Dual Switch in Lipid Metabolism in Cervical Epithelial Cells during Dysplasia Development Observed Using Raman Microscopy and Molecular Methods
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
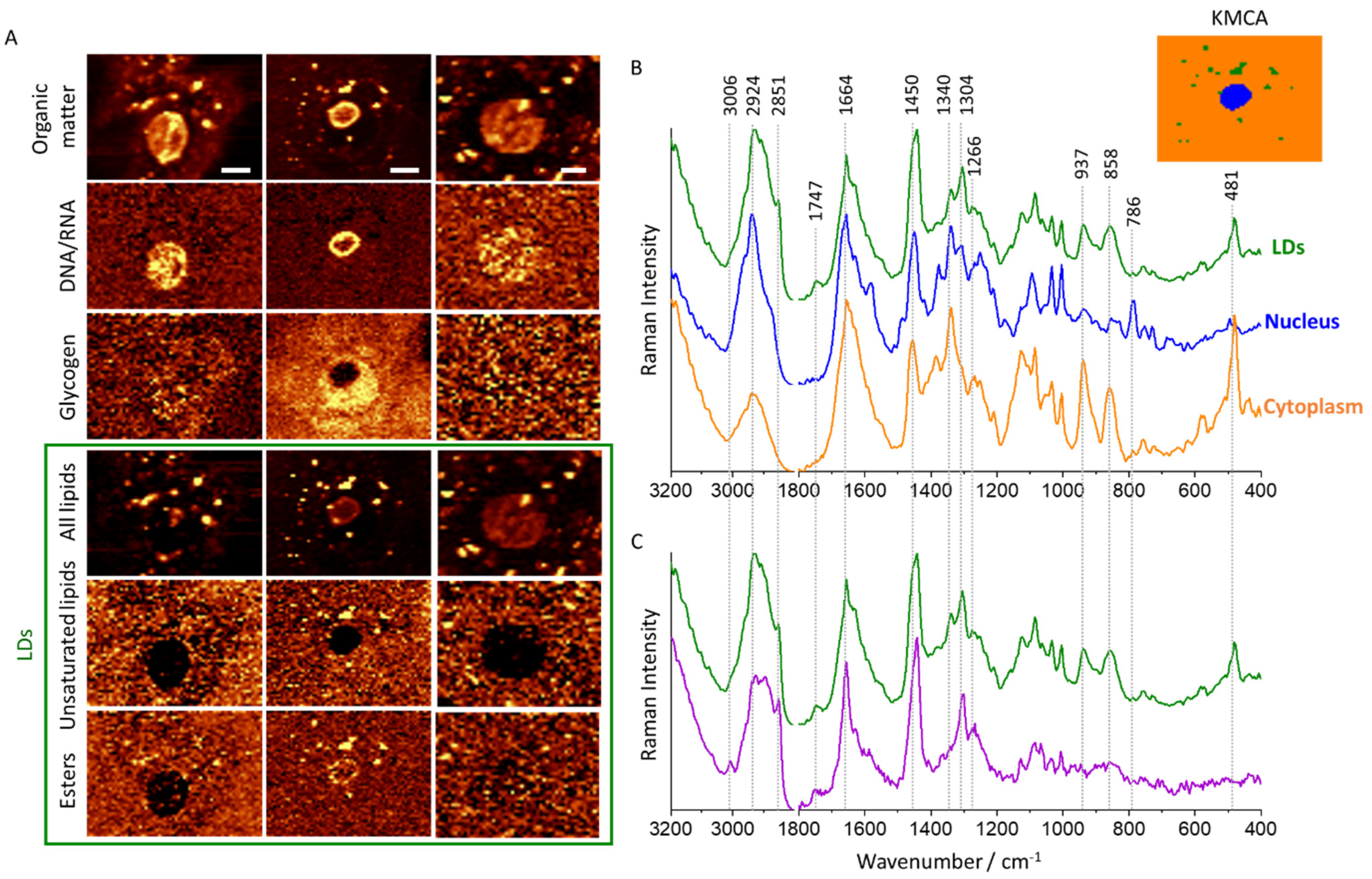
2.2. Raman Microscopy
2.3. HPV Testing
2.4. Counting Leukocytes in Samples
2.5. Determination of mtDNA Copy Number
2.6. Study of CpG Islands Methylation in SREBF1 Gene
2.7. Statistical Analysis
3. Results
3.1. Subcellular Distribution of Lipids in Cervical Epithelial Cells
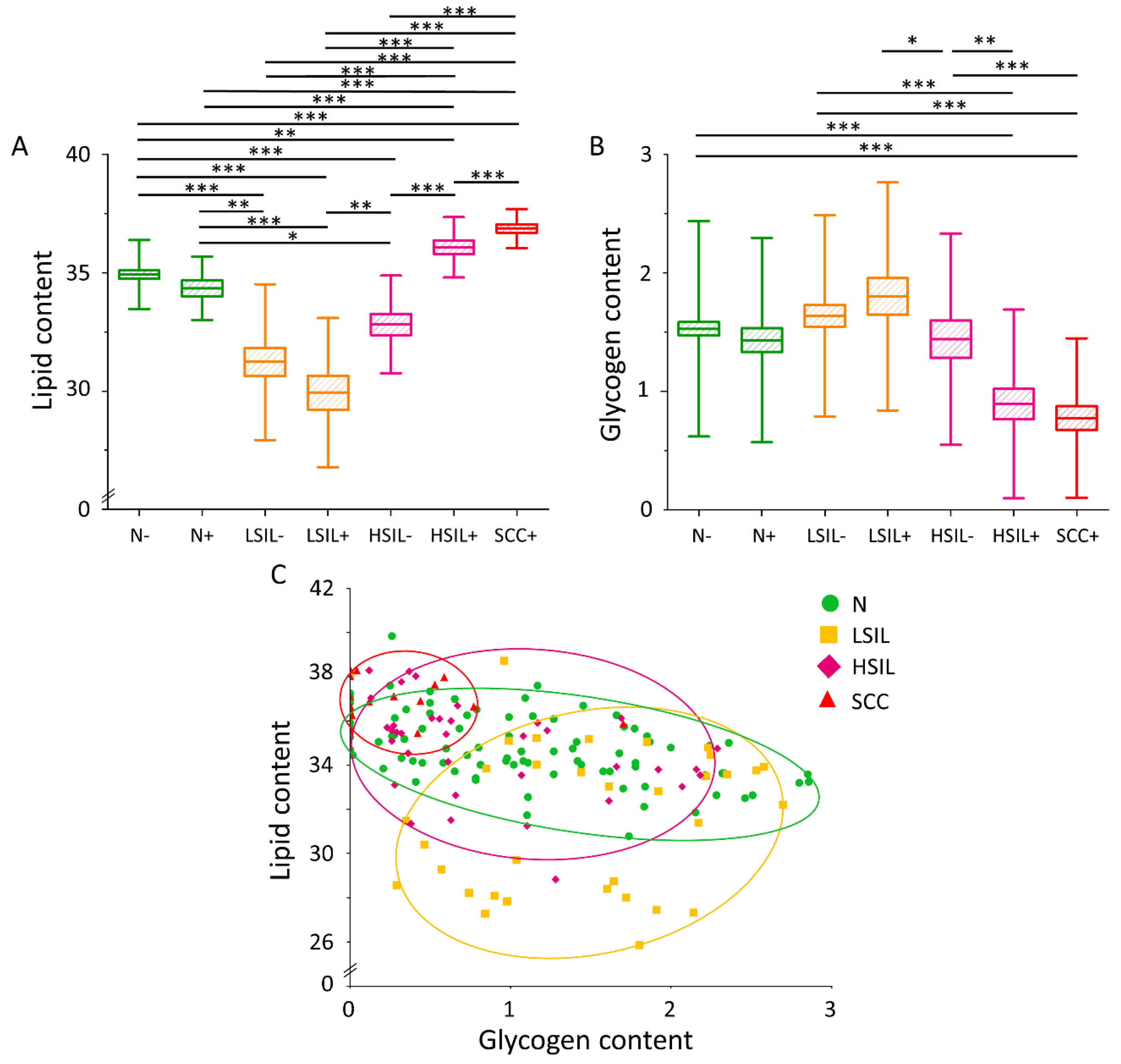
3.2. The Lipid Level Is Significantly Elevated in the Cytoplasm of Cervical Epithelial Cells in the HSIL+ and SCC+ Groups
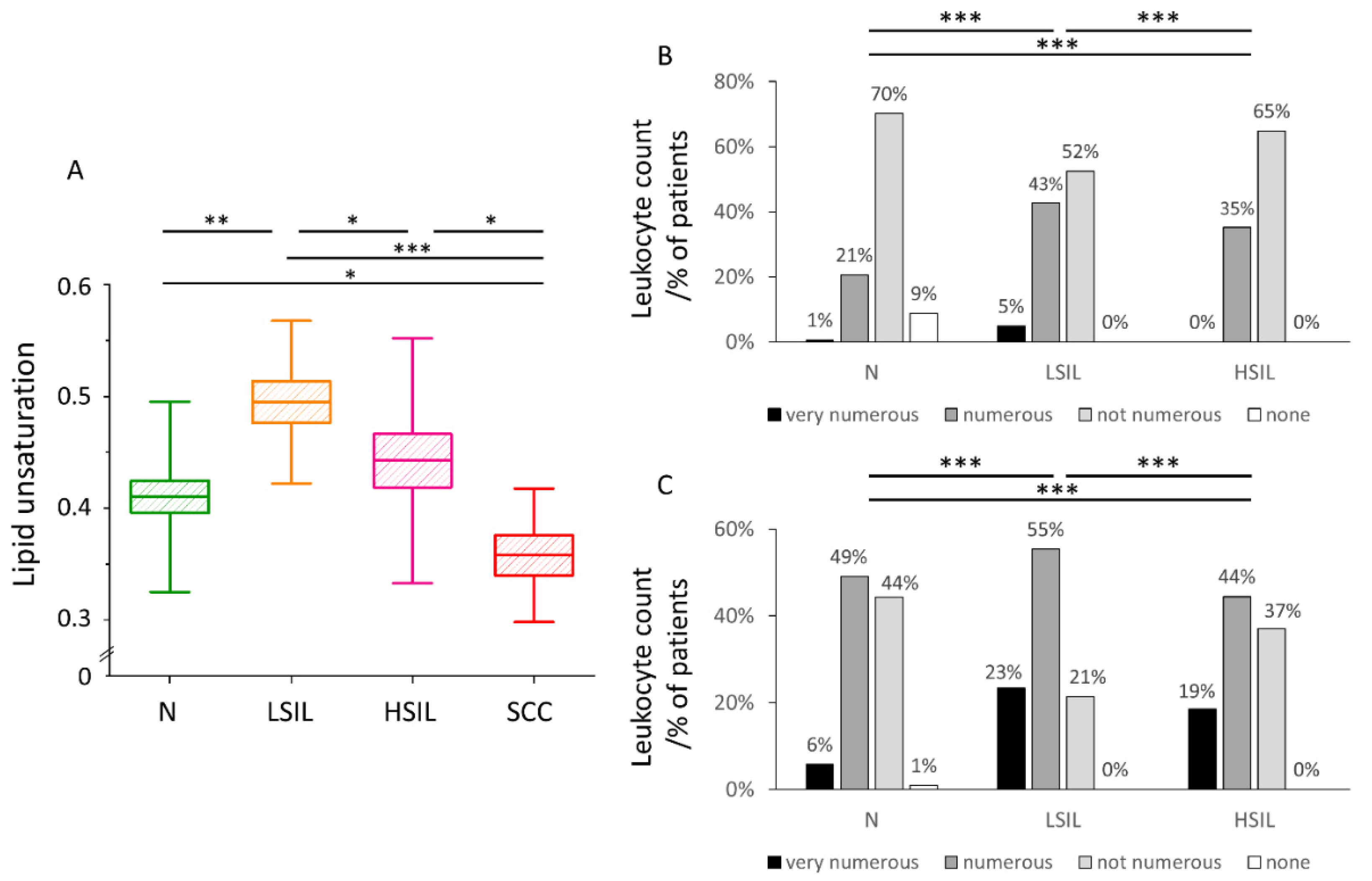
3.3. The Level of Lipid Unsaturation in Cervical Epithelial Cells Correlates with the Leucocyte Levels Confirming Increased Inflammation in the LSIL Group
3.4. The Level of CpG Island Methylation of the SREBF1 Gene Is Statistically the Highest in LSIL Group and the Lowest in SCC Group
3.5. The Level of Mitochondrial DNA Copies Is the Highest for the HSIL+ and SCC+ Groups
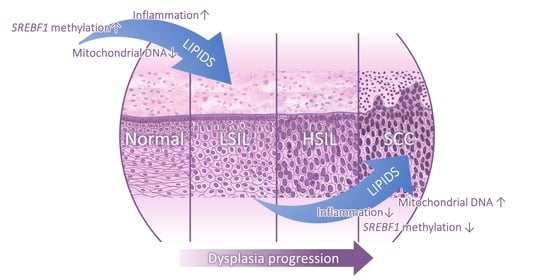
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | acetyl-CoA carboxylase |
| ACSS3 | acetyl-CoA synthetase 3 |
| Akt | protein kinase B |
| AMPK | AMP-activated protein kinase |
| BrdU | bromodeoxyuridine |
| FASN | fatty acid synthase |
| HPV | human papillomavirus |
| HPVhr | high-risk HPV |
| HSIL | high-grade squamous epithelial lesion |
| LDs | lipid droplets |
| LSIL | low-grade squamous epithelial lesion |
| MCT1 | monocarboxylate transporter 1 |
| MCT4 | monocarboxylate transporter 4 |
| mtDNA | mitochondrial DNA |
| mTOR | mammalian target of rapamycin kinase |
| mTORC1 | mammalian target of rapamycin complex 1 |
| MUFAs | monounsaturated fatty acids |
| NADPH | reduced form of nicotinamide adenine dinucleotide phosphate |
| PCR | polymerase chain reaction |
| PDHC | pyruvate dehydrogenase complex |
| PDK | pyruvate dehydrogenase kinase |
| PI3K | phosphatidylinositol 3-phosphate kinase |
| PUFAs | polyunsaturated fatty acids |
| qPCR | quantitative real-time polymerase reaction |
| SCC | squamous cell carcinoma |
| SCD1 | stearoyl-CoA desaturase |
| SFAs | saturated fatty acids |
| SREBF1 | sterol regulatory element-binding factor 1 |
| SREBP | sterol regulatory element-binding proteins |
| TCA | tricarboxylic acid |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [Green Version]
- Alexander, K.A.; Giuliano, A.R. HPV—beyond cervical cancer (online resource center). Am. J. Med. 2012, 125, S1. [Google Scholar] [CrossRef]
- Cline, B.J.; Simpson, M.C.; Gropler, M.; Bukatko, A.R.; Boakye, E.A.; Mohammed, K.A.; Osazuwa-Peters, N. Change in age at diagnosis of oropharyngeal cancer in the united states, 1975–2016. Cancers 2020, 12, 3191. [Google Scholar] [CrossRef]
- Bertoli, H.K.; Thomsen, L.T.; Iftner, T.; Dehlendorff, C.; Kjær, S.K. Risk of vulvar, vaginal and anal high-grade intraepithelial neoplasia and cancer according to cervical human papillomavirus (HPV) status: A population-based prospective cohort study. Gynecol. Oncol. 2020, 157, 456–462. [Google Scholar] [CrossRef]
- Faridi, R.; Zahra, A.; Khan, K.; Idrees, M. Oncogenic potential of human papillomavirus (HPV) and its relation with cervical cancer. Virol. J. 2011, 8, 269. [Google Scholar] [CrossRef] [Green Version]
- Kocjan, B.J.; Bzhalava, D.; Forslund, O.; Dillner, J.; Poljak, M. Molecular methods for identification and characterization of novel papillomaviruses. Clin. Microbiol. Infect. 2015, 21, 808–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef] [Green Version]
- Shanmugasundaram, S.; You, J. Targeting persistent human papillomavirus infection. Viruses 2017, 9, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, A.A.; Warburton, A. The role of integration in oncogenic progression of HPV—associated cancers. PLoS Pathog. 2017, 13, e1006211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petry, K.U.; Wörmann, B.; Schneider, A. Benefits and risks of cervical cancer screening. Oncol. Res. Treat. 2014, 37, 48–57. [Google Scholar] [CrossRef]
- European Medicines. HPV Vaccines: EMA Confirms Evidence Does Not Support That They Cause CRPS or Pots Reports after HPV Vaccination Consistent with What Would Be Expected in This; European Medicines Agency: London, UK, 2016. [Google Scholar]
- Lowy, D.R.; Solomon, D.; Hildesheim, A.; Schiller, J.T.; Schiffman, M. HPV infection and the primary and secondary prevention of cervical cancer. Cancer 2008, 113, 1980–1993. [Google Scholar] [CrossRef]
- Luo, X.; Zhao, X.; Cheng, C.; Li, N.; Liu, Y.; Cao, Y. The implications of signaling lipids in cancer metastasis. Exp. Mol. Med. 2018, 50, 127. [Google Scholar] [CrossRef] [Green Version]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beg, M.; Abdullah, N.; Thowfeik, F.S.; Altorki, N.K.; McGraw, T.E. Distinct Akt phosphorylation states are required for insulin regulated Glut4 and Glut1-mediated glucose uptake. eLife 2017, 6, e26896. [Google Scholar] [CrossRef]
- Krycer, J.R.; Sharpe, L.J.; Luu, W.; Brown, A.J. The Akt-SREBP nexus: Cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 2010, 21, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Espenshade, P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012, 16, 414–419. [Google Scholar] [CrossRef] [Green Version]
- Mashima, T.; Seimiya, H.; Tsuruo, T. De Novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br. J. Cancer 2009, 100, 1369–1372. [Google Scholar] [CrossRef] [Green Version]
- Livieri, A.L.; Navone, L.; Marcellin, E.; Gramajo, H.; Rodriguez, E. A Novel multidomain Acyl-CoA carboxylase in saccharopolyspora erythraea provides malonyl-CoA for de novo fatty acid biosynthesis. Sci. Rep. 2019, 9, 6725. [Google Scholar] [CrossRef]
- Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N. Lipid Metabolism in Cancer Cells under Metabolic Stress. Br. J. Cancer 2019, 120, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Young, R.M.; Ackerman, D.; Quinn, Z.L.; Mancuso, A.; Gruber, M.; Liu, L.; Giannoukos, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Keith, B.; et al. Dysregulated MTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013, 27, 1115–1131. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Condello, S.; Thomes-Pepin, J.; Ma, X.; Xia, Y.; Hurley, T.D.; Matei, D.; Cheng, J.X. Lipid Desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell 2017, 20, 303–314.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igal, R.A. Stearoyl-coa desaturase-1: A novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis 2010, 31, 1509–1515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Cui, S.; Ke, K.; Zhao, B.; Liu, X.; Yue, S.; Wang, P. Hyperspectral stimulated Raman scattering microscopy unravels aberrant accumulation of saturated fat in human liver cancer. Anal. Chem. 2018, 90, 6362–6366. [Google Scholar] [CrossRef]
- Wang, Y.P.; Lei, Q.Y. Metabolic recoding of epigenetics in cancer. Cancer Commun. 2018, 38, 25. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Herman, J.G.; Baylin, S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef]
- Zong, W.X.; Rabinowitz, J.D.; White, E. Mitochondria and cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, S.; Birkaya, B.; Rideout, T.C.; Patel, M.S. Lack of Mitochondria-generated acetyl-CoA by pyruvate dehydrogenase complex downregulates gene expression in the hepatic de novo lipogenic pathway. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E117–E127. [Google Scholar] [CrossRef] [Green Version]
- Vishwanath, V.A. Fatty acid beta-oxidation disorders: A brief review. Ann. Neurosci. 2016, 23, 51–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browsney, R.W.; Boone, A.N.; Elliott, J.E.; Kulpa, J.E.; Lee, W.M. Regulation of Acetyl-CoA carboxylase. Biochem. Soc. Trans. 2006, 34, 223–226. [Google Scholar] [CrossRef]
- Holness, M.J.; Sugden, M.C. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 2003, 31, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Prag, H.A.; Murphy, M.P. mtDNA mutations help support cancer cells. Nat. Cancer 2020, 1, 941–942. [Google Scholar] [CrossRef]
- Warowicka, A.; Kwasniewska, A.; Gozdzicka-Jozefiak, A. Alterations in mtDNA: A qualitative and quantitative study associated with cervical cancer development. Gynecol. Oncol. 2013, 129, 193–198. [Google Scholar] [CrossRef]
- Klein, K.; Gigler, A.M.; Aschenbrenner, T.; Monetti, R.; Bunk, W.; Jamitzky, F.; Morfill, G.; Stark, R.W.; Schlegel, J. Label-free live-cell imaging with confocal Raman microscopy. Biophys. J. 2012, 102, 360–368. [Google Scholar] [CrossRef] [Green Version]
- Sitarz, K.; Czamara, K.; Bialecka, J.; Klimek, M.; Zawilinska, B.; Szostek, S.; Kaczor, A. HPV infection significantly accelerates glycogen metabolism in cervical cells with large nuclei: Raman microscopic study with subcellular resolution. Int. J. Mol. Sci. 2020, 21, 2667. [Google Scholar] [CrossRef]
- Lyng, F.M.; Traynor, D.; Ramos, I.R.M.; Bonnier, F.; Byrne, H.J. Raman spectroscopy for screening and diagnosis of cervical cancer. Anal. Bioanal. Chem. 2015, 407, 8279–8289. [Google Scholar] [CrossRef] [Green Version]
- Duraipandian, S.; Traynor, D.; Kearney, P.; Martin, C.; O’Leary, J.J.; Lyng, F.M. Raman spectroscopic detection of high-grade cervical cytology: Using morphologically normal appearing cells. Sci. Rep. 2018, 8, 15048. [Google Scholar] [CrossRef] [Green Version]
- Bonnier, F.; Traynor, D.; Kearney, P.; Clarke, C.; Knief, P.; Martin, C.; O’Leary, J.J.; Byrne, H.J.; Lyng, F. Processing ThinPrep cervical cytological samples for Raman spectroscopic analysis. Anal. Methods 2014, 6, 7831–7841. [Google Scholar] [CrossRef]
- Wood, B.R.; Quinn, M.A.; Burden, F.R.; McNaughton, D. An investigation into FTIR spectroscopy as a biodiagnostic tool for cervical cancer. Biospectroscopy 1996, 2, 143–153. [Google Scholar] [CrossRef]
- Nayar, R.; Wilbur, D.C. The Pap Test and Bethesda 2014. Acta Cytol. 2015, 59, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Guzik, K.; Bzowska, M.; Dobrucki, J.; Pryjma, J. Heat-shocked monocytes are resistant to Staphylococcus aureus-induced apoptotic DNA fragmentation due to expression of HSP72. Infect. Immun. 1999, 67, 4216–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, H.; Le, F.; Zheng, Y.; Li, L.; Wang, L.; Wang, N.; Zhu, Y.; Huang, H.; Jin, F. Assisted reproductive technologies impair the expression and methylation of insulin-induced gene 1 and sterol regulatory element-binding factor 1 in the fetus and placenta. Fertil. Steril. 2014, 101, 974–980.e2. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Selmi, A.; Baranska, M.; Ozaki, Y.; Kaczor, A. Unsaturated lipid bodies as a hallmark of inflammation studied by Raman 2D and 3D microscopy. Sci. Rep. 2017, 7, 40889. [Google Scholar] [CrossRef]
- Fritsche, K.L. The science of fatty acids and inflammation. Adv. Nutr. 2015, 6, 293S–301S. [Google Scholar] [CrossRef]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. DMM Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Carroll, R.G.; Zasłona, Z.; Galván-Peña, S.; Koppe, E.L.; Sévin, D.C.; Angiari, S.; Triantafilou, M.; Triantafilou, K.; Modis, L.K.; O’Neill, L.A. An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J. Biol. Chem. 2018, 293, 5509–5521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras-Paredes, A.; De la Cruz-Hernández, E.; Martínez-Ramírez, I.; Dueñas-González, A.; Lizano, M. E6 Variants of human papillomavirus 18 differentially modulate the protein kinase B/phosphatidylinositol 3-Kinase (Akt/PI3K) signaling pathway. Virology 2009, 383, 78–85. [Google Scholar] [CrossRef] [Green Version]
- Menges, C.W.; Baglia, L.A.; Lapoint, R.; McCance, D.J. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 2006, 66, 5555–5559. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Curtis, M.; Kenny, H.A.; Ashcroft, B.; Mukherjee, A.; Johnson, A.; Zhang, Y.; Helou, Y.; Batlle, R.; Liu, X.; Gutierrez, N.; et al. Fibroblasts mobilize tumor cell glycogen to promote proliferation and metastasis. Cell Metab. 2019, 29, 141–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.; Jeong, S.; Jang, C.; Bae, H.; Kim, Y.H.; Park, I.; Kim, S.K.; Koh, G.Y. Tumor metastasis to lymph nodes requires YAP-dependent metabolic adaptation. Science 2019, 363, 644–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci. Rep. 2014, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, W.M.; Yu, S.; Venglarik, M.; Schwebke, J.R. Vaginal leucocyte counts in women with bacterial vaginosis: Relation to vagina and cervical infections. Sex. Transm. Infect. 2004, 80, 401–405. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.A.; Anderson, D.J. Human vaginal leukocytes and the effects of vaginal fluid on lymphocyte and macrophage defense functions. Am. J. Obstet. Gynecol. 1992, 166, 720–726. [Google Scholar] [CrossRef]
- Sekiya, M.; Yahagi, N.; Matsuzaka, T.; Takeuchi, Y.; Nakagawa, Y.; Takahashi, H.; Okazaki, H.; Iizuka, Y.; Ohashi, K.; Gotoda, T.; et al. SREBP-1-independent regulation of lipogenic gene expression in adipocytes. J. Lipid Res. 2007, 48, 1581–1591. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Bullón-Vela, M.V.; Abete, I.; Martínez, J.A.; Zulet, M.A. Obesity and nonalcoholic fatty liver disease: Role of oxidative stress. Obes. Oxidative Stress Diet. Antioxid. 2018, 111–133. [Google Scholar] [CrossRef]
- Sun, Y.; He, W.; Luo, M.; Zhou, Y.; Chang, G.; Ren, W.; Wu, K.; Li, X.; Shen, J.; Zhao, X.; et al. SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumor Biol. 2015, 36, 4133–4141. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.A.; Xiong, X.; Zaytseva, Y.Y.; Napier, D.L.; Vallee, E.; Li, A.T.; Wang, C.; Weiss, H.L.; Evers, B.M.; Gao, T. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer article. Cell Death Dis. 2018, 9, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, J.; Zhu, L.; Zhu, Q.; Su, J.; Liu, M.; Huang, W. SREBP-1 is an independent prognostic marker and promotes invasion and migration in breast cancer. Oncol. Lett. 2016, 12, 2409–2416. [Google Scholar] [CrossRef] [Green Version]
- Walther, T.C.; Farese, R.V. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012, 81, 687–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Qin, X.; Zhou, J.; Xu, M.; Lyu, Z.; Li, X.; Zhang, K.; Dai, M.; Li, N.; Hang, D. Mitochondrial DNA copy number in cervical exfoliated cells and risk of cervical cancer among HPV-positive women. BMC Womens. Health 2020, 20, 139. [Google Scholar] [CrossRef]
- Ye, B.; Yin, L.; Wang, Q.; Cunshuan, X.U. ACC1 Is overexpressed in liver cancers and contributes to the proliferation of human hepatoma Hep G2 cells and the rat liver cell line BRL 3A. Mol. Med. Rep. 2019, 49, 3431–3440. [Google Scholar] [CrossRef] [Green Version]
- Chajès, V.; Cambot, M.; Moreau, K.; Lenoir, G.M.; Joulin, V. Acetyl-CoA carboxylase α is essential to breast cancer cell survival. Cancer Res. 2006, 66, 5287–5294. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Rajput, S.; Watabe, K.; Liao, D.F.; Cao, D. Acetyl-CoA carboxylase-α as a novel target for cancer therapy. Front. Biosci. Sch. 2010, S2, 515–526. [Google Scholar] [CrossRef]
- Ferriero, R.; Brunetti-Pierri, N. Phenylbutyrate increases activity of pyruvate dehydrogenase complex. Oncotarget 2013, 4, 804–805. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Duan, H.; Feng, Z.; Han, X.; Gu, C. Acetyl-CoA synthetase 3 promotes bladder cancer cell growth under metabolic stress. Oncogenesis 2020, 9. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitarz, K.; Czamara, K.; Bialecka, J.; Klimek, M.; Szostek, S.; Kaczor, A. Dual Switch in Lipid Metabolism in Cervical Epithelial Cells during Dysplasia Development Observed Using Raman Microscopy and Molecular Methods. Cancers 2021, 13, 1997. https://doi.org/10.3390/cancers13091997
Sitarz K, Czamara K, Bialecka J, Klimek M, Szostek S, Kaczor A. Dual Switch in Lipid Metabolism in Cervical Epithelial Cells during Dysplasia Development Observed Using Raman Microscopy and Molecular Methods. Cancers. 2021; 13(9):1997. https://doi.org/10.3390/cancers13091997
Chicago/Turabian StyleSitarz, Katarzyna, Krzysztof Czamara, Joanna Bialecka, Malgorzata Klimek, Slawa Szostek, and Agnieszka Kaczor. 2021. "Dual Switch in Lipid Metabolism in Cervical Epithelial Cells during Dysplasia Development Observed Using Raman Microscopy and Molecular Methods" Cancers 13, no. 9: 1997. https://doi.org/10.3390/cancers13091997