CD81 Enhances Radioresistance of Glioblastoma by Promoting Nuclear Translocation of Rad51
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
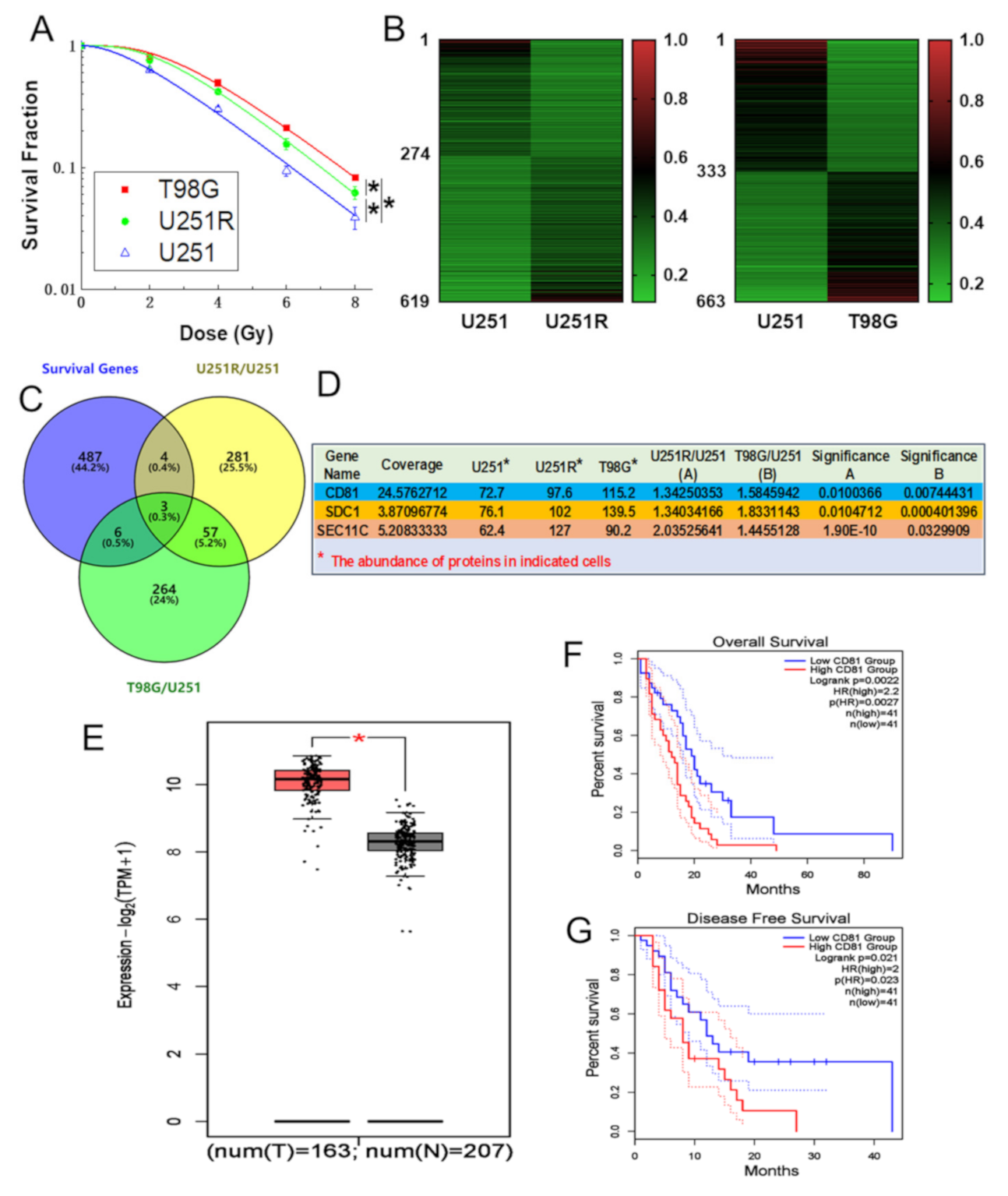
2.1. CD81 Had Close Relevance with GBM in Clinic
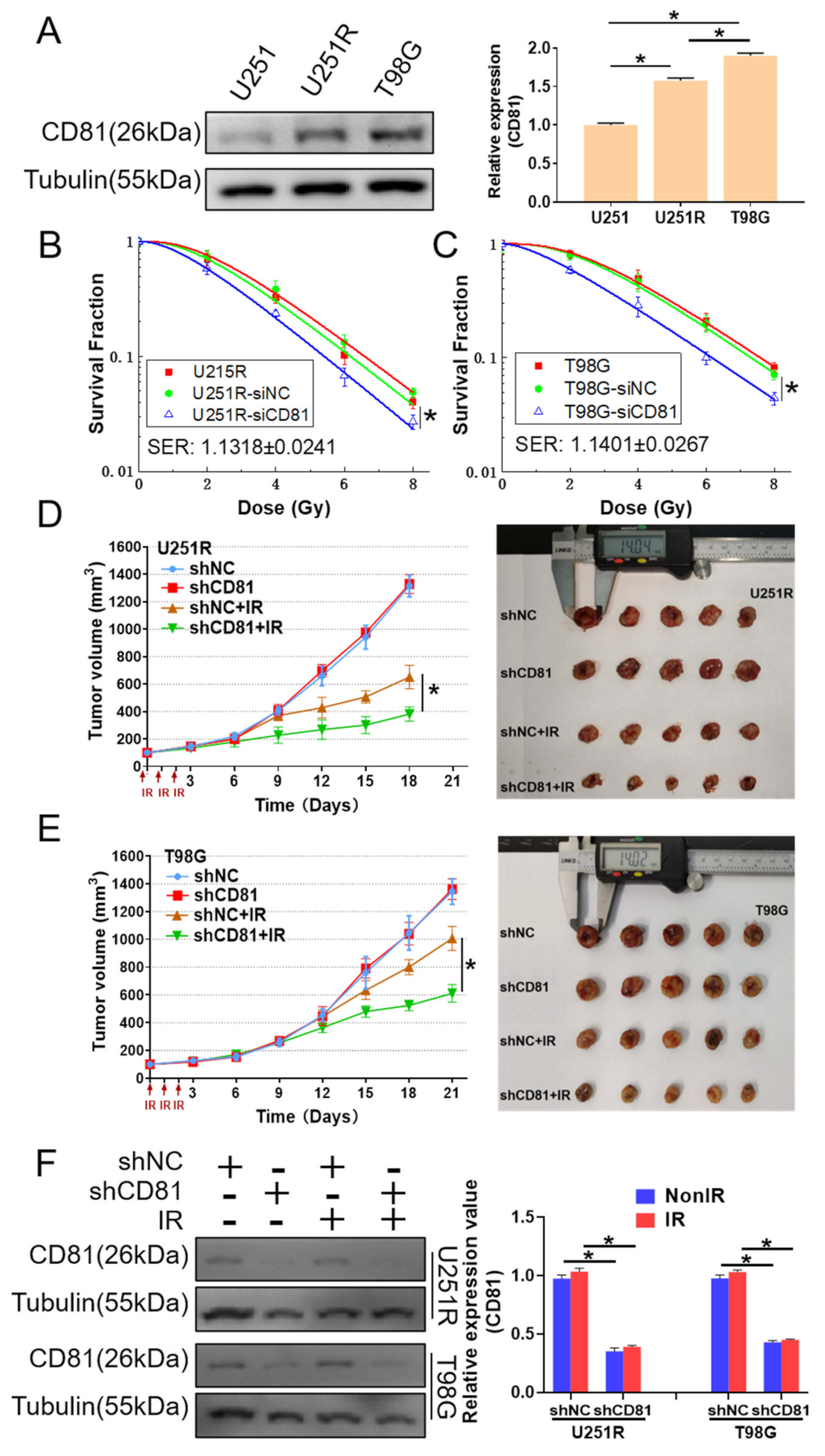
2.2. Inhibition of CD81 Enhanced Radiosensitivity of GBM Cells In Vitro and In Vivo
2.3. Downregulation of CD81 Increased DNA Damage and Decreased DNA Repair
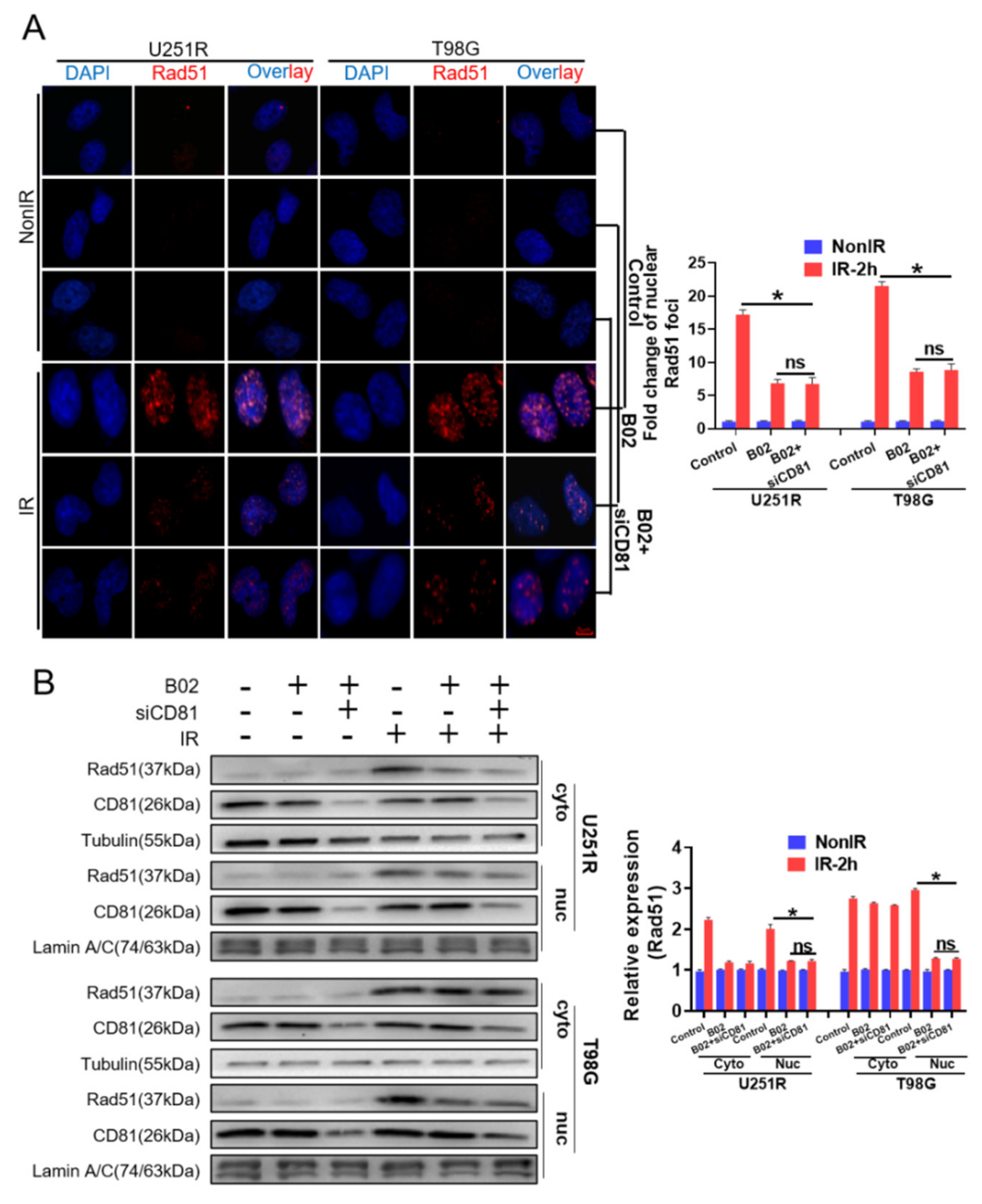
2.4. Suppress of CD81 Reduced Nuclear Translocation of Rad51
2.5. Rad51 Was Indispensable for CD81-Regulated Radioresistance
2.6. Nuclear Membrane CD81 Promoted Nuclear Transport of Rad51
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Irradiation
4.2. Tandem Mass Tag (TMT) Quantitative Proteomic Analysis
4.3. Clonogenic Assay
4.4. Transient Transfection of siRNA
4.5. Stable Transfection of shRNA
4.6. Subcellular Protein Extraction
4.7. Western Blotting Assay
4.8. In Vivo Tumor Growth and Irradiation
4.9. Immunofluorescence Staining of BrdU, γ-H2AX and Rad51 Foci
4.10. Dual Immunofluorescence Assay of CD81 and Rad51
4.11. Flow Cytometry Assay of Cell Cycle
4.12. Rad51 Inhibitor Treatment
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, R.; Smith-Cohn, M.; Cohen, A.L.; Colman, H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics 2017, 14, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Vredenburgh, J.J.; Desjardins, A.; Herndon, J.N.; Dowell, J.M.; Reardon, D.A.; Quinn, J.A.; Rich, J.N.; Sathornsumetee, S.; Gururangan, S.; Wagner, M.; et al. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin. Cancer Res. 2007, 13, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.C.; Veeravagu, A.; Hsu, A.R.; Tse, V.C. Recurrent glioblastoma multiforme: A review of natural history and management options. Neurosurg. Focus 2006, 20, E5. [Google Scholar] [CrossRef]
- Nguyen, H.S.; Shabani, S.; Awad, A.J.; Kaushal, M.; Doan, N. Molecular Markers of Therapy-Resistant Glioblastoma and Potential Strategy to Combat Resistance. Int. J. Mol. Sci. 2018, 19, 1765. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Xu, R.; Ji, J.; Xu, Y.; Han, M.; Wei, Y.; Huang, B.; Chen, A.; Zhang, Q.; et al. YM155 decreases radiation-induced invasion and reverses epithelial-mesenchymal transition by targeting STAT3 in glioblastoma. J. Transl. Med. 2018, 16, 79. [Google Scholar] [CrossRef]
- Palumbo, S.; Tini, P.; Toscano, M.; Allavena, G.; Angeletti, F.; Manai, F.; Miracco, C.; Comincini, S.; Pirtoli, L. Combined EGFR and autophagy modulation impairs cell migration and enhances radiosensitivity in human glioblastoma cells. J. Cell. Physiol. 2014, 229, 1863–1873. [Google Scholar] [CrossRef]
- Bruening, J.; Lasswitz, L.; Banse, P.; Kahl, S.; Marinach, C.; Vondran, F.W.; Kaderali, L.; Silvie, O.; Pietschmann, T.; Meissner, F.; et al. Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLOS Pathog. 2018, 14, e1007111. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef]
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 2005, 5, 726–734. [Google Scholar] [CrossRef]
- Yuan, X.; Du, J.; Hua, S.; Zhang, H.; Gu, C.; Wang, J.; Yang, L.; Huang, J.; Yu, J.; Liu, F. Suppression of autophagy augments the radiosensitizing effects of STAT3 inhibition on human glioma cells. Exp. Cell Res. 2015, 330, 267–276. [Google Scholar] [CrossRef]
- Ye, H.; Chen, M.; Cao, F.; Huang, H.; Zhan, R.; Zheng, X. Chloroquine, an autophagy inhibitor, potentiates the radiosensitivity of glioma initiating cells by inhibiting autophagy and activating apoptosis. BMC Neurol. 2016, 16, 178. [Google Scholar] [CrossRef]
- Shi, F.; Guo, H.; Zhang, R.; Liu, H.; Wu, L.; Wu, Q.; Liu, J.; Liu, T.; Zhang, Q. The PI3K inhibitor GDC-0941 enhances radiosensitization and reduces chemoresistance to temozolomide in GBM cell lines. Neuroscience 2017, 346, 298–308. [Google Scholar] [CrossRef]
- Kim, B.M.; Hong, Y.; Lee, S.; Liu, P.; Lim, J.H.; Lee, Y.H.; Lee, T.H.; Chang, K.T.; Hong, Y. Therapeutic Implications for Overcoming Radiation Resistance in Cancer Therapy. Int. J. Mol. Sci. 2015, 16, 26880–26913. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef]
- Chartron, E.; Theillet, C.; Guiu, S.; Jacot, W. Targeting homologous repair deficiency in breast and ovarian cancers: Biological pathways, preclinical and clinical data. Crit. Rev. Oncol. Hematol. 2019, 133, 58–73. [Google Scholar] [CrossRef]
- Zheng, W.; Chen, Q.; Wang, C.; Yao, D.; Zhu, L.; Pan, Y.; Zhang, J.; Bai, Y.; Shao, C. Inhibition of Cathepsin D (CTSD) enhances radiosensitivity of glioblastoma cells by attenuating autophagy. Mol. Carcinog. 2020, 59, 651–660. [Google Scholar] [CrossRef]
- Schlegel, B.P.; Jodelka, F.M.; Nunez, R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006, 66, 5181–5189. [Google Scholar] [CrossRef]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of hepatitis C virus to CD81. Science 1998, 282, 938–941. [Google Scholar] [CrossRef]
- Paiva, B.; Puig, N.; Cedena, M.T.; de Jong, B.G.; Ruiz, Y.; Rapado, I.; Martinez-Lopez, J.; Cordon, L.; Alignani, D.; Delgado, J.A.; et al. Differentiation stage of myeloma plasma cells: Biological and clinical significance. Leukemia 2017, 31, 382–392. [Google Scholar] [CrossRef]
- Uretmen, K.Z.; Sanal, E.; Karayel, O.; Polat, A.N.; Saatci, O.; Ersan, P.G.; Trappe, K.; Renard, B.Y.; Onder, T.T.; Tuncbag, N.; et al. Systems-level Analysis Reveals Multiple Modulators of Epithelial-mesenchymal Transition and Identifies DNAJB4 and CD81 as Novel Metastasis Inducers in Breast Cancer. Mol. Cell. Proteom. 2019, 18, 1756–1771. [Google Scholar] [CrossRef]
- Vences-Catalan, F.; Rajapaksa, R.; Srivastava, M.K.; Marabelle, A.; Kuo, C.C.; Levy, R.; Levy, S. Tetraspanin CD81 promotes tumor growth and metastasis by modulating the functions of T regulatory and myeloid-derived suppressor cells. Cancer Res. 2015, 75, 4517–4526. [Google Scholar] [CrossRef]
- Hazan, I.; Monin, J.; Bouwman, B.; Crosetto, N.; Aqeilan, R.I. Activation of Oncogenic Super-Enhancers Is Coupled with DNA Repair by RAD51. Cell Rep. 2019, 29, 560–572. [Google Scholar] [CrossRef]
- Haaf, T.; Golub, E.I.; Reddy, G.; Radding, C.M.; Ward, D.C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 1995, 92, 2298–2302. [Google Scholar] [CrossRef]
- Prakash, R.; Zhang, Y.; Feng, W.; Jasin, M. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015, 7, a16600. [Google Scholar] [CrossRef]
- Roberti, M.; Schipani, F.; Bagnolini, G.; Milano, D.; Giacomini, E.; Falchi, F.; Balboni, A.; Manerba, M.; Farabegoli, F.; De Franco, F.; et al. Rad51/BRCA2 disruptors inhibit homologous recombination and synergize with olaparib in pancreatic cancer cells. Eur. J. Med. Chem. 2019, 165, 80–92. [Google Scholar] [CrossRef]
- Callen, E.; Zong, D.; Wu, W.; Wong, N.; Stanlie, A.; Ishikawa, M.; Pavani, R.; Dumitrache, L.C.; Byrum, A.K.; Mendez-Dorantes, C.; et al. 53BP1 Enforces Distinct Pre- and Post-resection Blocks on Homologous Recombination. Mol. Cell 2020, 77, 26–38. [Google Scholar] [CrossRef]
- Maranto, C.; Udhane, V.; Hoang, D.T.; Gu, L.; Alexeev, V.; Malas, K.; Cardenas, K.; Brody, J.R.; Rodeck, U.; Bergom, C.; et al. STAT5A/B Blockade Sensitizes Prostate Cancer to Radiation through Inhibition of RAD51 and DNA Repair. Clin. Cancer Res. 2018, 24, 1917–1931. [Google Scholar] [CrossRef]
- Pearl, L.H.; Schierz, A.C.; Ward, S.E.; Al-Lazikani, B.; Pearl, F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 2015, 15, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Jeggo, P.A.; Lobrich, M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem. J. 2015, 471, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; He, Y.; Hong, T.S.; Corcoran, R.B.; Clark, J.W.; Ryan, D.P.; Zou, L.; Ting, D.T.; Catenacci, D.V.; Chao, J.; et al. Analysis of DNA Damage Response Gene Alterations and Tumor Mutational Burden Across 17,486 Tubular Gastrointestinal Carcinomas: Implications for Therapy. Oncologist 2019, 24, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Wei, F.; Wu, Y.; He, Y.; Shi, L.; Xiong, F.; Gong, Z.; Guo, C.; Li, X.; Deng, H.; et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J. Exp. Clin. Cancer Res. 2018, 37, 87. [Google Scholar] [CrossRef] [PubMed]
- Vendrely, V.; Amintas, S.; Noel, C.; Moranvillier, I.; Lamrissi, I.; Rousseau, B.; Coulibaly, S.; Bedel, A.; Moreau-Gaudry, F.; Buscail, E.; et al. Combination treatment of resveratrol and capsaicin radiosensitizes pancreatic tumor cells by unbalancing DNA repair response to radiotherapy towards cell death. Cancer Lett. 2019, 451, 1–10. [Google Scholar] [CrossRef]
- Efimova, E.V.; Ricco, N.; Labay, E.; Mauceri, H.J.; Flor, A.C.; Ramamurthy, A.; Sutton, H.G.; Weichselbaum, R.R.; Kron, S.J. HMG-CoA Reductase Inhibition Delays DNA Repair and Promotes Senescence After Tumor Irradiation. Mol. Cancer Ther. 2018, 17, 407–418. [Google Scholar] [CrossRef]
- Sun, L.L.; Ellerman, D.; Mathieu, M.; Hristopoulos, M.; Chen, X.; Li, Y.; Yan, X.; Clark, R.; Reyes, A.; Stefanich, E.; et al. Anti-CD20/CD3 T cell-dependent bispecific antibody for the treatment of B cell malignancies. Sci. Transl. Med. 2015, 7, 270r–287r. [Google Scholar] [CrossRef]
- Witkowska, M.; Smolewski, P.; Robak, T. Investigational therapies targeting CD37 for the treatment of B-cell lymphoid malignancies. Expert Opin. Investig. Drugs 2018, 27, 171–177. [Google Scholar] [CrossRef]
- Hirayama, A.V.; Turtle, C.J. Toxicities of CD19 CAR-T cell immunotherapy. Am. J. Hematol. 2019, 94, S42–S49. [Google Scholar] [CrossRef]
- Vexler, V.; Yu, L.; Pamulapati, C.; Garrido, R.; Grimm, H.P.; Sriraman, P.; Bohini, S.; Schraeml, M.; Singh, U.; Brandt, M.; et al. Target-mediated drug disposition and prolonged liver accumulation of a novel humanized anti-CD81 monoclonal antibody in cynomolgus monkeys. Mabs 2013, 5, 776–786. [Google Scholar] [CrossRef]
- Vences-Catalan, F.; Kuo, C.C.; Rajapaksa, R.; Duault, C.; Andor, N.; Czerwinski, D.K.; Levy, R.; Levy, S. CD81 is a novel immunotherapeutic target for B cell lymphoma. J. Exp. Med. 2019, 216, 1497–1508. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Chen, Q.; Liu, H.; Hu, S.; Zhou, Y.; Bai, Y.; Zhang, J.; Pan, Y.; Shao, C. CD81 Enhances Radioresistance of Glioblastoma by Promoting Nuclear Translocation of Rad51. Cancers 2021, 13, 1998. https://doi.org/10.3390/cancers13091998
Zheng W, Chen Q, Liu H, Hu S, Zhou Y, Bai Y, Zhang J, Pan Y, Shao C. CD81 Enhances Radioresistance of Glioblastoma by Promoting Nuclear Translocation of Rad51. Cancers. 2021; 13(9):1998. https://doi.org/10.3390/cancers13091998
Chicago/Turabian StyleZheng, Wang, Qianping Chen, Hongxia Liu, Songling Hu, Yuchuan Zhou, Yang Bai, Jianghong Zhang, Yan Pan, and Chunlin Shao. 2021. "CD81 Enhances Radioresistance of Glioblastoma by Promoting Nuclear Translocation of Rad51" Cancers 13, no. 9: 1998. https://doi.org/10.3390/cancers13091998
APA StyleZheng, W., Chen, Q., Liu, H., Hu, S., Zhou, Y., Bai, Y., Zhang, J., Pan, Y., & Shao, C. (2021). CD81 Enhances Radioresistance of Glioblastoma by Promoting Nuclear Translocation of Rad51. Cancers, 13(9), 1998. https://doi.org/10.3390/cancers13091998







