The Role of lncRNAs in the Pathobiology and Clinical Behavior of Multiple Myeloma
Abstract
Simple Summary
Abstract
1. Introduction
2. Features of lncRNAs
3. Role of lncRNAs in the Pathobiology of MM
4. Impact of lncRNAs on the Response of MM
5. lncRNAs as Biomarkers for Clinical Stratification of MM Patients
6. Epigenetic Drugs can Modulate lncRNA Expression in MM
7. lncRNA-Targeted Therapies for MM Treatment
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Morgan, G.J.; Walker, B.A.; Davies, F.E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer 2012, 12, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Rajkumar, V.; Kyle, R.A.; Van Duin, M.; Sonneveld, P.; Mateos, M.-V.; Gay, F.; Anderson, K.C. Multiple myeloma. Nat. Rev. Dis. Prim. 2017, 3, 17046. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, O.; Baz, R. Multiple Myeloma Genomics—A Concise Review. Acta Med. Acad. 2019, 48, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Medical Masterclass Contributors; Firth, J. Haematology: Multiple myeloma. Clin. Med. 2019, 19, 58–60. [Google Scholar] [CrossRef]
- Agirre, X.; Castellano, G.; Pascual, M.; Heath, S.; Kulis, M.; Segura, V.; Bergmann, A.; Esteve, A.; Merkel, A.; Raineri, E.; et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015, 25, 478–487. [Google Scholar] [CrossRef]
- Manier, S.; Salem, K.Z.; Park, J.; Landau, D.A.; Getz, G.; Ghobrial, I.M. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 100–113. [Google Scholar] [CrossRef]
- Prideaux, S.M.; O’Brien, E.C.; Chevassut, T.J. The Genetic Architecture of Multiple Myeloma. Adv. Hematol. 2014, 2014, 864058. [Google Scholar] [CrossRef]
- Barwick, B.G.; Neri, P.; Bahlis, N.J.; Nooka, A.K.; Dhodapkar, M.V.; Jaye, D.L.; Hofmeister, C.C.; Kaufman, J.L.; Gupta, V.A.; Auclair, D.; et al. Multiple myeloma immunoglobulin lambda translocations portend poor prognosis. Nat. Commun. 2019, 10, 1911. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple myeloma: Every year a new standard? Hematol. Oncol. 2019, 37 (Suppl. 1), 62–65. [Google Scholar] [CrossRef]
- Ordoñez, R.; Kulis, M.; Russiñol, N.; Chapaprieta, V.; Carrasco-Leon, A.; García-Torre, B.; Charalampopoulou, S.; Clot, G.; Beekman, R.; Meydan, C.; et al. Chromatin activation as a unifying principle underlying pathogenic mechanisms in multiple myeloma. Genome Res. 2020, 30, 1217–1227. [Google Scholar] [CrossRef]
- Beekman, R.; Chapaprieta, V.; Russiñol, N.; Vilarrasa-Blasi, R.; Verdaguer-Dot, N.; Martens, J.H.A.; Duran-Ferrer, M.; Kulis, M.; Serra, F.; Javierre, B.M.; et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 2018, 24, 868–880. [Google Scholar] [CrossRef]
- Ernst, J.; Kheradpour, P.; Mikkelsen, T.S.; Shoresh, N.; Ward, L.D.; Epstein, C.B.; Zhang, X.; Wang, L.; Issner, R.; Coyne, M.; et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011, 473, 43–49. [Google Scholar] [CrossRef]
- Carrasco-Leon, A.; Ezponda, T.; Meydan, C.; Valcárcel, L.V.; Ordoñez, R.; Kulis, M.; Garate, L.; Miranda, E.; Segura, V.; Guruceaga, E.; et al. Characterization of complete lncRNAs transcriptome reveals the functional and clinical impact of lncRNAs in multiple myeloma. Leukemia 2021, 1–13. [Google Scholar] [CrossRef]
- Ng, M.; Heckl, D.; Klusmann, J.-H. The Regulatory Roles of Long Noncoding RNAs in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 570. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Garitano-Trojaola, A.; José-Enériz, E.S.; Ezponda, T.; Unfried, J.P.; Carrasco-León, A.; Razquin, N.; Barriocanal, M.; Vilas-Zornoza, A.; Sangro, B.; Segura, V.; et al. Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget 2018, 9, 12842–12852. [Google Scholar] [CrossRef]
- Robinson, E.K.; Covarrubias, S.; Carpenter, S. The how and why of lncRNA function: An innate immune perspective. Biochim. Biophys. Acta Bioenerg. 2020, 1863, 194419. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Laurent, G.S.; Vyatkin, Y.; Antonets, D.; Ri, M.; Qi, Y.; Saik, O.; Shtokalo, D.; De Hoon, M.J.; Kawaji, H.; Itoh, M.; et al. Functional annotation of the vlinc class of non-coding RNAs using systems biology approach. Nucleic Acids Res. 2016, 44, 3233–3252. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta Bioenerg. 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Yan, P.; Luo, S.; Lu, J.Y.; Shen, X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 170–178. [Google Scholar] [CrossRef]
- Guenzl, P.M.; Barlow, D.P. Macro lncRNAs: A new layer of cis-regulatory information in the mammalian genome. RNA Biol. 2012, 9, 731–741. [Google Scholar] [CrossRef]
- Agirre, X.; Meydan, C.; Jiang, Y.; Garate, L.; Doane, A.S.; Li, Z.; Verma, A.; Paiva, B.; Martín-Subero, J.I.; Elemento, O.; et al. Long non-coding RNAs discriminate the stages and gene regulatory states of human humoral immune response. Nat. Commun. 2019, 10, 821. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Shi, W.; Wang, Q.; Bian, Y.; Fan, Y.; Zhou, Y.; Feng, T.; Li, Z.; Cao, X. Long noncoding RNA PANDA promotes esophageal squamous carcinoma cell progress by dissociating from NF-YA but interact with SAFA. Pathol. Res. Pract. 2019, 215, 152604. [Google Scholar] [CrossRef]
- Calle, A.S.; Kawamura, Y.; Yamamoto, Y.; Takeshita, F.; Ochiya, T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018, 109, 2093–2100. [Google Scholar] [CrossRef]
- Akhade, V.S.; Pal, D.; Kanduri, C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv. Exp. Med. Biol. 2017, 1008, 47–74. [Google Scholar] [CrossRef]
- Chen, C.-L.; Tseng, Y.-W.; Wu, J.-C.; Chen, G.-Y.; Lin, K.-C.; Hwang, S.-M.; Hu, Y.-C. Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials 2015, 44, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Hermans-Beijnsberger, S.; van Bilsen, M.; Schroen, B. Long non-coding RNAs in the failing heart and vasculature. Non-Coding RNA Res. 2018, 3, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yang, W.; Zhang, L.; Liu, K.; Luo, Z. Long non-coding RNA ANRIL and its target microRNAs (microRNA-34a, microRNA-125a and microRNA-186) relate to risk stratification and prognosis in multiple myeloma. Hematology 2021, 26, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, H.-Y.; Zhang, J.-L.; Wan, D.-M.; Li, Y.-M.; Jiang, Z.-X. Dysregulation of LncRNA ANRIL mediated by miR-411–3p inhibits the malignant proliferation and tumor stem cell like property of multiple myeloma via hypoxia-inducible factor 1α. Exp. Cell Res. 2020, 396, 112280. [Google Scholar] [CrossRef]
- Li, Z.; Kumar, S.; Jin, D.-Y.; Calin, G.A.; Chng, W.-J.; Siu, K.-L.; Poon, M.-W.; Chim, C.S. Epigenetic silencing of long non-coding RNA BM742401 in multiple myeloma: Impact on prognosis and myeloma dissemination. Cancer Cell Int. 2020, 20, 403. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Wu, J.; Khadka, B.; Fang, Z.; Gu, J.; Tang, B.; Xiao, R.; Pan, G.; Liu, J. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 54. [Google Scholar] [CrossRef]
- Chen, S.; Li, T.; Zhao, Q.; Xiao, B.; Guo, J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta 2017, 466, 167–171. [Google Scholar] [CrossRef]
- Luo, Y.-H.; Yang, Y.-P.; Chien, C.-S.; Yarmishyn, A.A.; Ishola, A.A.; Chien, Y.; Chen, Y.-M.; Huang, T.-W.; Lee, K.-Y.; Huang, W.-C.; et al. Plasma Level of Circular RNA hsa_circ_0000190 Correlates with Tumor Progression and Poor Treatment Response in Advanced Lung Cancers. Cancers 2020, 12, 1740. [Google Scholar] [CrossRef]
- Meng, Y.-B.; He, X.; Huang, Y.-F.; Wu, Q.-N.; Zhou, Y.-C.; Hao, D.-J. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 1207–1214. [Google Scholar] [CrossRef]
- David, A.; Zocchi, S.; Talbot, A.; Choisy, C.; Ohnona, A.; Lion, J.; Cuccuini, W.; Soulier, J.; Arnulf, B.; Bories, J.-C.; et al. The long non-coding RNA CRNDE regulates growth of multiple myeloma cells via an effect on IL6 signalling. Leukemia 2020. [Google Scholar] [CrossRef]
- Tong, J.; Xu, X.; Zhang, Z.; Ma, C.; Xiang, R.; Liu, J.; Xu, W.; Wu, C.; Li, J.; Zhan, F.; et al. Hypoxia-induced long non-coding RNA DARS-AS1 regulates RBM39 stability to promote myeloma malignancy. Haematologica 2020, 105, 1630–1640. [Google Scholar] [CrossRef]
- Li, Q.-Y.; Chen, L.; Hu, N.; Zhao, H. Long non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma via miR-610/Akt3 axis. Biomed. Pharmacother. 2018, 103, 1727–1732. [Google Scholar] [CrossRef]
- Isin, M.; Ozgur, E.; Cetin, G.; Erten, N.; Aktan, M.; Gezer, U.; Dalay, N. Investigation of circulating lncRNAs in B-cell neoplasms. Clin. Chim. Acta 2014, 431, 255–259. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, Y.; Liu, W.; Huang, Y.; Shen, X.; Jing, R.; Pu, J.; Wang, X.; Ju, S.; Cong, H.; et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019, 10, 106. [Google Scholar] [CrossRef]
- Zheng, J.-F.; Guo, N.-H.; Zi, F.-M.; Cheng, J. Long Noncoding RNA H19 Promotes Tumorigenesis of Multiple Myeloma by Activating BRD4 Signaling by Targeting MicroRNA 152-3p. Mol. Cell. Biol. 2019, 40. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, H.; Shen, X.; Wang, X.; Ju, S.; Lu, M.; Cong, H. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin. Chim. Acta 2018, 480, 199–205. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell. Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef]
- Zhu, B.Z.; Lin, L. Effects of lncRNA HOTAIR on proliferation and apoptosis of myeloma cells through NF-κB pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10042–10048. [Google Scholar] [CrossRef]
- Guan, R.; Wang, W.; Fu, B.; Pang, Y.; Lou, Y.; Li, H. Increased lncRNA HOTAIR expression promotes the chemoresistance of multiple myeloma to dexamethasone by regulating cell viability and apoptosis by mediating the JAK2/STAT3 signaling pathway. Mol. Med. Rep. 2019, 20, 3917–3923. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, X.; Wang, C. LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J. Cell. Biochem. 2019, 121, 4043–4051. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, J.; Chen, G. Long noncoding RNA IRAIN acts as tumor suppressor via miR-125b in multiple myeloma. Oncol. Lett. 2019, 18, 6787–6794. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Sun, Y.; Zhang, E.-B.; Lu, X.-Y.; Jin, S.-D.; Guo, R.-H. A novel long noncoding RNA IRAIN regulates cell proliferation in non small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12268–12275. [Google Scholar] [PubMed]
- Yu, T.; Xu, Z.; Zhang, X.; Men, L.; Nie, H. Long intergenic non-protein coding RNA 152 promotes multiple myeloma progression by negatively regulating microRNA-497. Oncol. Rep. 2018, 40, 3763–3771. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Wang, Y.; Li, H.; Sun, B.; Luo, X. Long non-coding RNA LINC00461/miR-149-5p/LRIG2 axis regulates hepatocellular carcinoma progression. Biochem. Biophys. Res. Commun. 2019, 512, 176–181. [Google Scholar] [CrossRef]
- Meng, Q.; Liu, M.; Cheng, R. LINC00461/miR-4478/E2F1 feedback loop promotes non-small cell lung cancer cell proliferation and migration. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Deng, M.; Yuan, H.; Liu, S.; Hu, Z.; Xiao, H. Exosome-transmitted LINC00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microRNA/BCL-2 expression. Cytotherapy 2019, 21, 96–106. [Google Scholar] [CrossRef]
- Lu, D.; Yang, C.; Zhang, Z.; Cong, Y.; Xiao, M. Knockdown of Linc00515 Inhibits Multiple Myeloma Autophagy and Chemoresistance by Upregulating miR-140-5p and Downregulating ATG14. Cell. Physiol. Biochem. 2018, 48, 2517–2527. [Google Scholar] [CrossRef]
- Wang, C.; Li, M.; Wang, S.; Jiang, Z.; Liu, Y. LINC00665 Promotes the Progression of Multiple Myeloma by Adsorbing miR-214-3p and Positively Regulating the Expression of PSMD10 and ASF1B. OncoTargets Ther. 2020, 13, 6511–6522. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Yang, Z.; Zhang, J.; Chen, S.; Cheng, J. LINC01234 promotes multiple myeloma progression by regulating miR-124-3p/GRB2 axis. Am. J. Transl. Res. 2019, 11, 6600–6618. [Google Scholar]
- Nian, F.; Zhu, J.; Chang, H. Long non-coding RNA ANGPTL1-3 promotes multiple myeloma bortezomib resistance by sponging miR-30a-3p to activate c-Maf expression. Biochem. Biophys. Res. Commun. 2019, 514, 1140–1146. [Google Scholar] [CrossRef]
- Zhang, C.; Chu, M.; Fan, Y.; Wu, L.; Li, Z.; Ma, X.; Zhuang, W. Long non-coding RNA T-cell factor 7 in multiple myeloma: A potential biomarker for deteriorated clinical features and poor prognosis. J. Clin. Lab. Anal. 2020, 34, e23400. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, H.; Peng, Q.; Yang, Y. Long Noncoding RNA LUCAT1 Promotes Multiple Myeloma Cell Growth by Regulating the TGF-β Signaling Pathway. Technol. Cancer Res. Treat. 2020, 19, 1533033820945770. [Google Scholar] [CrossRef]
- Handa, H.; Kuroda, Y.; Kimura, K.; Masuda, Y.; Hattori, H.; Alkebsi, L.; Matsumoto, M.; Kasamatsu, T.; Kobayashi, N.; Tahara, K.-I.; et al. Long non-coding RNAMALAT1is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br. J. Haematol. 2017, 179, 449–460. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, S.; Li, Q.; Ji, Q.; Guo, P.; Liu, X. MALAT1: A long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018, 16, 19–26. [Google Scholar] [CrossRef]
- Li, Z.-X.; Zhu, Q.-N.; Zhang, H.-B.; Hu, Y.; Wang, G.; Zhu, Y.-S. MALAT1: A potential biomarker in cancer. Cancer Manag. Res. 2018, 10, 6757–6768. [Google Scholar] [CrossRef]
- Gu, Y.; Xiao, X.; Yang, S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget 2017, 8, 101984–101993. [Google Scholar] [CrossRef]
- Liu, N.; Feng, S.; Li, H.; Chen, X.; Bai, S.; Liu, Y. Long non-coding RNA MALAT1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13 axis. J. Cancer Res. Clin. Oncol. 2020, 146, 367–379. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, T.; Jia, Y.; Zou, J.; Wang, X.; Gu, W. LncRNA MALAT1/miR-181a-5p affects the proliferation and adhesion of myeloma cells via regulation of Hippo-YAP signaling pathway. Cell Cycle 2019, 18, 2509–2523. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef]
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262. [Google Scholar] [CrossRef]
- Zhuang, W.; Ge, X.; Yang, S.; Huang, M.; Zhuang, W.; Chen, P.; Zhang, X.; Fu, J.; Qu, J.; Li, B. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells from Multiple Myeloma Patients by Targeting BMP4 Transcription. Stem Cells 2015, 33, 1985–1997. [Google Scholar] [CrossRef]
- Shen, X.; Bai, H.; Zhu, H.; Yan, Q.; Yang, Y.; Yu, W.; Shi, Q.; Wang, J.; Li, J.; Chen, L. Long Non-Coding RNA MEG3 Functions as a Competing Endogenous RNA to Regulate HOXA11 Expression by Sponging miR-181a in Multiple Myeloma. Cell. Physiol. Biochem. 2018, 49, 87–100. [Google Scholar] [CrossRef]
- Benetatos, L.; Dasoula, A.; Hatzimichael, E.; Georgiou, I.; Syrrou, M.; Bourantas, K.L. Promoter Hypermethylation of the MEG3 (DLK1/MEG3) Imprinted Gene in Multiple Myeloma. Clin. Lymphoma Myeloma 2008, 8, 171–175. [Google Scholar] [CrossRef]
- Yu, W.; Shi, Q.; Wu, C.; Shen, X.; Chen, L.; Xu, J. Promoter hypermethylation influences the suppressive role of long non-coding RNA MEG3 in the development of multiple myeloma. Exp. Ther. Med. 2020, 20, 637–645. [Google Scholar] [CrossRef]
- Ronchetti, D.; Agnelli, L.; Pietrelli, A.; Todoerti, K.; Manzoni, M.; Taiana, E.; Neri, A. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci. Rep. 2018, 8, 6557. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, X.; Zhang, F.; Jiang, S.; Liu, J.; Luo, Y. Bortezomib-inducible long non-coding RNA myocardial infarction associated transcript is an oncogene in multiple myeloma that suppresses miR-29b. Cell Death Dis. 2019, 10, 319. [Google Scholar] [CrossRef]
- Yu, H.; Peng, S.; Chen, X.; Han, S.; Luo, J. Long non-coding RNA NEAT1 serves as a novel biomarker for treatment response and survival profiles via microRNA-125a in multiple myeloma. J. Clin. Lab. Anal. 2020, 34, e23399. [Google Scholar] [CrossRef]
- Taiana, E.; Favasuli, V.; Ronchetti, D.; Todoerti, K.; Pelizzoni, F.; Manzoni, M.; Barbieri, M.; Fabris, S.; Silvestris, I.; Cantafio, M.E.G.; et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia 2020, 34, 234–244. [Google Scholar] [CrossRef]
- Taiana, E.; Ronchetti, M.; Favasuli, V.; Todoerti, K.; Manzoni, M.; Amodio, N.; Tassone, P.; Agnelli, L.; Neri, A. Long non-coding RNA NEAT1 shows high expression unrelated to molecular features and clinical outcome in multiple myeloma. Haematologica 2019, 104, e72–e76. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, P.; Li, W.-J.; Zhang, J.; Wang, G.-P.; Jiang, D.-F.; Chen, F.-P. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol. Immunol. 2020, 117, 20–28. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Ruan, J.; Zheng, W.; Yang, Z.; Pan, W. Long non-coding RNA OIP5-AS1 suppresses multiple myeloma progression by sponging miR-27a-3p to activate TSC1 expression. Cancer Cell Int. 2020, 20, 155. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chen, J.; Zhang, H.; Wanggang, Z.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Kong, S.; Yang, Q.; Yin, Q.; Cong, H.; Wang, X.; Ju, S. PCAT-1 promotes cell growth by sponging miR-129 via MAP3K7/NF-κB pathway in multiple myeloma. J. Cell. Mol. Med. 2020, 24, 3492–3503. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Shen, P.; Yang, Q.; Yin, Q.; Wang, F.; Cong, H.; Wang, X.; Ju, S. Knockdown of long non-coding RNA PCAT-1 inhibits myeloma cell growth and drug resistance via p38 and JNK MAPK pathways. J. Cancer 2019, 10, 6502–6510. [Google Scholar] [CrossRef]
- Yang, X.; Ye, H.; He, M.; Zhou, X.; Sun, N.; Guo, W.; Lin, X.; Huang, H.; Lin, Y.; Yao, R.; et al. LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem. Biophys. Res. Commun. 2018, 498, 207–213. [Google Scholar] [CrossRef]
- Xiao, G.; Li, Y.; Wang, Y.; Zhao, B.; Zou, Z.; Hou, S.; Jia, X.; Liu, X.; Yao, Y.; Wan, J.; et al. LncRNA PRAL is closely related to clinical prognosis of multiple myeloma and the bortezomib sensitivity. Exp. Cell Res. 2018, 370, 254–263. [Google Scholar] [CrossRef]
- Wen, Y.-Y.; Bai, B.; Hu, X.-S. Expression and Clinical Significance of Long Non-Coding RNA PRAL in Patients with Multiple Myeloma. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2020, 28, 185–190. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, L.; Wang, X.; Zhou, Y.; Wu, S. Down-regulation of miR-203a by lncRNA PVT1 in multiple myeloma promotes cell proliferation. Arch. Med Sci. 2018, 14, 1333–1339. [Google Scholar] [CrossRef]
- Handa, H.; Honma, K.; Oda, T.; Kobayashi, N.; Kuroda, Y.; Kimura-Masuda, K.; Watanabe, S.; Ishihara, R.; Murakami, Y.; Masuda, Y.; et al. Long Noncoding RNA PVT1 Is Regulated by Bromodomain Protein BRD4 in Multiple Myeloma and Is Associated with Disease Progression. Int. J. Mol. Sci. 2020, 21, 7121. [Google Scholar] [CrossRef]
- Yang, X.; Huang, H.; Wang, X.; Liu, H.; Liu, H.; Lin, Z. Knockdown of lncRNA SNHG16 suppresses multiple myeloma cell proliferation by sponging miR-342-3p. Cancer Cell Int. 2020, 20, 38. [Google Scholar] [CrossRef]
- Tianhua, Y.; Dianqiu, L.; Xuanhe, Z.; Zhe, Z.; Dongmei, G. Long non-coding RNA Sox2 overlapping transcript (SOX2OT) promotes multiple myeloma progression via microRNA-143-3p/c-MET axis. J. Cell. Mol. Med. 2020, 24, 5185–5194. [Google Scholar] [CrossRef]
- Ronchetti, D.; Todoerti, K.; Vinci, C.; Favasuli, V.; Agnelli, L.; Manzoni, M.; Pelizzoni, F.; Chiaramonte, R.; Platonova, N.; Giuliani, N.; et al. Expression Pattern and Biological Significance of the lncRNA ST3GAL6-AS1 in Multiple Myeloma. Cancers 2020, 12, 782. [Google Scholar] [CrossRef]
- Dong, H.; Jiang, S.; Fu, Y.; Luo, Y.; Gui, R.; Liu, J. Upregulation of lncRNA NR_046683 Serves as a Prognostic Biomarker and Potential Drug Target for Multiple Myeloma. Front. Pharmacol. 2019, 10, 45. [Google Scholar] [CrossRef]
- Liu, D.; Wang, J.; Liu, M. Long noncoding RNA TUG1 promotes proliferation and inhibits apoptosis in multiple myeloma by inhibiting miR-29b-3p. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, L. Downregulation of lncRNA UCA1 facilitates apoptosis and reduces proliferation in multiple myeloma via regulation of the miR-1271-5p/HGF axis. J. Chin. Med. Assoc. 2019, 82, 699–709. [Google Scholar] [CrossRef]
- Li, J.-L.; Liu, X.-L.; Guo, S.-F.; Yang, Y.; Zhu, Y.-L.; Li, J.-Z. Long noncoding RNA UCA1 regulates proliferation and apoptosis in multiple myeloma by targeting miR-331-3p/IL6R axis for the activation of JAK2/STAT3 pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9238–9250. [Google Scholar] [CrossRef]
- Pu, J.; Huang, H.; Su, J.; Yuan, J.; Cong, H.; Wang, X.; Ju, S. Decreased expression of long noncoding RNA XLOC_013703 promotes cell growth via NF-κB pathway in multiple myeloma. IUBMB Life 2019, 71, 1240–1251. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Gupta, S.C.; Tripathi, Y.N. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int. J. Cancer 2017, 140, 1955–1967. [Google Scholar] [CrossRef]
- Yang, X.; Yang, B. lncRNA PDIA3P regulates cell proliferation and invasion in non-small cell lung cancer. Exp. Ther. Med. 2019, 18, 3184–3190. [Google Scholar] [CrossRef]
- Zhou, C.-C.; Yang, F.; Yuan, S.-X.; Ma, J.-Z.; Liu, F.; Yuan, J.-H.; Bi, F.-R.; Lin, K.-Y.; Yin, J.-H.; Cao, G.-W.; et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 2016, 63, 850–863. [Google Scholar] [CrossRef]
- Su, P.; Wang, F.; Qi, B.; Wang, T.; Zhang, S. P53 Regulation-Association Long Non-Coding RNA (LncRNA PRAL) Inhibits Cell Proliferation by Regulation of P53 in Human Lung Cancer. Med. Sci. Monit. 2017, 23, 1751–1758. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7, e2123. [Google Scholar] [CrossRef]
- Yu, X.; Li, Z.; Zheng, H.; Chan, M.T.V.; Wu, W.K.K. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017, 50, e12329. [Google Scholar] [CrossRef]
- Xiong, D.-D.; Li, Z.-Y.; Liang, L.; He, R.-Q.; Ma, F.-C.; Luo, D.-Z.; Hu, X.-H.; Chen, G. The LncRNA NEAT1 Accelerates Lung Adenocarcinoma Deterioration and Binds to Mir-193a-3p as a Competitive Endogenous RNA. Cell. Physiol. Biochem. 2018, 48, 905–918. [Google Scholar] [CrossRef]
- Gao, D.; Lv, A.-E.; Li, H.-P.; Han, D.-H.; Zhang, Y.-P. LncRNA MALAT-1 Elevates HMGB1 to Promote Autophagy Resulting in Inhibition of Tumor Cell Apoptosis in Multiple Myeloma. J. Cell. Biochem. 2017, 118, 3341–3348. [Google Scholar] [CrossRef]
- Samur, M.K.; Minvielle, S.; Gulla, A.; Fulciniti, M.; Cleynen, A.; Samur, A.A.; Szalat, R.; Shammas, M.; Magrangeas, F.; Tai, Y.-T.; et al. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia 2018, 32, 2626–2635. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef]
- Arman, K.; Möröy, T. Crosstalk Between MYC and lncRNAs in Hematological Malignancies. Front. Oncol. 2020, 10, 579940. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Chen, S.; Tan, Y.; Li, Y.; Tang, F. Oncogenic super-enhancer formation in tumorigenesis and its molecular mechanisms. Exp. Mol. Med. 2020, 52, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Pang, P.; Jiang, M.; Cao, Q.; Li, H.; Xu, Y.; Li, Y.; Chen, X.; Han, J. eRNAs and Superenhancer lncRNAs Are Functional in Human Prostate Cancer. Dis. Markers 2020, 2020, 8847986. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zheng, Y.-H.; Xu, L.; Feng, J.; Tang, H.-L.; Luo, C.; Song, Y.-P.; Chen, X.-Q. BRD4 inhibitor nitroxoline enhances the sensitivity of multiple myeloma cells to bortezomib in vitro and in vivo by promoting mitochondrial pathway-mediated cell apoptosis. Ther. Adv. Hematol. 2020, 11, 2040620720932686. [Google Scholar] [CrossRef] [PubMed]
- Díaz, T.; Rodríguez, V.; Lozano, E.; Mena, M.-P.; Calderón, M.; Rosiñol, L.; Martínez, A.; Tovar, N.; Pérez-Galán, P.; Bladé, J.; et al. The BET bromodomain inhibitor CPI203 improves lenalidomide and dexamethasone activity in in vitro and in vivo models of multiple myeloma by blockade of Ikaros and MYC signaling. Haematologica 2017, 102, 1776–1784. [Google Scholar] [CrossRef]
- Stubbs, M.C.; Burn, T.C.; Sparks, R.; Maduskuie, T.; Diamond, S.; Rupar, M.; Wen, X.; Volgina, A.; Zolotarjova, N.; Waeltz, P.; et al. The Novel Bromodomain and Extraterminal Domain Inhibitor INCB054329 Induces Vulnerabilities in Myeloma Cells That Inform Rational Combination Strategies. Clin. Cancer Res. 2018, 25, 300–311. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Chaidos, A.; Caputo, V.; Gouvedenou, K.; Liu, B.; Marigo, I.; Chaudhry, M.S.; Rotolo, A.; Tough, D.F.; Smithers, N.N.; Bassil, A.K.; et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood 2014, 123, 697–705. [Google Scholar] [CrossRef]
- Abruzzese, M.P.; Bilotta, M.T.; Fionda, C.; Zingoni, A.; Soriani, A.; Petrucci, M.T.; Ricciardi, M.R.; Molfetta, R.; Paolini, R.; Santoni, A.; et al. The homeobox transcription factor MEIS2 is a regulator of cancer cell survival and IMiDs activity in Multiple Myeloma: Modulation by Bromodomain and Extra-Terminal (BET) protein inhibitors. Cell Death Dis. 2019, 10, 324. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Gilboa-Geffen, A.; Hamar, P.; Le, M.T.; Wheeler, L.A.; Trifonova, R.; Petrocca, F.; Wittrup, A.; Lieberman, J. Gene Knockdown by EpCAM Aptamer–siRNA Chimeras Suppresses Epithelial Breast Cancers and Their Tumor-Initiating Cells. Mol. Cancer Ther. 2015, 14, 2279–2291. [Google Scholar] [CrossRef]
- Wen, J.; Tao, W.; Hao, S.; Iyer, S.P.; Zu, Y. A unique aptamer-drug conjugate for targeted therapy of multiple myeloma. Leukemia 2015, 30, 987–991. [Google Scholar] [CrossRef]
- Yoon, S.; Wu, X.; Armstrong, B.; Habib, N.; Rossi, J.J. An RNA Aptamer Targeting the Receptor Tyrosine Kinase PDGFRα Induces Anti-tumor Effects through STAT3 and p53 in Glioblastoma. Mol. Ther. Nucleic Acids 2019, 14, 131–141. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]

| Gene | Location | Gene Type | Expression in MM | Molecular Mechanism | Molecular Interaction in MM | Biological Effect after lncRNA KD | Biological Effect after lncRNA UR | Prognosis in MM | References |
|---|---|---|---|---|---|---|---|---|---|
| ANRIL | 9p21.3 | Antisense | Up | Decoy | Binds to miR-34a, miR-125a, miR-186 and miR- 411–3p | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse PFS and OS | [33,34] |
| BM742401 | 18q11.2 | LincRNA | Down | NA | NA | NA | Decreases cell migration | Methylated lncRNA associated with worse OS | [35] |
| Circ_0000190 | 1q42.12 | Circular lncRNA | Down | NA | NA | NA | NA | High expression levels associated with better PFS and OS | [36,37,38] |
| CRNDE | 16q12.2 | LincRNA | Up | Decoy | Binds to miR-451 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse OS | [39,40] |
| DARS-AS1 | 2q21.3 | Antisense | Up under hypoxia | Decoy | Interacts with RBM39 and HIP-1α, suppressing mTOR pathway | Decreases cellular proliferation and increases apoptosis. Decreases tumorigenesis in vivo. Its upregulation reduces the sensitivity to bortezomib in vitro | NA | NA | [41] |
| ENSG00000249988 | 4p15.33 | LincRNA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS andbetter OS | [13] |
| ENSG00000254343 | 8q24.12 | LincRNA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS | [13] |
| FEZF1-AS1 | 7q31.32 | Antisense | Up | Decoy | Binds to miR-610 and regulates AKT3 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | NA | [42] |
| GAS5 | 1q25.1 | Processed transcript | Down | NA | NA | NA | Decreases cellular proliferation | NA | [43] |
| H19 | 11p15.5 | Processed transcript | Up | Decoy | Binds to miR-152-3p and miR-29b-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse PFS | [44,45,46] |
| HOTAIR | 12q13.31 | Antisense | Up | NA | Activates NF-κB pathway | Decreases cellular proliferation, triggers cell cycle arrest and decreases chemoresistance to dexamethasone | NA | NA | [21,47,48,49] |
| HOXB-AS1 | 17q21.32 | Antisense | Up | Scaffold | Scaffold for ELAVL1. Interacts with FUT4-mediated Wnt/β-catenin pathway | Decreases cellular proliferation and increases apoptosis | NA | NA | [50] |
| IRAIN | 15q26.3 | Antisense | Down | Decoy | Binds to miR-125b and regulates IGF-1 signaling | NA | Increases apoptosis | NA | [51,52] |
| LINC00152 | 2p11.2 | LincRNA | Up | Decoy | Binds to miR-497 | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest. Decreases tumorigenesis in vivo | NA | High expression levels associated with worse OS | [53] |
| LINC00461 | 5q14.3 | LincRNA | Up | NA | NA | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse OS | [54,55,56] |
| LINC00515 | 21q21.3 | LincRNA | Up | Decoy | Binds to miR-140-5p | Increases apoptosis | NA | NA | [57] |
| LINC00665 | 19q13.12 | LincRNA | Up | Decoy | Binds to miR-214-3p | Decreases cellular proliferation and increases apoptosis | NA | NA | [58] |
| LINC01234 | 12q24.13 | LincRNA | Up | Decoy | Binds to miR-124-3p | Decreases cellular proliferation and increases apoptosis. Decreases cell proliferation and tumor growth in vivo | NA | High expression levels associated with worse OS | [59] |
| lnc-ANGPTL1-3 | 1q25.2 | Antisense | Up | Decoy | Binds to miR-30a-3p | Increases the sensitivity to bortezomib | NA | High expression levels associated with worse OS | [60] |
| lnc-TCF7 | 5q31.1 | NA | Up | NA | NA | NA | NA | High expression levels associated with worse PFS and OS | [61] |
| LUCAT1 | 5q14.3 | LincRNA | Up | NA | Activates the TGF-β signaling pathway | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with shorter five-year survival | [62] |
| MALAT1 | 11q13.1 | LincRNA | Up | Decoy and Scaffold | Binds to miR-1271-5p, miR-181a-5p and miR-509- 5p. Scaffold for PARP1 | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse PFS and OS | [63,64,65,66,67,68,69,70] |
| MEG3 | 14q32.2 | LincRNA | Down | Decoy | Binds to miR-181a | NA | Decreases cellular proliferation and increases apoptosis | High expression levels associated with better PFS and OS | [71,72,73,74] |
| MIAT | 22q12.1 | LincRNA | Up | Decoy | Binds to miR-29b | Sensitizes MM cells to bortezomib | NA | High expression levels associated with worse PFS and OS | [75,76] |
| NEAT1 | 11q13.1 | LincRNA | Up | Decoy | Binds to miR-214 and miR-125a | Decreases cellular proliferation | NA | High expression levels associated with worse PFS and OS | [77,78,79,80] |
| OIP5-AS1 | 15q15.1 | Processed transcript | Down | Decoy | Binds to miR-410 and miR-27a-3p | NA | Decreases cellular proliferation and increases apoptosis | NA | [81,82] |
| PCAT-1 | 8q24.21 | LincRNA | Up | Decoy | Binds to miR-129 | Increases apoptosis and sensitizes MM cells to bortezomib | NA | NA | [83,84] |
| PDIA3P | 1q21.1 | Pseudogene | Up | NA | NA | Decreases cellular proliferation. Increases the sensitivity to bortezomib | NA | High expression levels associated with worse OS | [85] |
| PDLIM1P4 | 3q12.1 | Pseudogene | Up | NA | NA | NA | NA | High expression levels associated with worse PFS and OS | [13] |
| PRAL | 17p13.1 | NA | Down | Decoy | Binds to miR-210 | NA | Decreases cellular proliferation and increases apoptosis. Increases the anti-tumor effect of bortezomib | High expression levels associated with better PFS and OS | [86,87] |
| PVT1 | 8q24.21 | Processed transcript | Up | Decoy | Binds to miR-203a. It is inhibited by BRD4 | Decreases cellular proliferation and increases apoptosis | NA | NA | [88,89] |
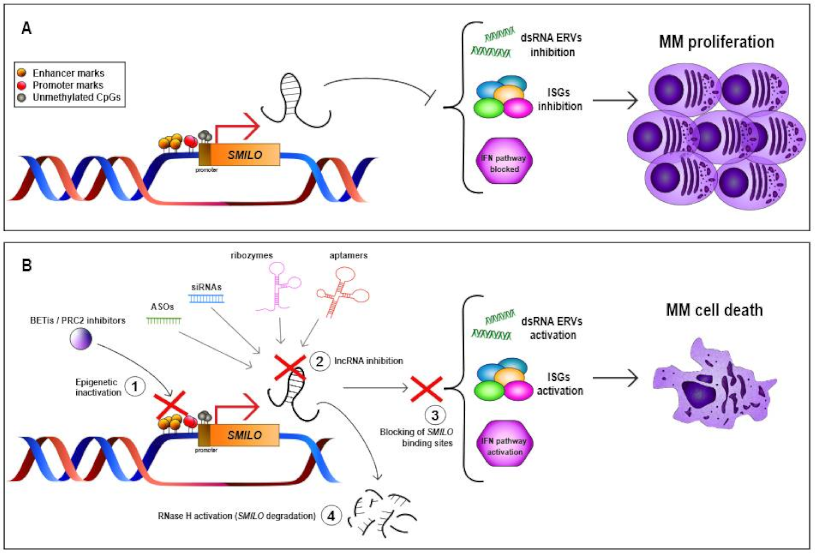
| SMILO | 1q42.2 | LincRNA | Up | NA | Regulates IFN pathway | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with better OS | [13] |
| SNHG16 | 17q25.1 | Processed transcript | Up | Decoy | Binds to miR-342-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | NA | [90] |
| SOX2OT | 3q26.3 | Sense overlapping | Up | Decoy | Binds to miR-144-3p | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest. Decreases tumor growth in vivo | NA | NA | [91] |
| ST3GAL6-AS1 | 3q12.1 | Antisense | Up | NA | NA | Decreases cellular proliferation, increases apoptosis and triggers cell cycle arrest | NA | High expression levels associated with worse PFS | [92,93] |
| TUG1 | 22q12.2 | Antisense | Up | Decoy | Binds to miR-29b-3p and targets HDAC4 | Decreases cellular proliferation and increases apoptosis | NA | NA | [43,94] |
| UCA1 | 19p13.12 | Processed transcript | Up | Decoy | Binds to miR-1271-5p and miR-331-3p | Decreases cellular proliferation and increases apoptosis | NA | High expression levels associated with worse OS | [95,96] |
| XLOC_013703 | 20p11.21 | NA | Down | NA | Involved in NF-κB signaling activation | NA | Decreases cellular proliferation and increases apoptosis | NA | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrasco-León, A.; Amundarain, A.; Gómez-Echarte, N.; Prósper, F.; Agirre, X. The Role of lncRNAs in the Pathobiology and Clinical Behavior of Multiple Myeloma. Cancers 2021, 13, 1976. https://doi.org/10.3390/cancers13081976
Carrasco-León A, Amundarain A, Gómez-Echarte N, Prósper F, Agirre X. The Role of lncRNAs in the Pathobiology and Clinical Behavior of Multiple Myeloma. Cancers. 2021; 13(8):1976. https://doi.org/10.3390/cancers13081976
Chicago/Turabian StyleCarrasco-León, Arantxa, Ane Amundarain, Nahia Gómez-Echarte, Felipe Prósper, and Xabier Agirre. 2021. "The Role of lncRNAs in the Pathobiology and Clinical Behavior of Multiple Myeloma" Cancers 13, no. 8: 1976. https://doi.org/10.3390/cancers13081976
APA StyleCarrasco-León, A., Amundarain, A., Gómez-Echarte, N., Prósper, F., & Agirre, X. (2021). The Role of lncRNAs in the Pathobiology and Clinical Behavior of Multiple Myeloma. Cancers, 13(8), 1976. https://doi.org/10.3390/cancers13081976






