression Represents a Tumour-Autonomous Phenotype and Predicts Response to Anti-PD-1/PD-L1 Therapy. Nat. Commun. 2016, 7, 10582. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Bordeaux, J.; Kim, J.Y.; Vaupel, C.; Rimm, D.L.; Ho, T.H.; Joseph, R.W.; Daud, A.I.; Conry, R.M.; Gaughan, E.M.; et al. Quantitative Spatial Profiling of PD-1/PD-L1 Interaction and HLA-DR/IDO-1 Predicts Improved Outcomes of Anti-PD-1 Therapies in Metastatic Melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 5250–5260. [Google Scholar] [CrossRef] [PubMed]
- Rodig, S.J.; Gusenleitner, D.; Jackson, D.G.; Gjini, E.; Giobbie-Hurder, A.; Jin, C.; Chang, H.; Lovitch, S.B.; Horak, C.; Weber, J.S.; et al. MHC Proteins Confer Differential Sensitivity to CTLA-4 and PD-1 Blockade in Untreated Metastatic Melanoma. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.-P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to Checkpoint Blockade Therapy through Inactivation of Antigen Presentation. Nat. Commun. 2017, 8, 1136. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated with Ipilimumab. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.-M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic Short Chain Fatty Acids Limit Antitumor Effect of CTLA-4 Blockade in Hosts with Cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Nagatomo, R.; Doi, K.; Shimizu, J.; Baba, K.; Saito, T.; Matsumoto, S.; Inoue, K.; Muto, M. Association of Short-Chain Fatty Acids in the Gut Microbiome With Clinical Response to Treatment With Nivolumab or Pembrolizumab in Patients With Solid Cancer Tumors. JAMA Netw. Open 2020, 3, e202895. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Schmidt, H.; Nissan, A.; Ridolfi, L.; Aamdal, S.; Hansson, J.; Guida, M.; Hyams, D.M.; Gómez, H.; Bastholt, L.; et al. A Prospective Phase II Trial Exploring the Association between Tumor Microenvironment Biomarkers and Clinical Activity of Ipilimumab in Advanced Melanoma. J. Transl. Med. 2011, 9, 204. [Google Scholar] [CrossRef]
- Daud, A.I.; Loo, K.; Pauli, M.L.; Sanchez-Rodriguez, R.; Sandoval, P.M.; Taravati, K.; Tsai, K.; Nosrati, A.; Nardo, L.; Alvarado, M.D.; et al. Tumor Immune Profiling Predicts Response to Anti–PD-1 Therapy in Human Melanoma. J. Clin. Investig. 2016, 126, 3447–3452. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 Blockade Induces Responses by Inhibiting Adaptive Immune Resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Orlowski, R.J.; Xu, X.; Mick, R.; George, S.M.; Yan, P.K.; Manne, S.; Kraya, A.A.; Wubbenhorst, B.; Dorfman, L.; et al. A Single Dose of Neoadjuvant PD-1 Blockade Predicts Clinical Outcomes in Resectable Melanoma. Nat. Med. 2019, 25, 454–461. [Google Scholar] [CrossRef]
- Amaria, R.N.; Reddy, S.M.; Tawbi, H.A.; Davies, M.A.; Ross, M.I.; Glitza, I.C.; Cormier, J.N.; Lewis, C.; Hwu, W.-J.; Hanna, E.; et al. Neoadjuvant Immune Checkpoint Blockade in High-Risk Resectable Melanoma. Nat. Med. 2018, 24, 1649–1654. [Google Scholar] [CrossRef]
- Uryvaev, A.; Passhak, M.; Hershkovits, D.; Sabo, E.; Bar-Sela, G. The Role of Tumor-Infiltrating Lymphocytes (TILs) as a Predictive Biomarker of Response to Anti-PD1 Therapy in Patients with Metastatic Non-Small Cell Lung Cancer or Metastatic Melanoma. Med. Oncol. Northwood Lond. Engl. 2018, 35, 25. [Google Scholar] [CrossRef] [PubMed]
- Bifulco, C.; Capone, M.; Feng, Z.; Madonna, G.; Simeone, E.; Curvietto, M.; Mozzillo, N.; Ciliberto, G.; Botti, G.; Fox, B.A.; et al. MISIPI Study: Melanoma ImmunoScore Evaluation in Patients Treated with IPIlimumab. J. Transl. Med. 2014, 12, P11. [Google Scholar] [CrossRef]
- Galon, J.; Fox, B.A.; Bifulco, C.B.; Masucci, G.; Rau, T.; Botti, G.; Marincola, F.M.; Ciliberto, G.; Pages, F.; Ascierto, P.A.; et al. Immunoscore and Immunoprofiling in Cancer: An Update from the Melanoma and Immunotherapy Bridge 2015. J. Transl. Med. 2016, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, N.; Gonzalez-Cao, M.; Crespo, G.; Drozdowskyj, A.; Aldeguer, E.; Gimenez-Capitan, A.; Teixido, C.; Molina-Vila, M.A.; Viteri, S.; De Los Llanos Gil, M.; et al. Interferon Gamma, an Important Marker of Response to Immune Checkpoint Blockade in Non-Small Cell Lung Cancer and Melanoma Patients. Ther. Adv. Med. Oncol. 2018, 10, 1758834017749748. [Google Scholar] [CrossRef] [PubMed]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ-Related MRNA Profile Predicts Clinical Response to PD-1 Blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-L.; Roh, W.; Reuben, A.; Cooper, Z.A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Bassett, R.L.; Gopalakrishnan, V.; Wani, K.; et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016, 6, 827–837. [Google Scholar] [CrossRef]
- Gide, T.N.; Quek, C.; Menzies, A.M.; Tasker, A.T.; Shang, P.; Holst, J.; Madore, J.; Lim, S.Y.; Velickovic, R.; Wongchenko, M.; et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 2019, 35, 238–255.e6. [Google Scholar] [CrossRef] [PubMed]
- Diem, S.; Kasenda, B.; Spain, L.; Martin-Liberal, J.; Marconcini, R.; Gore, M.; Larkin, J. Serum Lactate Dehydrogenase as an Early Marker for Outcome in Patients Treated with Anti-PD-1 Therapy in Metastatic Melanoma. Br. J. Cancer 2016, 114, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.B.; Forschner, A.; Leiter, U.; Garbe, C.; Eigentler, T.K. S100B and LDH as Early Prognostic Markers for Response and Overall Survival in Melanoma Patients Treated with Anti-PD-1 or Combined Anti-PD-1 plus Anti-CTLA-4 Antibodies. Br. J. Cancer 2018, 119, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Simeone, E.; Gentilcore, G.; Giannarelli, D.; Grimaldi, A.M.; Caracò, C.; Curvietto, M.; Esposito, A.; Paone, M.; Palla, M.; Cavalcanti, E.; et al. Immunological and Biological Changes during Ipilimumab Treatment and Their Potential Correlation with Clinical Response and Survival in Patients with Advanced Melanoma. Cancer Immunol. Immunother. 2014, 63, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Laino, A.S.; Woods, D.; Vassallo, M.; Qian, X.; Tang, H.; Wind-Rotolo, M.; Weber, J. Serum Interleukin-6 and C-Reactive Protein Are Associated with Survival in Melanoma Patients Receiving Immune Checkpoint Inhibition. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Wagstaff, J.; Schadendorf, D.; Ferrucci, P.F.; et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2017, 377, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, F.; Lamant, L.; Gerard, E.; Torossian, N.; Chaltiel, L.; Filleron, T.; Beylot-Barry, M.; Dutriaux, C.; Prey, S.; Gros, A.; et al. Clinical, Histological and Molecular Predictors of Metastatic Melanoma Responses to Anti-PD-1 Immunotherapy. Br. J. Cancer 2018, 119, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Atkinson, V.; Cebon, J.S.; Jameson, M.B.; Fitzharris, B.M.; McNeil, C.M.; Hill, A.G.; Ribas, A.; Atkins, M.B.; Thompson, J.A.; et al. Standard-Dose Pembrolizumab in Combination with Reduced-Dose Ipilimumab for Patients with Advanced Melanoma (KEYNOTE-029): An Open-Label, Phase 1b Trial. Lancet Oncol. 2017, 18, 1202–1210. [Google Scholar] [CrossRef]
- Pérottet, J.; Le Goff, E.; Legoupil, D.; Quéré, G.; Schick, U.; Marcorelles, P.; Uguen, A. PD-L1 Copy Number Variation Does Not Correlate With PD-L1 Expression or Response to Anti-PD-1 Immunotherapy In Patients With Advanced Melanomas. Appl. Immunohistochem. Mol. Morphol. AIMM 2020, 28, 161–165. [Google Scholar] [CrossRef]
- Puzanov, I.; Dummer, R.; Schachter, J.; Pavlick, A.C.; Gonzalez, R.; Ascierto, P.A.; Margolin, K.A.; Hamid, O.; Agarwala, S.S.; Carlino, M.S.; et al. Efficacy Based on Tumor PD-L1 Expression in KEYNOTE-002, a Randomized Comparison of Pembrolizumab (Pembro; MK-3475) versus Chemotherapy in Patients (Pts) with Ipilimumab-Refractory (IPI-R) Advanced Melanoma (MEL). J. Clin. Oncol. 2015, 33, 3012. [Google Scholar] [CrossRef]
- Valpione, S.; Pasquali, S.; Campana, L.G.; Piccin, L.; Mocellin, S.; Pigozzo, J.; Chiarion-Sileni, V. Sex and Interleukin-6 Are Prognostic Factors for Autoimmune Toxicity Following Treatment with Anti-CTLA4 Blockade. J. Transl. Med. 2018, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Perez-Gracia, J.L.; Schalper, K.A.; Fusco, J.P.; Gonzalez, A.; Rodriguez-Ruiz, M.E.; Oñate, C.; Perez, G.; Alfaro, C.; Martín-Algarra, S.; et al. Changes in Serum Interleukin-8 (IL-8) Levels Reflect and Predict Response to Anti-PD-1 Treatment in Melanoma and Non-Small-Cell Lung Cancer Patients. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, T.; Sato, Y.; Tanita, K.; Kambayashi, Y.; Otsuka, A.; Fujisawa, Y.; Yoshino, K.; Matsushita, S.; Funakoshi, T.; Hata, H.; et al. Serum Levels of Soluble CD163 and CXCL5 May Be Predictive Markers for Immune-Related Adverse Events in Patients with Advanced Melanoma Treated with Nivolumab: A Pilot Study. Oncotarget 2018, 9, 15542–15551. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, T.; Sato, Y.; Tanita, K.; Lyu, C.; Kambayashi, Y.; Amagai, R.; Otsuka, A.; Fujisawa, Y.; Yoshino, K.; Matsushita, S.; et al. Association of Baseline Serum Levels of CXCL5 With the Efficacy of Nivolumab in Advanced Melanoma. Front. Med. 2019, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- Dronca, R.S.; Mansfield, A.S.; Liu, X.; Harrington, S.; Enninga, E.A.; Kottschade, L.A.; Koo, C.W.; McWilliams, R.R.; Block, M.S.; Nevala, W.K.; et al. Bim and Soluble PD-L1 (SPD-L1) as Predictive Biomarkers of Response to Anti-PD-1 Therapy in Patients with Melanoma and Lung Carcinoma. J. Clin. Oncol. 2017, 35, 11534. [Google Scholar] [CrossRef]
- Zhou, J.; Mahoney, K.M.; Giobbie-Hurder, A.; Zhao, F.; Lee, S.; Liao, X.; Rodig, S.; Li, J.; Wu, X.; Butterfield, L.H.; et al. Soluble PD-L1 as a Biomarker in Malignant Melanoma Treated with Checkpoint Blockade. Cancer Immunol. Res. 2017, 5, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Tucci, M.; Passarelli, A.; Mannavola, F.; Stucci, L.S.; Ascierto, P.A.; Capone, M.; Madonna, G.; Lopalco, P.; Silvestris, F. Serum Exosomes as Predictors of Clinical Response to Ipilimumab in Metastatic Melanoma. Oncoimmunology 2018, 7, e1387706. [Google Scholar] [CrossRef]
- Cordonnier, M.; Nardin, C.; Chanteloup, G.; Derangere, V.; Algros, M.-P.; Arnould, L.; Garrido, C.; Aubin, F.; Gobbo, J. Tracking the Evolution of Circulating Exosomal-PD-L1 to Monitor Melanoma Patients. J. Extracell. Vesicles 2020, 9, 1710899. [Google Scholar] [CrossRef]
- Pistillo, M.P.; Fontana, V.; Morabito, A.; Dozin, B.; Laurent, S.; Carosio, R.; Banelli, B.; Ferrero, F.; Spano, L.; Tanda, E.; et al. Soluble CTLA-4 as a Favorable Predictive Biomarker in Metastatic Melanoma Patients Treated with Ipilimumab: An Italian Melanoma Intergroup Study. Cancer Immunol. Immunother. 2019, 68, 97–107. [Google Scholar] [CrossRef]
- Weide, B.; Martens, A.; Hassel, J.C.; Berking, C.; Postow, M.A.; Bisschop, K.; Simeone, E.; Mangana, J.; Schilling, B.; Di Giacomo, A.M.; et al. Baseline Biomarkers for Outcome of Melanoma Patients Treated with Pembrolizumab. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5487–5496. [Google Scholar] [CrossRef]
- Delyon, J.; Mateus, C.; Lefeuvre, D.; Lanoy, E.; Zitvogel, L.; Chaput, N.; Roy, S.; Eggermont, A.M.M.; Routier, E.; Robert, C. Experience in Daily Practice with Ipilimumab for the Treatment of Patients with Metastatic Melanoma: An Early Increase in Lymphocyte and Eosinophil Counts Is Associated with Improved Survival. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 1697–1703. [Google Scholar] [CrossRef]
- Martens, A.; Wistuba-Hamprecht, K.; Geukes Foppen, M.; Yuan, J.; Postow, M.A.; Wong, P.; Romano, E.; Khammari, A.; Dreno, B.; Capone, M.; et al. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 2908–2918. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kitano, S.; Takahashi, A.; Tsutsumida, A.; Namikawa, K.; Tanese, K.; Abe, T.; Funakoshi, T.; Yamamoto, N.; Amagai, M.; et al. Nivolumab for Advanced Melanoma: Pretreatment Prognostic Factors and Early Outcome Markers during Therapy. Oncotarget 2016, 7, 77404–77415. [Google Scholar] [CrossRef]
- Ferrucci, P.F.; Gandini, S.; Battaglia, A.; Alfieri, S.; Di Giacomo, A.M.; Giannarelli, D.; Cappellini, G.C.A.; De Galitiis, F.; Marchetti, P.; Amato, G.; et al. Baseline Neutrophil-to-Lymphocyte Ratio Is Associated with Outcome of Ipilimumab-Treated Metastatic Melanoma Patients. Br. J. Cancer 2015, 112, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.F.; Ascierto, P.A.; Pigozzo, J.; Del Vecchio, M.; Maio, M.; Antonini Cappellini, G.C.; Guidoboni, M.; Queirolo, P.; Savoia, P.; Mandalà, M.; et al. Baseline Neutrophils and Derived Neutrophil-to-Lymphocyte Ratio: Prognostic Relevance in Metastatic Melanoma Patients Receiving Ipilimumab. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 732–738. [Google Scholar] [CrossRef]
- Capone, M.; Giannarelli, D.; Mallardo, D.; Madonna, G.; Festino, L.; Grimaldi, A.M.; Vanella, V.; Simeone, E.; Paone, M.; Palmieri, G.; et al. Baseline Neutrophil-to-Lymphocyte Ratio (NLR) and Derived NLR Could Predict Overall Survival in Patients with Advanced Melanoma Treated with Nivolumab. J. Immunother. Cancer 2018, 6, 74. [Google Scholar] [CrossRef]
- Bartlett, E.K.; Flynn, J.R.; Panageas, K.S.; Ferraro, R.A.; Sta Cruz, J.M.; Postow, M.A.; Coit, D.G.; Ariyan, C.E. High Neutrophil-to-Lymphocyte Ratio (NLR) Is Associated with Treatment Failure and Death in Patients Who Have Melanoma Treated with PD-1 Inhibitor Monotherapy. Cancer 2020, 126, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, J.; Caille, A.; Beneton, N.; Bens, G.; Christiann, F.; Maillard, H.; Machet, L. High Neutrophil to Lymphocyte Ratio Measured before Starting Ipilimumab Treatment Is Associated with Reduced Overall Survival in Patients with Melanoma. Br. J. Dermatol. 2016, 174, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, M.R.; Wolchok, R.E.; Zheng, J.; Panageas, K.S.; Wolchok, J.D.; Coit, D.; Postow, M.A.; Ariyan, C. Neutrophil to Lymphocyte Ratio Is Associated With Outcome During Ipilimumab Treatment. EBioMedicine 2017, 18, 56–61. [Google Scholar] [CrossRef]
- Fujisawa, Y.; Yoshino, K.; Otsuka, A.; Funakoshi, T.; Fujimura, T.; Yamamoto, Y.; Hata, H.; Tanaka, R.; Yamaguchi, K.; Nonomura, Y.; et al. Baseline Neutrophil to Lymphocyte Ratio Combined with Serum Lactate Dehydrogenase Level Associated with Outcome of Nivolumab Immunotherapy in a Japanese Advanced Melanoma Population. Br. J. Dermatol. 2018, 179, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Wistuba-Hamprecht, K.; Martens, A.; Heubach, F.; Romano, E.; Geukes Foppen, M.; Yuan, J.; Postow, M.; Wong, P.; Mallardo, D.; Schilling, B.; et al. Peripheral CD8 Effector-Memory Type 1 T-Cells Correlate with Outcome in Ipilimumab-Treated Stage IV Melanoma Patients. Eur. J. Cancer Oxf. Engl. 1990 2017, 73, 61–70. [Google Scholar] [CrossRef]
- Tietze, J.K.; Angelova, D.; Heppt, M.V.; Ruzicka, T.; Berking, C. Low Baseline Levels of NK Cells May Predict a Positive Response to Ipilimumab in Melanoma Therapy. Exp. Dermatol. 2017, 26, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Kudchadkar, R.R.; Yu, B.; Gallenstein, D.; Horak, C.E.; Inzunza, H.D.; Zhao, X.; Martinez, A.J.; Wang, W.; Gibney, G.; et al. Safety, Efficacy, and Biomarkers of Nivolumab with Vaccine in Ipilimumab-Refractory or -Naive Melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 4311–4318. [Google Scholar] [CrossRef]
- Bochem, J.; Zelba, H.; Amaral, T.; Spreuer, J.; Soffel, D.; Eigentler, T.; Wagner, N.B.; Uslu, U.; Terheyden, P.; Meier, F.; et al. Peripheral PD-1+CD56+ T-Cell Frequencies Correlate with Outcome in Stage IV Melanoma under PD-1 Blockade. PLoS ONE 2019, 14, e0221301. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanyam, P.B.; Dong, Z.; Gusenleitner, D.; Giobbie-Hurder, A.; Severgnini, M.; Zhou, J.; Manos, M.; Eastman, L.M.; Maecker, H.T.; Hodi, F.S. Distinct Predictive Biomarker Candidates for Response to Anti-CTLA-4 and Anti-PD-1 Immunotherapy in Melanoma Patients. J. Immunother. Cancer 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Pirozyan, M.R.; McGuire, H.M.; Emran, A.A.; Tseng, H.-Y.; Tiffen, J.C.; Lee, J.H.; Carlino, M.S.; Menzies, A.M.; Long, G.V.; Scolyer, R.A.; et al. Pretreatment Innate Cell Populations and CD4 T Cells in Blood Are Associated With Response to Immune Checkpoint Blockade in Melanoma Patients. Front. Immunol. 2020, 11, 372. [Google Scholar] [CrossRef] [PubMed]
- Cha, E.; Klinger, M.; Hou, Y.; Cummings, C.; Ribas, A.; Faham, M.; Fong, L. Improved Survival with T Cell Clonotype Stability after Anti-CTLA-4 Treatment in Cancer Patients. Sci. Transl. Med. 2014, 6, 238ra70. [Google Scholar] [CrossRef] [PubMed]
- Postow, M.A.; Chesney, J.; Pavlick, A.C.; Robert, C.; Grossmann, K.; McDermott, D.; Linette, G.P.; Meyer, N.; Giguere, J.K.; Agarwala, S.S.; et al. Nivolumab and Ipilimumab versus Ipilimumab in Untreated Melanoma. N. Engl. J. Med. 2015, 372, 2006–2017. [Google Scholar] [CrossRef] [PubMed]
- Krieg, C.; Nowicka, M.; Guglietta, S.; Schindler, S.; Hartmann, F.J.; Weber, L.M.; Dummer, R.; Robinson, M.D.; Levesque, M.P.; Becher, B. High-Dimensional Single-Cell Analysis Predicts Response to Anti-PD-1 Immunotherapy. Nat. Med. 2018, 24, 144–153. [Google Scholar] [CrossRef]
- de Coaña, Y.P.; Wolodarski, M.; Poschke, I.; Yoshimoto, Y.; Yang, Y.; Nyström, M.; Edbäck, U.; Brage, S.E.; Lundqvist, A.; Masucci, G.V.; et al. Ipilimumab Treatment Decreases Monocytic MDSCs and Increases CD8 Effector Memory T Cells in Long-Term Survivors with Advanced Melanoma. Oncotarget 2017, 8, 21539–21553. [Google Scholar] [CrossRef]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of Circulating MDSC Correlate with Clinical Outcome of Melanoma Patients Treated with Ipilimumab. Cancer Immunol. Immunother. 2014, 63, 247–257. [Google Scholar] [CrossRef]
- Lee, J.H.; Long, G.V.; Boyd, S.; Lo, S.; Menzies, A.M.; Tembe, V.; Guminski, A.; Jakrot, V.; Scolyer, R.A.; Mann, G.J.; et al. Circulating Tumour DNA Predicts Response to Anti-PD1 Antibodies in Metastatic Melanoma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1130–1136. [Google Scholar] [CrossRef]
- Forschner, A.; Battke, F.; Hadaschik, D.; Schulze, M.; Weißgraeber, S.; Han, C.-T.; Kopp, M.; Frick, M.; Klumpp, B.; Tietze, N.; et al. Tumor Mutation Burden and Circulating Tumor DNA in Combined CTLA-4 and PD-1 Antibody Therapy in Metastatic Melanoma - Results of a Prospective Biomarker Study. J. Immunother. Cancer 2019, 7, 180. [Google Scholar] [CrossRef]
- Marsavela, G.; Lee, J.; Calapre, L.; Wong, S.Q.; Pereira, M.R.; McEvoy, A.C.; Reid, A.L.; Robinson, C.; Warburton, L.; Abed, A.; et al. Circulating Tumor DNA Predicts Outcome from First-, but Not Second-Line Treatment and Identifies Melanoma Patients Who May Benefit from Combination Immunotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 5926–5933. [Google Scholar] [CrossRef] [PubMed]
- Rowe, S.P.; Luber, B.; Makell, M.; Brothers, P.; Santmyer, J.; Schollenberger, M.D.; Quinn, H.; Edelstein, D.L.; Jones, F.S.; Bleich, K.B.; et al. From Validity to Clinical Utility: The Influence of Circulating Tumor DNA on Melanoma Patient Management in a Real-World Setting. Mol. Oncol. 2018, 12, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Long, G.V.; Menzies, A.M.; Lo, S.; Guminski, A.; Whitbourne, K.; Peranec, M.; Scolyer, R.; Kefford, R.F.; Rizos, H.; et al. Association Between Circulating Tumor DNA and Pseudoprogression in Patients With Metastatic Melanoma Treated With Anti-Programmed Cell Death 1 Antibodies. JAMA Oncol. 2018, 4, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Galuppini, F.; Dal Pozzo, C.A.; Deckert, J.; Loupakis, F.; Fassan, M.; Baffa, R. Tumor Mutation Burden: From Comprehensive Mutational Screening to the Clinic. Cancer Cell Int. 2019, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Meléndez, B.; Van Campenhout, C.; Rorive, S.; Remmelink, M.; Salmon, I.; D’Haene, N. Methods of Measurement for Tumor Mutational Burden in Tumor Tissue. Transl. Lung Cancer Res. 2018, 7, 661–667. [Google Scholar] [CrossRef]
- Vilimas, T. Measuring Tumor Mutational Burden Using Whole-Exome Sequencing. In Biomarkers for Immunotherapy of Cancer; Humana: New York, NY, USA, 2020; Volume 2055, pp. 63–91. [Google Scholar] [CrossRef]
- Campesato, L.F.; Barroso-Sousa, R.; Jimenez, L.; Correa, B.R.; Sabbaga, J.; Hoff, P.M.; Reis, L.F.L.; Galante, P.A.F.; Camargo, A.A. Comprehensive Cancer-Gene Panels Can Be Used to Estimate Mutational Load and Predict Clinical Benefit to PD-1 Blockade in Clinical Practice. Oncotarget 2015, 6, 34221–34227. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.-H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor Mutational Load Predicts Survival after Immunotherapy across Multiple Cancer Types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Davis, A.A.; Chae, Y.K.; Agte, S.; Pan, A.; Simon, N.I.; Taxter, T.J.; Behdad, A.; Carneiro, B.A.; Cristofanilli, M.; Giles, F.J. Comparison of Tumor Mutational Burden (TMB) across Tumor Tissue and Circulating Tumor DNA (CtDNA). J. Clin. Oncol. 2017, 35, e23028. [Google Scholar] [CrossRef]
- Peters, S.; Cho, B.C.; Reinmuth, N.; Lee, K.H.; Luft, A.; Ahn, M.-J.; Baas, P.; Dols, M.C.; Smolin, A.; Vicente, D.; et al. Abstract CT074: Tumor Mutational Burden (TMB) as a Biomarker of Survival in Metastatic Non-Small Cell Lung Cancer (MNSCLC): Blood and Tissue TMB Analysis from MYSTIC, a Phase III Study of First-Line Durvalumab ± Tremelimumab vs Chemotherapy. Cancer Res. 2019, 79, CT074. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-Based Tumor Mutational Burden as a Predictor of Clinical Benefit in Non-Small-Cell Lung Cancer Patients Treated with Atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Vokes, N.I.; Liu, D.; Ricciuti, B.; Jimenez-Aguilar, E.; Rizvi, H.; Dietlein, F.; He, M.X.; Margolis, C.A.; Elmarakeby, H.A.; Girshman, J.; et al. Harmonization of Tumor Mutational Burden Quantification and Association With Response to Immune Checkpoint Blockade in Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef] [PubMed]
- TMB Harmonization Working Group Meeting | Friends of Cancer Research. Available online: https://friendsofcancerresearch.org/events/tmb-harmonization-working-group-meeting (accessed on 4 November 2020).
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Garrido, F.; Ruiz-Cabello, F.; Aptsiauri, N. Rejection versus Escape: The Tumor MHC Dilemma. Cancer Immunol. Immunother. 2017, 66, 259–271. [Google Scholar] [CrossRef]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8+ Cytotoxic T Lymphocytes in Cancer Immunotherapy: A Review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Tay, R.E.; Richardson, E.K.; Toh, H.C. Revisiting the Role of CD4 + T Cells in Cancer Immunotherapy—New Insights into Old Paradigms. Cancer Gene Ther. 2020, 1–13. [Google Scholar] [CrossRef]
- Olbryt, M.; Rajczykowski, M.; Widłak, W. Biological Factors behind Melanoma Response to Immune Checkpoint Inhibitors. Int. J. Mol. Sci. 2020, 21, 4071. [Google Scholar] [CrossRef]
- Merelli, B.; Massi, D.; Cattaneo, L.; Mandalà, M. Targeting the PD1/PD-L1 Axis in Melanoma: Biological Rationale, Clinical Challenges and Opportunities. Crit. Rev. Oncol. Hematol. 2014, 89, 140–165. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Albacker, L.A.; Hopkins, A.C.; Montesion, M.; Murugesan, K.; Vithayathil, T.T.; Zaidi, N.; Azad, N.S.; Laheru, D.A.; Frampton, G.M.; et al. PD-L1 Expression and Tumor Mutational Burden Are Independent Biomarkers in Most Cancers. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Emancipator, K. Keytruda and PD-L1: A Real-World Example of Co-Development of a Drug with a Predictive Biomarker. AAPS J. 2020, 23, 5. [Google Scholar] [CrossRef] [PubMed]
- FDA; U.F.; D.A. Companion Diagnostics. Available online: https://www.fda.gov/medical-devices/vitro-diagnostics/companion-diagnostics (accessed on 29 October 2020).
- Krigsfeld, G.S.; Prince, E.A.; Pratt, J.; Chizhevsky, V.; William Ragheb, J.; Novotny, J.; Huron, D. Analysis of Real-World PD-L1 IHC 28-8 and 22C3 PharmDx Assay Utilisation, Turnaround Times and Analytical Concordance across Multiple Tumour Types. J. Clin. Pathol. 2020, 73, 656–664. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.P.; Yang, Y.; Boisot, S.; Sudarsanam, S.; Wang, J.-F.; Chizhevsky, V.; Zhao, G.; Arain, S.; Weiss, L.M. Immunohistochemical Detection of PD-L1 among Diverse Human Neoplasms in a Reference Laboratory: Observations Based upon 62,896 Cases. Mod. Pathol. 2019, 32, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.-J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.M.; Kefford, R.; Hersey, P.; et al. Programmed Death-Ligand 1 Expression and Response to the Anti-Programmed Death 1 Antibody Pembrolizumab in Melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef] [PubMed]
- Carbognin, L.; Pilotto, S.; Milella, M.; Vaccaro, V.; Brunelli, M.; Caliò, A.; Cuppone, F.; Sperduti, I.; Giannarelli, D.; Chilosi, M.; et al. Differential Activity of Nivolumab, Pembrolizumab and MPDL3280A According to the Tumor Expression of Programmed Death-Ligand-1 (PD-L1): Sensitivity Analysis of Trials in Melanoma, Lung and Genitourinary Cancers. PLoS ONE 2015, 10, e0130142. [Google Scholar] [CrossRef]
- Yamazaki, N.; Takenouchi, T.; Fujimoto, M.; Ihn, H.; Uchi, H.; Inozume, T.; Kiyohara, Y.; Uhara, H.; Nakagawa, K.; Furukawa, H.; et al. Phase 1b Study of Pembrolizumab (MK-3475; Anti-PD-1 Monoclonal Antibody) in Japanese Patients with Advanced Melanoma (KEYNOTE-041). Cancer Chemother. Pharmacol. 2017, 79, 651–660. [Google Scholar] [CrossRef]
- Madonna, G.; Ballesteros-Merino, C.; Feng, Z.; Bifulco, C.; Capone, M.; Giannarelli, D.; Mallardo, D.; Simeone, E.; Grimaldi, A.M.; Caracò, C.; et al. PD-L1 Expression with Immune-Infiltrate Evaluation and Outcome Prediction in Melanoma Patients Treated with Ipilimumab. Oncoimmunology 2018, 7, e1405206. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef]
- Cho, J.; Ahn, S.; Yoo, K.H.; Kim, J.H.; Choi, S.-H.; Jang, K.-T.; Lee, J. Treatment Outcome of PD-1 Immune Checkpoint Inhibitor in Asian Metastatic Melanoma Patients: Correlative Analysis with PD-L1 Immunohistochemistry. Investig. New Drugs 2016, 34, 677–684. [Google Scholar] [CrossRef]
- Weber, J.S.; D’Angelo, S.P.; Minor, D.; Hodi, F.S.; Gutzmer, R.; Neyns, B.; Hoeller, C.; Khushalani, N.I.; Miller, W.H.; Lao, C.D.; et al. Nivolumab versus Chemotherapy in Patients with Advanced Melanoma Who Progressed after Anti-CTLA-4 Treatment (CheckMate 037): A Randomised, Controlled, Open-Label, Phase 3 Trial. Lancet Oncol. 2015, 16, 375–384. [Google Scholar] [CrossRef]
- Larkin, J.; Lao, C.D.; Urba, W.J.; McDermott, D.F.; Horak, C.; Jiang, J.; Wolchok, J.D. Efficacy and Safety of Nivolumab in Patients With BRAF V600 Mutant and BRAF Wild-Type Advanced Melanoma: A Pooled Analysis of 4 Clinical Trials. JAMA Oncol. 2015, 1, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Daud, A.; Hamid, O.; Robert, C.; Hodi, F.S.; Wolchok, J.D.; Hwu, W.J.; Weber, J.S.; Kefford, R.; Hersey, P.; Joshua, A.M.; et al. Relationship between Programmed Death Ligand 1 (PD-L1) Expression and Clinical Outcome in Patients (Pts) with Melanoma (MEL) Treated with Pembrolizumab (Pembro; MK-3475). Eur. J. Cancer 2014, 50, 48–49. [Google Scholar] [CrossRef]
- Long, G.V.; Dummer, R.; Hamid, O.; Gajewski, T.F.; Caglevic, C.; Dalle, S.; Arance, A.; Carlino, M.S.; Grob, J.-J.; Kim, T.M.; et al. Epacadostat plus Pembrolizumab versus Placebo plus Pembrolizumab in Patients with Unresectable or Metastatic Melanoma (ECHO-301/KEYNOTE-252): A Phase 3, Randomised, Double-Blind Study. Lancet Oncol. 2019, 20, 1083–1097. [Google Scholar] [CrossRef]
- Robert, C.; Ribas, A.; Schachter, J.; Arance, A.; Grob, J.-J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma (KEYNOTE-006): Post-Hoc 5-Year Results from an Open-Label, Multicentre, Randomised, Controlled, Phase 3 Study. Lancet Oncol. 2019, 20, 1239–1251. [Google Scholar] [CrossRef]
- Robert, C.; Ribas, A.; Hamid, O.; Daud, A.; Wolchok, J.D.; Joshua, A.M.; Hwu, W.-J.; Weber, J.S.; Gangadhar, T.C.; Joseph, R.W.; et al. Durable Complete Response After Discontinuation of Pembrolizumab in Patients With Metastatic Melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 1668–1674. [Google Scholar] [CrossRef]
- Diggs, L.P.; Hsueh, E.C. Utility of PD-L1 Immunohistochemistry Assays for Predicting PD-1/PD-L1 Inhibitor Response. Biomark. Res. 2017, 5, 12. [Google Scholar] [CrossRef]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-Related Inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Carlino, M.S.; Long, G.V.; Schadendorf, D.; Robert, C.; Ribas, A.; Richtig, E.; Nyakas, M.; Caglevic, C.; Tarhini, A.; Blank, C.; et al. Outcomes by Line of Therapy and Programmed Death Ligand 1 Expression in Patients with Advanced Melanoma Treated with Pembrolizumab or Ipilimumab in KEYNOTE-006: A Randomised Clinical Trial. Eur. J. Cancer Oxf. Engl. 1990 2018, 101, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.H.; From, L.; Bernardino, E.A.; Mihm, M.C. The Histogenesis and Biologic Behavior of Primary Human Malignant Melanomas of the Skin. Cancer Res. 1969, 29, 705–727. [Google Scholar] [PubMed]
- Clemente, C.G.; Mihm, M.C.; Bufalino, R.; Zurrida, S.; Collini, P.; Cascinelli, N. Prognostic Value of Tumor Infiltrating Lymphocytes in the Vertical Growth Phase of Primary Cutaneous Melanoma. Cancer 1996, 77, 1303–1310. [Google Scholar] [CrossRef]
- Taylor, R.C.; Patel, A.; Panageas, K.S.; Busam, K.J.; Brady, M.S. Tumor-Infiltrating Lymphocytes Predict Sentinel Lymph Node Positivity in Patients with Cutaneous Melanoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 869–875. [Google Scholar] [CrossRef]
- Tuthill, R.J.; Unger, J.M.; Liu, P.Y.; Flaherty, L.E.; Sondak, V.K. Southwest Oncology Group Risk Assessment in Localized Primary Cutaneous Melanoma: A Southwest Oncology Group Study Evaluating Nine Factors and a Test of the Clark Logistic Regression Prediction Model. Am. J. Clin. Pathol. 2002, 118, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Bindea, G.; Pagès, F.; Galon, J. Tumor Immunosurveillance in Human Cancers. Cancer Metastasis Rev. 2011, 30, 5–12. [Google Scholar] [CrossRef]
- Weiss, S.A.; Han, S.W.; Lui, K.; Tchack, J.; Shapiro, R.; Berman, R.; Zhong, J.; Krogsgaard, M.; Osman, I.; Darvishian, F. Immunologic Heterogeneity of Tumor-Infiltrating Lymphocyte Composition in Primary Melanoma. Hum. Pathol. 2016, 57, 116–125. [Google Scholar] [CrossRef]
- Antohe, M.; Nedelcu, R.I.; Nichita, L.; Popp, C.G.; Cioplea, M.; Brinzea, A.; Hodorogea, A.; Calinescu, A.; Balaban, M.; Ion, D.A.; et al. Tumor Infiltrating Lymphocytes: The Regulator of Melanoma Evolution. Oncol. Lett. 2019, 17, 4155–4161. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Capone, M.; Urba, W.J.; Bifulco, C.B.; Botti, G.; Lugli, A.; Marincola, F.M.; Ciliberto, G.; Galon, J.; Fox, B.A. The Additional Facet of Immunoscore: Immunoprofiling as a Possible Predictive Tool for Cancer Treatment. J. Transl. Med. 2013, 11, 54. [Google Scholar] [CrossRef]
- Nanda, V.G.Y.; Peng, W.; Hwu, P.; Davies, M.A.; Ciliberto, G.; Fattore, L.; Malpicci, D.; Aurisicchio, L.; Ascierto, P.A.; Croce, C.M.; et al. Melanoma and Immunotherapy Bridge 2015. J. Transl. Med. 2016, 14, 65. [Google Scholar] [CrossRef]
- Spranger, S.; Spaapen, R.M.; Zha, Y.; Williams, J.; Meng, Y.; Ha, T.T.; Gajewski, T.F. Up-Regulation of PD-L1, IDO, and T(Regs) in the Melanoma Tumor Microenvironment Is Driven by CD8(+) T Cells. Sci. Transl. Med. 2013, 5, 200ra116. [Google Scholar] [CrossRef] [PubMed]
- Meireson, A.; Devos, M.; Brochez, L. IDO Expression in Cancer: Different Compartment, Different Functionality? Front. Immunol. 2020, 11, 531491. [Google Scholar] [CrossRef] [PubMed]
- Radvanyi, L.G. Tumor-Infiltrating Lymphocyte Therapy: Addressing Prevailing Questions. Cancer J. Sudbury Mass 2015, 21, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Mastracci, L.; Fontana, V.; Queirolo, P.; Carosio, R.; Grillo, F.; Morabito, A.; Banelli, B.; Tanda, E.; Boutros, A.; Dozin, B.; et al. Response to Ipilimumab Therapy in Metastatic Melanoma Patients: Potential Relevance of CTLA-4+ Tumor Infiltrating Lymphocytes and Their in Situ Localization. Cancer Immunol. Immunother. 2020, 69, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Oble, D.A.; Loewe, R.; Yu, P.; Mihm, M.C. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human melanoma. Cancer Immun. 2009, 9, 3. [Google Scholar]
- Parigi, S.M.; Czarnewski, P.; Das, S.; Steeg, C.; Brockmann, L.; Fernandez-Gaitero, S.; Yman, V.; Forkel, M.; Höög, C.; Mjösberg, J.; et al. Flt3 Ligand Expands Bona Fide Innate Lymphoid Cell Precursors in Vivo. Sci. Rep. 2018, 8, 154. [Google Scholar] [CrossRef]
- Barry, K.C.; Hsu, J.; Broz, M.L.; Cueto, F.J.; Binnewies, M.; Combes, A.J.; Nelson, A.E.; Loo, K.; Kumar, R.; Rosenblum, M.D.; et al. A Natural Killer-Dendritic Cell Axis Defines Checkpoint Therapy-Responsive Tumor Microenvironments. Nat. Med. 2018, 24, 1178–1191. [Google Scholar] [CrossRef]
- de Andrade, L.F.; Lu, Y.; Luoma, A.; Ito, Y.; Pan, D.; Pyrdol, J.W.; Yoon, C.H.; Yuan, G.-C.; Wucherpfennig, K.W. Discovery of Specialized NK Cell Populations Infiltrating Human Melanoma Metastases. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Kitamura, T. Targeting Tumor-Associated Macrophages as a Potential Strategy to Enhance the Response to Immune Checkpoint Inhibitors. Front. Cell Dev. Biol. 2018, 6, 38. [Google Scholar] [CrossRef]
- Cassetta, L.; Kitamura, T. Macrophage Targeting: Opening New Possibilities for Cancer Immunotherapy. Immunology 2018, 155, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Chen, D.S. Immune Escape to PD-L1/PD-1 Blockade: Seven Steps to Success (or Failure). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Angell, H.K.; Bedognetti, D.; Marincola, F.M. The Continuum of Cancer Immunosurveillance: Prognostic, Predictive, and Mechanistic Signatures. Immunity 2013, 39, 11–26. [Google Scholar] [CrossRef]
- Sobhani, N.; Corona, S.P.; Roviello, G.; Bagby, S.; D’Angelo, A.; Iezzi, G.; Generali, D. Immune-Gene Signature: A New Tool for Patient Selection for Checkpoint Inhibitors? Future Oncol. Lond. Engl. 2020, 16, 1327–1330. [Google Scholar] [CrossRef]
- Rozeman, E.A.; Menzies, A.M.; van Akkooi, A.C.J.; Adhikari, C.; Bierman, C.; van de Wiel, B.A.; Scolyer, R.A.; Krijgsman, O.; Sikorska, K.; Eriksson, H.; et al. Identification of the Optimal Combination Dosing Schedule of Neoadjuvant Ipilimumab plus Nivolumab in Macroscopic Stage III Melanoma (OpACIN-Neo): A Multicentre, Phase 2, Randomised, Controlled Trial. Lancet Oncol. 2019, 20, 948–960. [Google Scholar] [CrossRef]
- Blank, C.U.; Rozeman, E.A.; Fanchi, L.F.; Sikorska, K.; van de Wiel, B.; Kvistborg, P.; Krijgsman, O.; van den Braber, M.; Philips, D.; Broeks, A.; et al. Neoadjuvant versus Adjuvant Ipilimumab plus Nivolumab in Macroscopic Stage III Melanoma. Nat. Med. 2018, 24, 1655–1661. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.-R.; Chasalow, S.D.; Wang, L.; Hamid, O.; Schmidt, H.; Cogswell, J.; Alaparthy, S.; Berman, D.; Jure-Kunkel, M.; Siemers, N.O.; et al. An Immune-Active Tumor Microenvironment Favors Clinical Response to Ipilimumab. Cancer Immunol. Immunother. 2012, 61, 1019–1031. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T Cell Dysfunction and Exclusion Predict Cancer Immunotherapy Response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Ardito, R.; Merelli, B.; Lonati, V.; Cabiddu, M.; Seghezzi, S.; Barni, S.; Ghidini, A. Prognostic and Predictive Role of Elevated Lactate Dehydrogenase in Patients with Melanoma Treated with Immunotherapy and BRAF Inhibitors: A Systematic Review and Meta-Analysis. Melanoma Res. 2019, 29, 1–12. [Google Scholar] [CrossRef]
- Balch, C.M.; Gershenwald, J.E.; Soong, S.-J.; Thompson, J.F.; Atkins, M.B.; Byrd, D.R.; Buzaid, A.C.; Cochran, A.J.; Coit, D.G.; Ding, S.; et al. Final Version of 2009 AJCC Melanoma Staging and Classification. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 6199–6206. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma Staging: Evidence-Based Changes in the American Joint Committee on Cancer Eighth Edition Cancer Staging Manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Van Wilpe, S.; Koornstra, R.; Den Brok, M.; De Groot, J.W.; Blank, C.; De Vries, J.; Gerritsen, W.; Mehra, N. Lactate Dehydrogenase: A Marker of Diminished Antitumor Immunity. Oncoimmunology 2020, 9, 1731942. [Google Scholar] [CrossRef] [PubMed]
- Singer, K.; Gottfried, E.; Kreutz, M.; Mackensen, A. Suppression of T-Cell Responses by Tumor Metabolites. Cancer Immunol. Immunother. 2011, 60, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, A.; Engel, G.; Brenner, W.; Gläser, R.; Mönig, H.; Henze, E.; Christophers, E. S100B Protein Detection in Serum Is a Significant Prognostic Factor in Metastatic Melanoma. Oncology 1999, 56, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Harpio, R.; Einarsson, R. S100 Proteins as Cancer Biomarkers with Focus on S100B in Malignant Melanoma. Clin. Biochem. 2004, 37, 512–518. [Google Scholar] [CrossRef]
- Fang, S.; Wang, Y.; Sui, D.; Liu, H.; Ross, M.I.; Gershenwald, J.E.; Cormier, J.N.; Royal, R.E.; Lucci, A.; Schacherer, C.W.; et al. C-Reactive Protein as a Marker of Melanoma Progression. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Nyakas, M.; Aamdal, E.; Jacobsen, K.D.; Guren, T.K.; Aamdal, S.; Hagene, K.T.; Brunsvig, P.; Yndestad, A.; Halvorsen, B.; Tasken, K.A.; et al. Prognostic Biomarkers for Immunotherapy with Ipilimumab in Metastatic Melanoma. Clin. Exp. Immunol. 2019, 197, 74–82. [Google Scholar] [CrossRef]
- Weber, J.S.; Tang, H.; Hippeli, L.; Qian, M.; Wind-Rotolo, M.; Larkin, J.M.G.; Wolchok, J.D.; Sznol, M.; Robert, C.; Woods, D.M.; et al. Serum IL-6 and CRP as Prognostic Factors in Melanoma Patients Receiving Single Agent and Combination Checkpoint Inhibition. J. Clin. Oncol. 2019, 37, 100. [Google Scholar] [CrossRef]
- Kubo, Y.; Fukushima, S.; Inamori, Y.; Tsuruta, M.; Egashira, S.; Yamada-Kanazawa, S.; Nakahara, S.; Tokuzumi, A.; Miyashita, A.; Aoi, J.; et al. Serum Concentrations of HGF Are Correlated with Response to Anti-PD-1 Antibody Therapy in Patients with Metastatic Melanoma. J. Dermatol. Sci. 2019, 93, 33–40. [Google Scholar] [CrossRef]
- Tobin, R.P.; Jordan, K.R.; Kapoor, P.; Spongberg, E.; Davis, D.; Vorwald, V.M.; Couts, K.L.; Gao, D.; Smith, D.E.; Borgers, J.S.W.; et al. IL-6 and IL-8 Are Linked With Myeloid-Derived Suppressor Cell Accumulation and Correlate With Poor Clinical Outcomes in Melanoma Patients. Front. Oncol. 2019, 9, 1223. [Google Scholar] [CrossRef]
- Chen, S.-H.; Gong, X.; Zhang, Y.; Van Horn, R.D.; Yin, T.; Huber, L.; Burke, T.F.; Manro, J.; Iversen, P.W.; Wu, W.; et al. RAF Inhibitor LY3009120 Sensitizes RAS or BRAF Mutant Cancer to CDK4/6 Inhibition by Abemaciclib via Superior Inhibition of Phospho-RB and Suppression of Cyclin D1. Oncogene 2018, 37, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Martens, A.; Wistuba-Hamprecht, K.; Yuan, J.; Postow, M.A.; Wong, P.; Capone, M.; Madonna, G.; Khammari, A.; Schilling, B.; Sucker, A.; et al. Increases in Absolute Lymphocytes and Circulating CD4+ and CD8+ T Cells Are Associated with Positive Clinical Outcome of Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 4848–4858. [Google Scholar] [CrossRef]
- Capone, M.; Fratangelo, F.; Giannarelli, D.; Sorrentino, C.; Turiello, R.; Zanotta, S.; Galati, D.; Madonna, G.; Tuffanelli, M.; Scarpato, L.; et al. Frequency of Circulating CD8+CD73+T Cells Is Associated with Survival in Nivolumab-Treated Melanoma Patients. J. Transl. Med. 2020, 18, 121. [Google Scholar] [CrossRef]
- Sen, D.R.; Kaminski, J.; Barnitz, R.A.; Kurachi, M.; Gerdemann, U.; Yates, K.B.; Tsao, H.-W.; Godec, J.; LaFleur, M.W.; Brown, F.D.; et al. The Epigenetic Landscape of T Cell Exhaustion. Science 2016, 354, 1165–1169. [Google Scholar] [CrossRef]
- Fuertes Marraco, S.A.; Neubert, N.J.; Verdeil, G.; Speiser, D.E. Inhibitory Receptors Beyond T Cell Exhaustion. Front. Immunol. 2015, 6, 310. [Google Scholar] [CrossRef]
- Postow, M.A.; Manuel, M.; Wong, P.; Yuan, J.; Dong, Z.; Liu, C.; Perez, S.; Tanneau, I.; Noel, M.; Courtier, A.; et al. Peripheral T Cell Receptor Diversity Is Associated with Clinical Outcomes Following Ipilimumab Treatment in Metastatic Melanoma. J. Immunother. Cancer 2015, 3, 23. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef]
- Peters, B.A.; Wilson, M.; Moran, U.; Pavlick, A.; Izsak, A.; Wechter, T.; Weber, J.S.; Osman, I.; Ahn, J. Relating the Gut Metagenome and Metatranscriptome to Immunotherapy Responses in Melanoma Patients. Genome Med. 2019, 11, 61. [Google Scholar] [CrossRef]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal Microbiome Analyses Identify Melanoma Patients at Risk for Checkpoint-Blockade-Induced Colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef]
- Hoffmann, F.; Zarbl, R.; Niebel, D.; Sirokay, J.; Fröhlich, A.; Posch, C.; Holderried, T.A.W.; Brossart, P.; Saavedra, G.; Kuster, P.; et al. Prognostic and Predictive Value of PD-L2 DNA Methylation and MRNA Expression in Melanoma. Clin. Epigenetics 2020, 12, 94. [Google Scholar] [CrossRef]
- Gupta, S.; McCann, L.; Chan, Y.G.Y.; Lai, E.W.; Wei, W.; Wong, P.F.; Smithy, J.W.; Weidler, J.; Rhees, B.; Bates, M.; et al. Closed System RT-QPCR as a Potential Companion Diagnostic Test for Immunotherapy Outcome in Metastatic Melanoma. J. Immunother. Cancer 2019, 7, 254. [Google Scholar] [CrossRef]
- Wang, X.; Chai, Z.; Li, Y.; Long, F.; Hao, Y.; Pan, G.; Liu, M.; Li, B. Identification of Potential Biomarkers for Anti-PD-1 Therapy in Melanoma by Weighted Correlation Network Analysis. Genes 2020, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Lovly, C.M.; Flavin, M.; Panageas, K.S.; Ayers, G.D.; Zhao, Z.; Iams, W.T.; Colgan, M.; DeNoble, S.; Terry, C.R.; et al. Impact of NRAS Mutations for Patients with Advanced Melanoma Treated with Immune Therapies. Cancer Immunol. Res. 2015, 3, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Havel, J.J.; Kendall, S.M.; Makarov, V.; Walsh, L.A.; Desrichard, A.; Weinhold, N.; Chan, T.A. Recurrent SERPINB3 and SERPINB4 Mutations in Patients Who Respond to Anti-CTLA4 Immunotherapy. Nat. Genet. 2016, 48, 1327–1329. [Google Scholar] [CrossRef]
- Chen, H.; Chong, W.; Wu, Q.; Yao, Y.; Mao, M.; Wang, X. Association of LRP1B Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-Small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front. Immunol. 2019, 10, 1113. [Google Scholar] [CrossRef]
- Galore-Haskel, G.; Nemlich, Y.; Greenberg, E.; Ashkenazi, S.; Hakim, M.; Itzhaki, O.; Shoshani, N.; Shapira-Fromer, R.; Ben-Ami, E.; Ofek, E.; et al. A Novel Immune Resistance Mechanism of Melanoma Cells Controlled by the ADAR1 Enzyme. Oncotarget 2015, 6, 28999–29015. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, W.; Li, A.; Chen, Y.; Ou, Q.; He, Z.; Zhang, Y.; Liu, R.; Yao, H.; Song, E. Association of Long Noncoding RNA Biomarkers With Clinical Immune Subtype and Prediction of Immunotherapy Response in Patients With Cancer. JAMA Netw. Open 2020, 3, e202149. [Google Scholar] [CrossRef]
- Bronte, V.; Brandau, S.; Chen, S.-H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for Myeloid-Derived Suppressor Cell Nomenclature and Characterization Standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Weber, J.; Gibney, G.; Kudchadkar, R.; Yu, B.; Cheng, P.; Martinez, A.J.; Kroeger, J.; Richards, A.; McCormick, L.; Moberg, V.; et al. Phase I/II Study of Metastatic Melanoma Patients Treated with Nivolumab Who Had Progressed after Ipilimumab. Cancer Immunol. Res. 2016, 4, 345–353. [Google Scholar] [CrossRef]
- Blank, C.U.; Haanen, J.B.; Ribas, A.; Schumacher, T.N. CANCER IMMUNOLOGY. The “Cancer Immunogram.”. Science 2016, 352, 658–660. [Google Scholar] [CrossRef] [PubMed]
- Anagnostou, V.; Niknafs, N.; Marrone, K.; Bruhm, D.C.; White, J.R.; Naidoo, J.; Hummelink, K.; Monkhorst, K.; Lalezari, F.; Lanis, M.; et al. Multimodal Genomic Features Predict Outcome of Immune Checkpoint Blockade in Non-Small-Cell Lung Cancer. Nat. Cancer 2020, 1, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Nabet, B.Y.; Esfahani, M.S.; Moding, E.J.; Hamilton, E.G.; Chabon, J.J.; Rizvi, H.; Steen, C.B.; Chaudhuri, A.A.; Liu, C.L.; Hui, A.B.; et al. Noninvasive Early Identification of Therapeutic Benefit from Immune Checkpoint Inhibition. Cell 2020, 183, 363–376.e13. [Google Scholar] [CrossRef]
- Auslander, N.; Zhang, G.; Lee, J.S.; Frederick, D.T.; Miao, B.; Moll, T.; Tian, T.; Wei, Z.; Madan, S.; Sullivan, R.J.; et al. Robust Prediction of Response to Immune Checkpoint Blockade Therapy in Metastatic Melanoma. Nat. Med. 2018, 24, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Teulings, H.-E.; Limpens, J.; Jansen, S.N.; Zwinderman, A.H.; Reitsma, J.B.; Spuls, P.I.; Luiten, R.M. Vitiligo-Like Depigmentation in Patients With Stage III-IV Melanoma Receiving Immunotherapy and Its Association With Survival: A Systematic Review and Meta-Analysis. J. Clin. Oncol. 2015, 33, 773–781. [Google Scholar] [CrossRef]
| Biomarker | Study Details & Reference | Patients | Clinical Utility |
|---|---|---|---|
| TMB (WES) | Ipilimumab Van Allen et al. [7] | 110 | CB * |
| TMB (WES) | Ipilimumab or tremelimumab Snyder et al. [8] | 64 | OS |
| TMB (WES) | Nivolumab or pembrolizumab Hugo et al. [9] | 38 | OS |
| TMB (NGS) | Nivolumab or pembrolizumab or atezolizumab Johnson et al. [10] | 65 | RR, PFS, OS |
| TMB (WES) | Nivolumab and ipilimumab Weber et al. [11] | 94 | RR for the sequence: Nivo → Ipi |
| TMB (WES) | Nivolumab Riaz et al. [12] | 68 | OS |
| TMB (WES) | Pembrolizumab Cristescu et al. [13] | 89 | RR |
| TMB (NGS) | Ipilimumab or adoptive T-cell therapy Rosizik et al. [14] | 76 | PFS, OS |
| TMB (WES) | Anti-PD-1 or anti-CTLA-4 Roh et al. [15] | 21 | RR (numerical) |
| TMB (NGS) | Anti-PD-1 and/or anti-CTLA-4 Morrison et al. [16] | 160 | RR |
| MHC-II | Nivolumab or pembrolizumab Liu et al. [17] | 32 | RR (in previously exposed to anti-CTLA-4) |
| MHC-II | Anti-PD-1 Johnson et al. [18] | 30 | RR, PFS, OS |
| MHC-II | Nivolumab or pembrolizumab Johnson et al. [19] | 166 | RR, PFS |
| MHC-I MHC-II | Ipilimumab and/or nivolumab Rodig et al. [20] | 181 | OS (MHC-I for ipilimumab; MHC-II for nivolumab) |
| B2M | Anti-CTLA-4 or anti-PD-1 Sade-Feldman et al. [21] | 143 | RR, OS |
| Gut Microbiota | Anti-CTLA-4 Vetizou et al. [22] | 25 | Antitumor response |
| Gut Microbiota | Anti-CTLA-4 Chaput et al. [23] | 26 | PFS, OS |
| Gut Microbiota | Anti-CTLA-4 Coutzac et al. [24] | 85 | PFS, OS |
| Gut microbiota | anti-PD-1 Gopalakrishnan et al. [25] | 112 | ORR, PFS |
| Gut microbiota | Anti-PD-1 Nomura et al. [26] | 52 | ORR, PFS |
| TILs | Anti-CTLA-4 Hamid et al. [27] | 82 | RR |
| TILs | Anti-PD-1 Daud et al. [28] | 20 | RR, PFS |
| TILs | Anti-PD-1 Tumeh et al. [29] | 46 | RR |
| TILs | Anti-PD-1 Huang et al. [30] | 27 | PCR, DFS |
| TILs | Anti-PD-1 or Anti-PD-1 + anti-CTLA-4 Amaria et al. [31] | 23 | ORR |
| TILs | Anti-PD-1 Uryvaev et al. [32] | 30 | RR, PFS |
| Immunoscore | Anti-CTLA-4 Bifulco et al., Galon et al. [33,34] | 190 | No relationship with RR |
| IFN signature | Anti-PD-1 Karachaliou et al. [35] | 21 | DFS, PFS, OS |
| IFN signature | Anti-PD-1 Ayers et al. [36] | 19 | RR, PFS |
| Expanded immune signature | Anti-PD-1 Ayers et al. [36] | 62 | RR, PFS |
| Immune signature | Anti-PD-1 Chen et al. [37] | 46 | RR |
| Immune signature | Anti-PD-1 Huang et al. [30] | 27 | RFS |
| Immune signature | Anti-PD-1 Gide et al. [38] | 158 (samples) | PFS |
| LDH | Anti-PD-1 Diem et al. [39] | 66 | OS, ORR |
| LDH | Anti PD-1 or anti-CTLA-4 Wagner et al. [40] | 238 | OS |
| LDH | Anti-CTLA-4 Simeone et al. [41] | 95 | OS, RR |
| S100B | Anti PD-1 or anti-CTLA-4 Wagner et al. [40] | 238 | OS |
| PCR | Anti-PD-1 end/or anti-CTLA-4 Laino et al. [42] Wagner et al. [40] | 1503 | OS, RR |
| PCR | Anti-CTLA-4 Simeone et al. [43] | 95 | OS, RR |
| PCR | Anti-CTLA-4 Nyakas et al. [44] | 69 | OS |
| HGF | Anti-PD-1 Kubo et al. [45] | 29 | OS, PFS |
| IL-6 | Anti-PD-1 end/or anti-CTLA-4 Laino et al. [46] Wagner et al. [47] | 1503 | OS, RR |
| IL-6 | Anti-CTLA-4 Valpione et al. [48] | 140 | OS |
| IL-8 | Anti-PD-1 ± anti-CTL-4 Sanmamed et al. [49] | 29 + 34 validation cohort NSCLC + melanoma pts) | OS, RR |
| CXCL-5 | Anti-PD-1 Fujimura et al. [50,51] | 46 | ORR |
| sPDL-1 | Anti-PD-1 Dronca et al. [52] | 60 | ORR |
| sPDL-1 | Anti-CTLA-4 or anti-PDL-1 Zhou et al. [53] | 90 | ORR |
| PD-L1 and CD28 exosomal expression | Anti-CTLA-4 Tucci et al. [54] | 59 | CR, PFS, OS |
| Exosomal PD-L1 | Anti-PDL-1 and/or anti-CTLA-4 Cordonnier et al. [55] | 100 | RR, PFS, OS |
| CTLA-4 | Anti-CTLA-4 Pistillo et al. [56] | 113 | ORR |
| Absolute lymphocyte count | Anti-CTLA-4 Simeone et al. [41] | 95 | OS, RR |
| Relative eosinophil and lymphocyte count | Anti-PDL-1 Weide et al. [57] | 616 | OS |
| Eosinophil and lymphocyte count | Anti-CTLA-4 Delyon et al. [58] | 73 | OS |
| Absolute lymphocyte count | Anti-CTLA-4 Martens et al. [59] | 82 | OS |
| Absolute monocyte/eosinophil and relative lymphocyte counts | Anti-CTLA-4 Martens et al. [59] | 209 | OS, RR |
| Absolute lymphocyte and neutrophil count | Anti-PDL-1 Nakamura et al. [60] | 98 | OS |
| NLR | Anti-PD-1 end/or anti-CTLA-4 Laino et al. [42] | 1503 | OS, RR |
| NLR | Anti-CTLA-4 Ferrucci et al. [61] | 69 | PFS, OS |
| NLR | Anti-CTLA-4 Ferrucci et al. [62] | 720 | PFS, OS |
| NLR | Anti-PD-1 Capone et al. [63] | 97 | PFS, OS |
| NLR | Anti-PD-1 Bartlett et al. [64] | 224 | TTF, OS |
| NLR | Anti-CTLA-4 Zaragoza et al. [65] | 58 | OS |
| NLR | Anti-CTLA-4 Cassidy et al. [66] | 197 | RR, PFS, OS |
| NLR | Anti-PD-1 Fujisawa et al. [67] | 90 | ORR |
| CD8 effector-memory type 1 T-cells | Anti-CTLA-4 Wistuba-Hamprecht et al. [68] | 137 | OS, RR |
| CD45RO+CD8+ T-cells | Anti-CTLA-4 or anti-PD-1 Tietze et al. [69] | 30 | OS |
| FoxP3-Tregs | Anti-CTLA-4 Simeone et al. [41] | 95 | OS, RR |
| FoxP3-Tregs | Anti-CTLA-4 Martens et al. [59] | 209 | OS, RR |
| FoxP3-Tregs | Nivolumab with vaccine Weber et al. [70] | 90 | ORR |
| CD37 | Anti-PD-1 Capone et al. [63] | 100 | ORR, OS |
| PD-1+CD56+ T-cells | Anti-PD-1 Bochem et al. [71] | 75 | PFS, OS, RR |
| CD4 and CD8 memory T- cells | Anti-CTLA-4 Martens et al. [59] | 82 | OS |
| NK T-cells | Anti-PD-1 Subrahmanyam et al. [72] | 67 | ORR |
| T-cell exhaustion markers | Anti-PD-1 Pirozyan et al. [73] | 42 | No differences in RR |
| TCR repertoire | Nivolumab with vaccine Weber et al. [70] | 90 | ORR |
| TCR repertoire | Anti-CTLA-4 Cha et al. [74] | 21 melanoma pts | ORR |
| TCR repertoire | Anti-CTLA-4 Postow et al. [75] | 12 | CB * |
| Monocyte frequency | Anti-PD-1 Weide et al. [57] | 616 | OS |
| Monocyte frequency | Anti-PD-1 Krieg et al. [76] | 60 samples | PFS, OS |
| MDSCSs | Anti-PD-1 Weide et al. [57] | 616 | OS |
| MDSCSs | Anti-PD-1 De Coana et al. [77] | 43 | ORR, OS |
| MDSCSs | Anti-CTLA-4 Meyer et al. [78] | 49 (+15 controls) | ORR |
| MDSCSs | Anti-CTLA-4 Weber et al. [11] | 92 | ORR, PFS, OS |
| ctDNA | Anti-PD-1 (± anti-CTLA-4) Lee et al. [79] | 86 | RR, PFS, OS |
| TMB, ctDNA, cell-free DNA | Anti-PD-1 or anti-CTLA-4 Forschner et al. [80] | 35 | ORR, OS |
| ctDNA | Anti-PD-1 Marsavela et al. [81] | 125 patients discovery cohort + 128 validation samples | PFS, OS |
| ctDNA | Anti-PD-L1 and/or anti-CTLA-4 or BRAF/MEK inhibitors Rowe et al. [82] | 127 | ctDNA mutant fraction and sum of tumor diameters relationship |
| ctDNA | Anti-PD-1 (± anti-CTLA-4) Lee et al. [83] | 125 | ORR, OS |
| References | Treatment | Mean ORR in PD-L1-Positive (SD, 95% CI) | Mean ORR in PD-L1-Negative (SD, 95% CI) | p * Value |
|---|---|---|---|---|
| [43,46,47,110,111,118,119,121,125] | Pembrolizumab | 41.3 (23.2, 24.8–57.9) | 23.0 (17.7, 10.4–35.7) | 0.009 |
| [75,112,118,125] | Ipilimumab | 22.6 (11.5, 4.4–40.9) | 10.0 (4.3, 3.2–16.8) | 0.1 |
| [43,46,70,110,113,116,117,120] | Nivolumab | 47.5 (9.6, 40.1–54.9) | 24.0 (19.5, 8.9–39.0) | 0.01 |
| [43,118] | Nivolumab + ipilimumab | 65.2 (9.7, -22.0–152.4) | 55.5 (0.3, 53.3–57.8) | 0.4 |
| § [16,19,44,45,110,114,115] | Combined therapy and atezolizumab (1 study) | 46.2 (19.8, 29.6–62.7) | 27.7 (18.6, 12.1–43.2) | 0.0008 |
| Overall results based on a total of 5069 patients | 44.1 (19.0, 37.3–50.9) | 24.5 (18.5, 18.0–31.0) | <0.0001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garutti, M.; Bonin, S.; Buriolla, S.; Bertoli, E.; Pizzichetta, M.A.; Zalaudek, I.; Puglisi, F. Find the Flame: Predictive Biomarkers for Immunotherapy in Melanoma. Cancers 2021, 13, 1819. https://doi.org/10.3390/cancers13081819
Garutti M, Bonin S, Buriolla S, Bertoli E, Pizzichetta MA, Zalaudek I, Puglisi F. Find the Flame: Predictive Biomarkers for Immunotherapy in Melanoma. Cancers. 2021; 13(8):1819. https://doi.org/10.3390/cancers13081819
Chicago/Turabian StyleGarutti, Mattia, Serena Bonin, Silvia Buriolla, Elisa Bertoli, Maria Antonietta Pizzichetta, Iris Zalaudek, and Fabio Puglisi. 2021. "Find the Flame: Predictive Biomarkers for Immunotherapy in Melanoma" Cancers 13, no. 8: 1819. https://doi.org/10.3390/cancers13081819
APA StyleGarutti, M., Bonin, S., Buriolla, S., Bertoli, E., Pizzichetta, M. A., Zalaudek, I., & Puglisi, F. (2021). Find the Flame: Predictive Biomarkers for Immunotherapy in Melanoma. Cancers, 13(8), 1819. https://doi.org/10.3390/cancers13081819










