The Tubulin Code and Tubulin-Modifying Enzymes in Autophagy and Cancer
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Microtubule Dynamic Instability
1.2. The α-/β-Tubulin Code
2. The Tubulin Code and Its Associated Enzymes in Cancer
| Tubulin Isotype | Alteration | Cancer Type | Effect | References |
|---|---|---|---|---|
| TubA1A | High levels | Gastric | Macrophage infiltration in tumor microenvironment | [50] |
| TubA1B | High level | Hepatocellular carcinoma | Poor overall survival and resistance to paclitaxel | [48] |
| TubA1C | High level | Glioma | Poor prognosis | [52] |
| High level | Lung | Immune cell infiltration | [51] | |
| TubB1 | High level | Breast | Taxane resistance | [54] |
| TubB2 | Depletion | Lung | Enhanced sensitivity to Vinca alkaloids | [58] |
| Low level | Ovarian and breast | Resistance to taxanes; correlated with advanced stages | [59,60] | |
| High level | Lung | Biomarker for tumor differentiation and aggressiveness | [61] | |
| Tub3 | High level | Ovarian | Correlated with advanced stages | [59] |
| High level | Clear cell renal cell carcinoma | Poor prognosis | [56] | |
| High level | Prostate | Poor overall survival | [65] | |
| High level | Urinary bladder cancer | Poor prognosis | [57] | |
| High level | Thyroid carcinoma | Invasive potential and poor prognosis | [62] | |
| TubB4A | High level | Lung | Resistance to paclitaxel | [63] |
| TubB4B | Depletion | Lung cancer cells | Enhanced sensitivity to Vinca alkaloids | [58] |
| Tub5 | High level | Lung | Biomarker for tumor differentiation and aggressiveness | [61] |
| High level | Lung | Treatment response to paclitaxel | [64] |
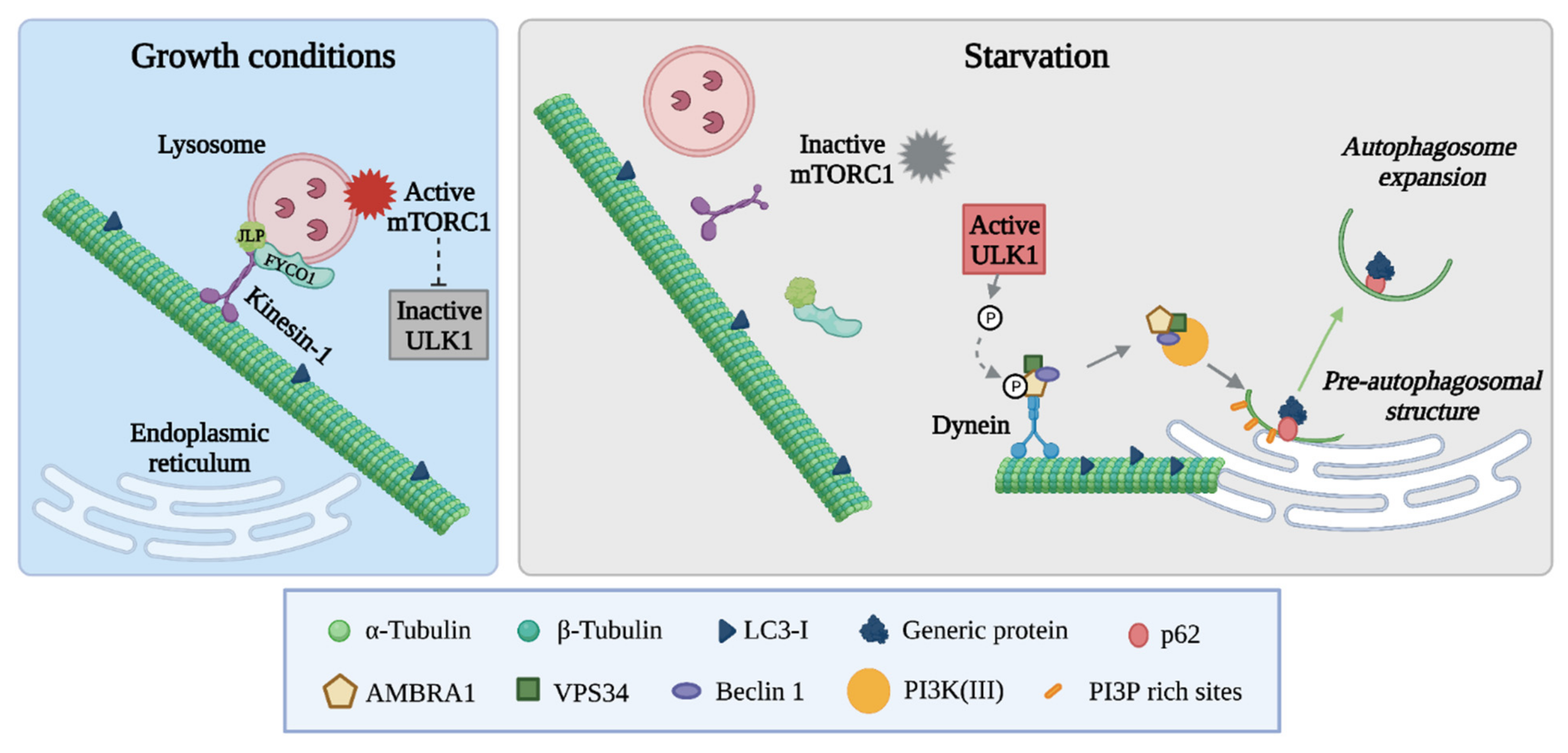
3. The Tubulin Code and Its Associated Enzymes in Autophagy
3.1. The Autophagic Machinery: Mechanisms and Regulation
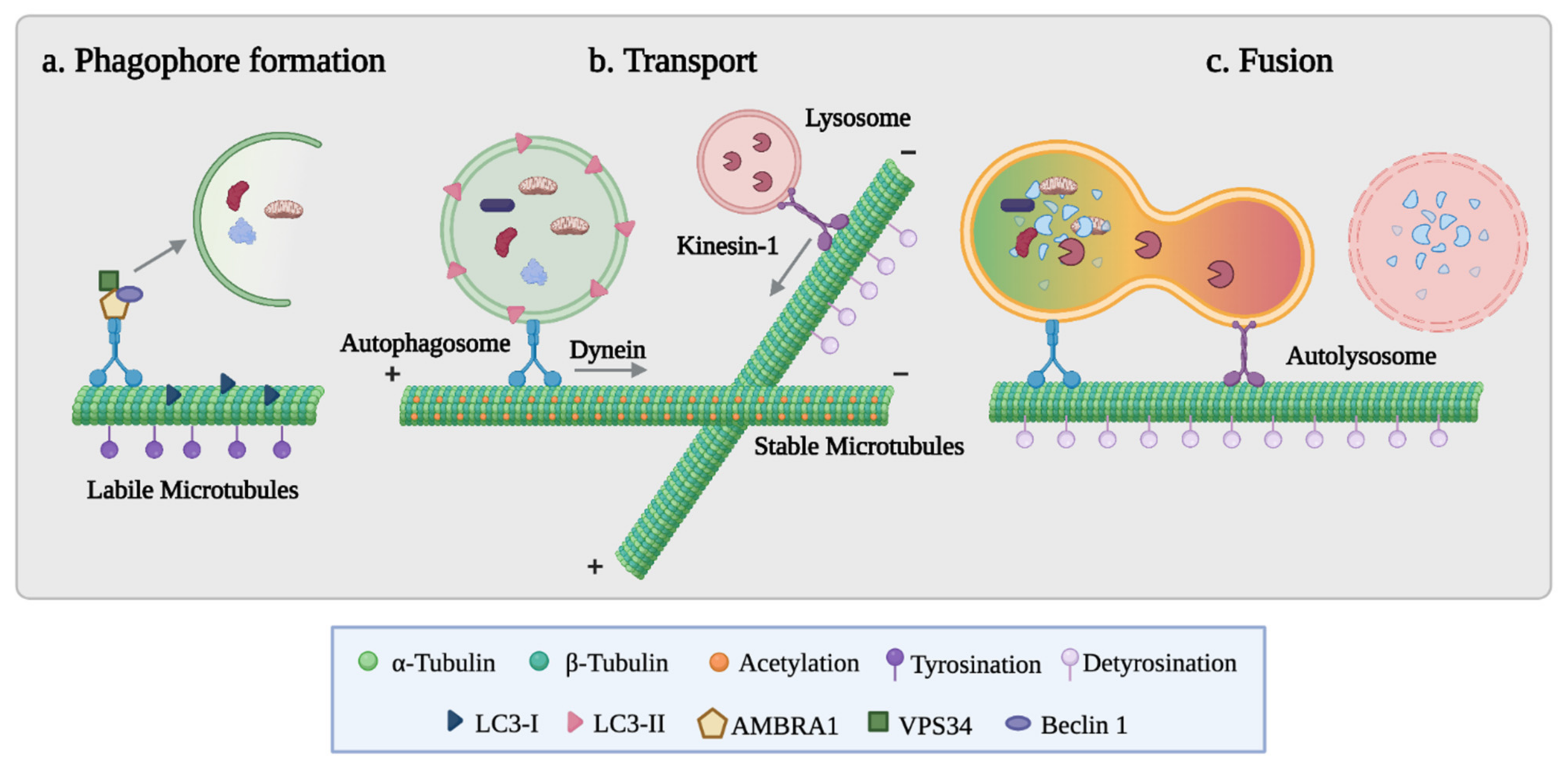
3.2. The Role of Microtubules in Autophagosome Formation and Fusion with Lysosomes
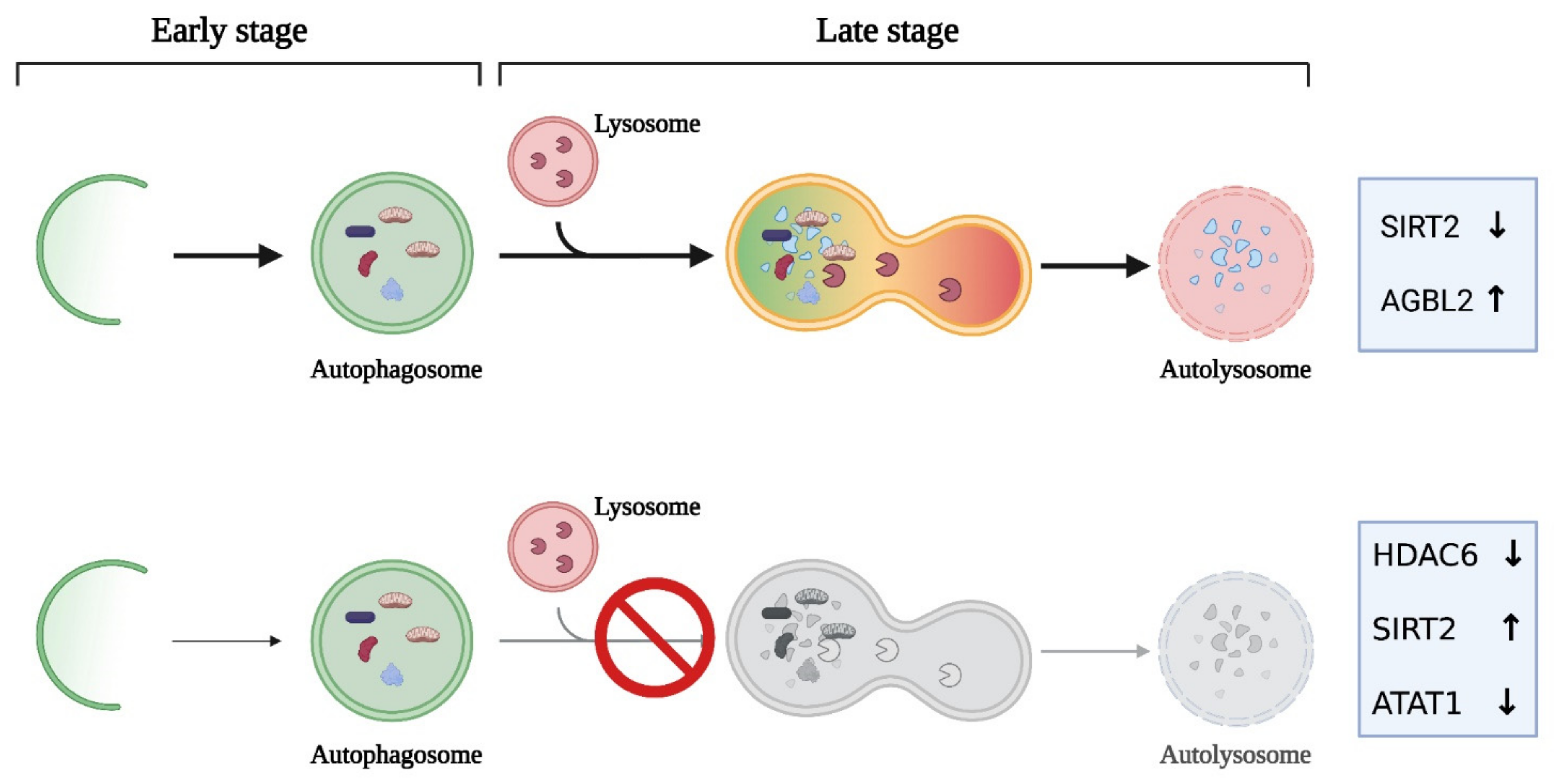
3.3. The Tubulin Code in the Autophagic Process
| Enzymes | Cancer Cells | Experimental Approach | Impact on Cancer Autophagy | References |
|---|---|---|---|---|
| Lysine Acetyltransferase | ||||
| ATAT1 | Lung | Downregulation | Autophagosome accumulation | [139] |
| Lysine Deacetylase | ||||
| HDAC6 | Glioblastoma, cancer stem-like | Downregulation | Induction of cancer stem cell differentiation by promoting autophagy | [135] |
| head and neck | Pharmacological inhibition | Autophagy inhibition | [130] | |
| SIRT2 | Neuroblastoma | Overexpression | Inhibition of autophagic flux | [137] |
| Colon | Downregulation | Autophagy and mitotic post-slippage death induction | [136] | |
| Leukemic lines | Pharmacological inhibition | Apoptosis and autophagic cell death | [140] | |
| Tubulin–tyrosine ligase | ||||
| TTL | Breast, pancreatic | Pharmacological inhibition | Apoptosis and autophagic cell death | [141,142] |
| Tubulin-specific carboxypeptidase | ||||
| AGBL2 | Hepatocellular carcinoma | Overexpression | Enhanced autophagy by upregulating immunity-related GTPase M | [95] |
4. Autophagy–Microtubule Crosstalk as a Possible Target for Cancer Growth Control
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Etienne-Manneville, S. From Signaling Pathways to Microtubule Dynamics: The Key Players. Curr. Opin. Cell Biol. 2010, 22, 104–111. [Google Scholar] [CrossRef]
- Mitchison, T.; Kirschner, M. Dynamic Instability of Microtubule Growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Compton, D.A. Chromosomal Instability and Cancer: A Complex Relationship with Therapeutic Potential. J. Clin. Investig. 2012, 122, 1138–1143. [Google Scholar] [CrossRef]
- Cirillo, L.; Gotta, M.; Meraldi, P. The elephant in the room: The role of microtubules in cancer. In Cell Division Machinery and Disease. Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2017; Volume 1002, pp. 93–124. [Google Scholar] [CrossRef]
- Lopes, D.; Maiato, H. The Tubulin Code in Mitosis and Cancer. Cells 2020, 9, 2356. [Google Scholar] [CrossRef]
- Dumontet, C.; Jordan, M.A. Microtubule-Binding Agents: A Dynamic Field of Cancer Therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef]
- Steinmetz, M.O.; Prota, A.E. Microtubule-Targeting Agents: Strategies To Hijack the Cytoskeleton. Trends Cell Biol. 2018, 28, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Kanakkanthara, A. Beyond the Paclitaxel and Vinca Alkaloids: Next Generation of Plant-Derived Microtubule-Targeting Agents with Potential Anticancer Activity. Cancers 2020, 12, 1721. [Google Scholar] [CrossRef] [PubMed]
- Matthew, S.; Chen, Q.Y.; Ratnayake, R.; Fermaintt, C.S.; Lucena-Agell, D.; Bonato, F.; Prota, A.E.; Lim, S.T.; Wang, X.; Díaz, J.F.; et al. Gatorbulin-1, a Distinct Cyclodepsipeptide Chemotype, Targets a Seventh Tubulin Pharmacological Site. Proc. Natl. Acad. Sci. USA 2021, 118, e2021847118. [Google Scholar] [CrossRef] [PubMed]
- Hussey, S.P.; Fritz-Laylin, L.K. “The Missing Link”: The Tubulin Mutation Database Connects Over 1500 Missense Mutations With Phenotypes Across Eukaryotes. Cytoskeleton 2019, 76, 175–176. [Google Scholar] [CrossRef]
- Breuss, M.W.; Leca, I.; Gstrein, T.; Hansen, A.H.; Keays, D.A. Tubulins and Brain Development—The Origins of Functional Specification. Mol. Cell. Neurosci. 2017, 84, 58–67. [Google Scholar] [CrossRef]
- Lewis, S.A.; Lee, M.G.; Cowan, N.J. Five Mouse Tubulin Isotypes and Their Regulated Expression during Development. J. Cell Biol. 1985, 101, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.C.; Cleveland, D.W. Differential Utilization of Beta-Tubulin Isotypes in Differentiating Neurites. J. Cell Biol. 1989, 109, 663–673. [Google Scholar] [CrossRef]
- Denoulet, P.; Eddé, B.; Gros, F. Differential Expression of Several Neurospecific β-Tubulin MRNAs in the Mouse Brain during Development. Gene 1986, 50, 289–297. [Google Scholar] [CrossRef]
- Wang, D.; Villasante, A.; Lewis, S.; Cowan, N. The Mammalian Beta-Tubulin Repertoire: Hematopoietic Expression of a Novel, Heterologous Beta-Tubulin Isotype. J. Cell Biol. 1986, 103, 1903–1910. [Google Scholar] [CrossRef]
- Stearns, T.; Evans, L.; Kirschner, M. γ-Tubulin Is a Highly Conserved Component of the Centrosome. Cell 1991, 65, 825–836. [Google Scholar] [CrossRef]
- Kollman, J.M.; Merdes, A.; Mourey, L.; Agard, D.A. Microtubule Nucleation by γ-Tubulin Complexes. Nat. Rev. Mol. Cell Biol. 2011, 12, 709–721. [Google Scholar] [CrossRef]
- Prosser, S.L.; Pelletier, L. Mitotic Spindle Assembly in Animal Cells: A Fine Balancing Act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef]
- Chang, P.; Stearns, T. δ-Tubulin and ε-Tubulin: Two New Human Centrosomal Tubulins Reveal New Aspects of Centrosome Structure and Function. Nat. Cell Biol. 1999, 2, 30–35. [Google Scholar] [CrossRef]
- Verhey, K.J.; Gaertig, J. The Tubulin Code. Cell Cycle 2007, 6, 2152–2160. [Google Scholar] [CrossRef]
- Bodakuntla, S.; Jijumon, A.; Villablanca, C.; Gonzalez-Billault, C.; Janke, C. Microtubule-Associated Proteins: Structuring the Cytoskeleton. Trends Cell Biol. 2019, 29, 804–819. [Google Scholar] [CrossRef]
- Roll-Mecak, A. The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cell 2020, 54, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Renthal, R.; Schneider, B.G.; Miller, M.M.; Ludueña, R.F. Beta IV Is the Major Beta-Tubulin Isotype in Bovine Cilia. Cell Motil. Cytoskelet. 1993, 25, 19–29. [Google Scholar] [CrossRef]
- Panda, D.; Miller, H.P.; Banerjee, A.; Ludueña, R.F.; Wilson, L. Microtubule Dynamics in Vitro Are Regulated by the Tubulin Isotype Composition. Proc. Natl. Acad. Sci. USA 1994, 91, 11358–11362. [Google Scholar] [CrossRef] [PubMed]
- Vemu, A.; Atherton, J.; Spector, J.O.; Moores, C.A.; Roll-Mecak, A. Tubulin Isoform Composition Tunes Microtubule Dynamics. Mol. Biol. Cell 2017, 28, 3564–3572. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The Tubulin Code and Its Role in Controlling Microtubule Properties and Functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Li, L.; Yang, X.-J. Tubulin Acetylation: Responsible Enzymes, Biological Functions and Human Diseases. Cell. Mol. Life Sci. 2015, 72, 4237–4255. [Google Scholar] [CrossRef]
- Palazzo, A.; Ackerman, B.; Gundersen, G.G. Tubulin Acetylation and Cell Motility. Nature 2003, 421, 230. [Google Scholar] [CrossRef] [PubMed]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, A.; Wang, X.F.; Yao, T.P. HDAC6 Is a Microtubule-Associated Deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef]
- Portran, D.; Schaedel, L.; Xu, Z.; Théry, M.; Nachury, M.V. Tubulin Acetylation Protects Long-Lived Microtubules against Mechanical Ageing. Nat. Cell Biol. 2017, 19, 391–398. [Google Scholar] [CrossRef]
- Eshun-Wilson, L.; Zhang, R.; Portran, D.; Nachury, M.V.; Toso, D.B.; Löhr, T.; Vendruscolo, M.; Bonomi, M.; Fraser, J.S.; Nogales, E. Effects of α-Tubulin Acetylation on Microtubule Structure and Stability. Proc. Natl. Acad. Sci. USA 2019, 116, 10366–10371. [Google Scholar] [CrossRef]
- Akella, J.S.; Wloga, D.; Kim, J.; Starostina, N.G.; Lyons-Abbott, S.; Morrissette, N.S.; Dougan, S.T.; Kipreos, E.T.; Gaertig, J. MEC-17 Is an α-Tubulin Acetyltransferase. Nature 2010, 467, 218–222. [Google Scholar] [CrossRef]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The Human Sir2 Ortholog, SIRT2, Is an NAD+-Dependent Tubulin Deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Ouyang, C.; Mu, J.; Lu, Q.; Li, J.; Zhu, H.; Wang, Q.; Zou, M.-H.; Xie, Z. Autophagic Degradation of KAT2A/GCN5 Promotes Directional Migration of Vascular Smooth Muscle Cells by Reducing TUBA/α-Tubulin Acetylation. Autophagy 2019, 16, 1753–1770. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-W.; Hou, F.; Zhang, J.; Phu, L.; Loktev, A.V.; Kirkpatrick, D.S.; Jackson, P.K.; Zhao, Y.; Zou, H. A Novel Acetylation of β-Tubulin by San Modulates Microtubule Polymerization via down-Regulating Tubulin Incorporation. Mol. Biol. Cell 2011, 22, 448. [Google Scholar] [CrossRef]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule Acetylation Promotes Kinesin-1 Binding and Transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Rice, L.M.; Vale, R.D. Regulation of Microtubule Motors by Tubulin Isotypes and Posttranslational Modifications. Nat. Cell Biol. 2014, 16, 335. [Google Scholar] [CrossRef]
- Nekooki-Machida, Y.; Hagiwara, H. Role of Tubulin Acetylation in Cellular Functions and Diseases. Med. Mol. Morphol. 2020, 53, 191–197. [Google Scholar] [CrossRef]
- Barra, H.S.; Arce, C.A.; Argaraña, C.E. Posttranslational Tyrosination/Detyrosination of Tubulin. Mol. Neurobiol. 1988, 2, 133–153. [Google Scholar] [CrossRef]
- Nieuwenhuis, J.; Brummelkamp, T.R. The Tubulin Detyrosination Cycle: Function and Enzymes. Trends Cell Biol. 2019, 29, 80–92. [Google Scholar] [CrossRef]
- Robison, P.; Caporizzo, M.A.; Ahmadzadeh, H.; Bogush, A.I.; Chen, C.Y.; Margulies, K.B.; Shenoy, V.B.; Prosser, B.L. Detyrosinated Microtubules Buckle and Bear Load in Contracting Cardiomyocytes. Science 2016, 352, aaf0659. [Google Scholar] [CrossRef]
- Erck, C.; Peris, L.; Andrieux, A.; Meissirel, C.; Gruber, A.D.; Vernet, M.; Schweitzer, A.; Saoudi, Y.; Pointu, H.; Bosc, C.; et al. A Vital Role of Tubulin-Tyrosine-Ligase for Neuronal Organization. Proc. Natl. Acad. Sci. USA 2005, 102, 7853–7858. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; Sousa, R.S.E.; Tripathy, S.K.; Magiera, M.M.; Zaytsev, A.V.; Pereira, A.L.; Janke, C.; Grishchuk, E.L.; Maiato, H. Microtubule Detyrosination Guides Chromosomes during Mitosis. Science 2015, 348, 799. [Google Scholar] [CrossRef]
- Nieuwenhuis, J.; Adamopoulos, A.; Bleijerveld, O.B.; Mazouzi, A.; Stickel, E.; Celie, P.; Altelaar, M.; Knipscheer, P.; Perrakis, A.; Blomen, V.A.; et al. Vasohibins Encode Tubulin Detyrosinating Activity. Science 2017, 358, 1453–1456. [Google Scholar] [CrossRef]
- Aillaud, C.; Bosc, C.; Peris, L.; Bosson, A.; Heemeryck, P.; van Dijk, J.; Friec, J.L.; Boulan, B.; Vossier, F.; Sanman, L.E.; et al. Vasohibins/SVBP Are Tubulin Carboxypeptidases (TCPs) That Regulate Neuron Differentiation. Science 2017, 358, 1448–1453. [Google Scholar] [CrossRef]
- Ersfeld, K.; Wehland, J.; Plessmann, U.; Dodemont, H.; Gerke, V.; Weber, K. Characterization of the Tubulin-Tyrosine Ligase. J. Cell Biol. 1993, 120, 725. [Google Scholar] [CrossRef]
- Field, J.J.; Kanakkanthara, A.; Miller, J.H. Microtubule-Targeting Agents Are Clinically Successful Due to Both Mitotic and Interphase Impairment of Microtubule Function. Bioorg. Med. Chem. 2014, 22, 5050–5059. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Zhang, J.; He, S.; Wan, C.; Shan, A.; Wang, Y.; Yu, L.; Liu, G.; Chen, K.; Shi, J.; et al. Increased α-Tubulin1b Expression Indicates Poor Prognosis and Resistance to Chemotherapy in Hepatocellular Carcinoma. Dig. Dis. Sci. 2013, 58, 2713–2720. [Google Scholar] [CrossRef]
- Gui, S.; Chen, P.; Liu, Y.; Chen, Q.; Cheng, T.; Lv, S.; Zhou, T.; Song, Z.; Xiao, J.; He, W.; et al. TUBA1C Expression Promotes Proliferation by Regulating the Cell Cycle and Indicates Poor Prognosis in Glioma. Biochem. Biophys. Res. Commun. 2021, 577, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Jiao, Z.; Ji, Y.; Zhang, S. Elevated TUBA1A Might Indicate the Clinical Outcomes of Patients with Gastric Cancer, Being Associated with the Infiltration of Macrophages in the Tumor Immune Microenvironment. J. Gastrointest. Liver Dis. JGLD 2020, 29, 509–522. [Google Scholar] [CrossRef]
- Bian, T.; Zheng, M.; Jiang, D.; Liu, J.; Sun, H.; Li, X.; Liu, L.; Zhang, J.; Liu, Y. Prognostic Biomarker TUBA1C Is Correlated to Immune Cell Infiltration in the Tumor Microenvironment of Lung Adenocarcinoma. Cancer Cell Int. 2021, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Hu, X.; Gu, L.; Jian, Z.; Li, L.; Hu, S.; Qiu, S.; Xiong, X. TUBA1C Is a Prognostic Marker in Low-Grade Glioma and Correlates with Immune Cell Infiltration in the Tumor Microenvironment. Front. Genet. 2021, 12, 759953. [Google Scholar] [CrossRef]
- Nicoletti, M.I.; Valoti, G.; Giannakakou, P.; Zhan, Z.; Kim, J.-H.; Lucchini, V.; Landoni, F.; Mayo, J.G.; Giavazzi, R.; Fojo, T. Expression Of Tubulin Isotypes in Human Ovarian Carcinoma Xenografts and in a Sub-Panel of Human Cancer Cell Lines from the NCI-Anticancer Drug Screen: Correlation with Sensitivity to Microtubule Active Agents. Clin. Cancer Res. 2001, 7, 2912–2922. [Google Scholar]
- Nami, B.; Wang, Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef]
- Kanakkanthara, A.; Miller, J.H. ΒIII-Tubulin Overexpression in Cancer: Causes, Consequences, and Potential Therapies. Biochim. Biophys. Acta (BBA) Rev. Cancer 2021, 1876, 188607. [Google Scholar] [CrossRef] [PubMed]
- Quaas, A.; Rahvar, A.-H.; Burdelski, C.; Koop, C.; Eichelberg, C.; Rink, M.; Dahlem, R.; Schlomm, T.; Tsourlakis, M.C.; Simon, R.; et al. ΒIII-Tubulin Overexpression Is Linked to Aggressive Tumor Features and Shortened Survival in Clear Cell Renal Cell Carcinoma. World J. Urol. 2014, 33, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, A.; Chaker, A.; Burdelski, C.; Koop, C.; Tsourlakis, M.C.; Steurer, S.; Rink, M.; Eichenauer, T.S.; Wilczak, W.; Wittmer, C.; et al. ΒIII-Tubulin Overexpression Is Linked to Aggressive Tumor Features and Genetic Instability in Urinary Bladder Cancer. Hum. Pathol. 2017, 61, 210–220. [Google Scholar] [CrossRef]
- Gan, P.P.; Kavallaris, M. Tubulin-Targeted Drug Action: Functional Significance of Class II and Class IVb β-Tubulin in Vinca Alkaloid Sensitivity. Cancer Res. 2008, 68, 9817–9824. [Google Scholar] [CrossRef]
- Ohishi, Y.; Oda, Y.; Basaki, Y.; Kobayashi, H.; Wake, N.; Kuwano, M.; Tsuneyoshi, M. Expression of Beta-Tubulin Isotypes in Human Primary Ovarian Carcinoma. Gynecol. Oncol. 2007, 105, 586–592. [Google Scholar] [CrossRef]
- Bernard-Marty, C.; Treilleux, I.; Dumontet, C.; Cardoso, F.; Fellous, A.; Gancberg, D.; Bissery, M.C.; Paesmans, M.; Larsimont, D.; Piccart, M.J.; et al. Microtubule-Associated Parameters as Predictive Markers of Docetaxel Activity in Advanced Breast Cancer Patients: Results of a Pilot Study. Clin. Breast Cancer 2002, 3, 341–345. [Google Scholar] [CrossRef]
- Cucchiarelli, V.; Hiser, L.; Smith, H.; Frankfurter, A.; Spano, A.; Correia, J.J.; Lobert, S. β-Tubulin Isotype Classes II and V Expression Patterns in Nonsmall Cell Lung Carcinomas. Cell Motil. Cytoskelet. 2008, 65, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Na, H.Y.; Park, M.; Kim, Y.A.; Won, J.K.; Park, Y.J.; Shin, S.A.; Lee, S.; Oh, S.; Kim, J.E. Expression of Class III Beta-Tubulin Is Associated with Invasive Potential and Poor Prognosis in Thyroid Carcinoma. J. Clin. Med. 2020, 9, 3830. [Google Scholar] [CrossRef] [PubMed]
- Atjanasuppat, K.; Lirdprapamongkol, K.; Jantaree, P.; Svasti, J. Non-Adherent Culture Induces Paclitaxel Resistance in H460 Lung Cancer Cells via ERK-Mediated up-Regulation of ΒIVa-Tubulin. Biochem. Biophys. Res. Commun. 2015, 466, 493–498. [Google Scholar] [CrossRef]
- Christoph, D.C.; Kasper, S.; Gauler, T.C.; Loesch, C.; Engelhard, M.; Theegarten, D.; Poettgen, C.; Hepp, R.; Peglow, A.; Loewendick, H.; et al. ΒV-Tubulin Expression Is Associated with Outcome Following Taxane-Based Chemotherapy in Non-Small Cell Lung Cancer. Br. J. Cancer 2012, 107, 823–830. [Google Scholar] [CrossRef]
- Sekino, Y.; Han, X.; Babasaki, T.; Miyamoto, S.; Kobatake, K.; Kitano, H.; Ikeda, K.; Goto, K.; Inoue, S.; Hayashi, T.; et al. TUBB3 Is Associated with PTEN, Neuroendocrine Differentiation, and Castration Resistance in Prostate Cancer. Urol. Oncol. Semin. Orig. Investig. 2021, 39, 368.e1–368.e9. [Google Scholar] [CrossRef] [PubMed]
- Sekino, Y.; Han, X.; Babasaki, T.; Miyamoto, S.; Kitano, H.; Kobayashi, G.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; et al. TUBB3 Is Associated with High-Grade Histology, Poor Prognosis, P53 Expression, and Cancer Stem Cell Markers in Clear Cell Renal Cell Carcinoma. Oncology 2020, 98, 689–698. [Google Scholar] [CrossRef]
- Bailey, J.M.; Alsina, J.; Rasheed, Z.A.; McAllister, F.M.; Fu, Y.Y.; Plentz, R.; Zhang, H.; Pasricha, P.J.; Bardeesy, N.; Matsui, W.; et al. DCLK1 Marks a Morphologically Distinct Subpopulation of Cells With Stem Cell Properties in Preinvasive Pancreatic Cancer. Gastroenterology 2014, 146, 245–256. [Google Scholar] [CrossRef]
- Boggs, A.E.; Vitolo, M.I.; Whipple, R.A.; Charpentier, M.S.; Goloubeva, O.G.; Ioffe, O.B.; Tuttle, K.C.; Slovic, J.; Lu, Y.; Mills, G.B.; et al. α-Tubulin Acetylation Elevated in Metastatic and Basal-like Breast Cancer Cells Promotes Microtentacle Formation, Adhesion, and Invasive Migration. Cancer Res. 2015, 75, 203–215. [Google Scholar] [CrossRef]
- Wattanathamsan, O.; Thararattanobon, R.; Rodsiri, R.; Chanvorachote, P.; Vinayanuwattikun, C.; Pongrakhananon, V. Tubulin Acetylation Enhances Lung Cancer Resistance to Paclitaxel-Induced Cell Death through Mcl-1 Stabilization. Cell Death Discov. 2021, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Levidou, G.; Gajdzis, P.; Cassoux, N.; Donizy, P.; Masaoutis, C.; Gajdzis, M.; Gardrat, S.; Pergaris, A.; Danas, E.; Klijanienko, J.; et al. Histone Deacetylase (HDAC)-1, -2, -4, and -6 in Uveal Melanomas: Associations with Clinicopathological Parameters and Patients’ Survival. Cancers 2021, 13, 4763. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, X.; Huang, A. Aggresome–Autophagy Associated Gene HDAC6 Is a Potential Biomarker in Pan-Cancer, Especially in Colon Adenocarcinoma. Front. Oncol. 2021, 11, 2625. [Google Scholar] [CrossRef] [PubMed]
- Saji, S.; Kawakami, M.; Hayashi, S.; Yoshida, N.; Hirose, M.; Horiguchi, S.; Itoh, A.; Funata, N.; Schreiber, S.L.; Yoshida, M.; et al. Significance of HDAC6 Regulation via Estrogen Signaling for Cell Motility and Prognosis in Estrogen Receptor-Positive Breast Cancer. Oncogene 2005, 24, 4531–4539. [Google Scholar] [CrossRef]
- Shi, P.; Hoang-Minh, L.B.; Tian, J.; Cheng, A.; Basrai, R.; Kalaria, N.; Lebowitz, J.J.; Khoshbouei, H.; Deleyrolle, L.P.; Sarkisian, M.R. HDAC6 Signaling at Primary Cilia Promotes Proliferation and Restricts Differentiation of Glioma Cells. Cancers 2021, 13, 1644. [Google Scholar] [CrossRef] [PubMed]
- Urdiciain, A.; Erausquin, E.; Zelaya, M.V.; Zazpe, I.; Lanciego, J.L.; Meléndez, B.; Rey, J.A.; Idoate, M.A.; Galdo, N.A.R.-D.; Castresana, J.S. Silencing of Histone Deacetylase 6 Decreases Cellular Malignancy and Contributes to Primary Cilium Restoration, Epithelial-to-Mesenchymal Transition Reversion, and Autophagy Inhibition in Glioblastoma Cell Lines. Biology 2021, 10, 467. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; He, D.; Zhang, X. Role of SIRT2 in Regulation of Stemness of Cancer Stem-Like Cells in Renal Cell Carcinoma. Cell. Physiol. Biochem. 2018, 49, 2348–2357. [Google Scholar] [CrossRef]
- Zhao, N.; Guo, Y.; Liu, P.; Chen, Y.; Wang, Y. Sirtuin 2 Promotes Cell Stemness and MEK/ERK Signaling Pathway While Reduces Chemosensitivity in Endometrial Cancer. Arch. Gynecol. Obstet. 2021, 1, 1–9. [Google Scholar] [CrossRef]
- Li, Z.; Xie, Q.R.; Chen, Z.; Lu, S.; Xia, W. Regulation of SIRT2 Levels for Human Non-Small Cell Lung Cancer Therapy. Lung Cancer 2013, 82, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; You, E.; Ko, P.; Jeong, J.; Keum, S.; Rhee, S. Genetic Disruption of Tubulin Acetyltransferase, ATAT1, Inhibits Proliferation and Invasion of Colon Cancer Cells through Decreases in Wnt1/β-Catenin Signaling. Biochem. Biophys. Res. Commun. 2017, 482, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-C.; Cheng, Y.-C.; Chang, C.-Y.; Lin, C.-M.; Chang, J.-Y. Alpha-Tubulin Acetyltransferase/MEC-17 Regulates Cancer Cell Migration and Invasion through Epithelial–Mesenchymal Transition Suppression and Cell Polarity Disruption. Sci. Rep. 2018, 8, 17477. [Google Scholar] [CrossRef]
- Chien, J.-Y.; Tsen, S.-D.; Chien, C.-C.; Liu, H.-W.; Tung, C.-Y.; Lin, C.-H. ATAT1 Downregulation Induces Mitotic Catastrophe in HeLa and A549 Cells. Cell Death Discov. 2016, 2, 16006. [Google Scholar] [CrossRef]
- Whipple, R.A.; Matrone, M.A.; Cho, E.H.; Balzer, E.M.; Vitolo, M.I.; Yoon, J.R.; Ioffe, O.B.; Tuttle, K.C.; Yang, J.; Martin, S.S. Epithelial-to-Mesenchymal Transition Promotes Tubulin Detyrosination and Microtentacles That Enhance Endothelial Engagement. Cancer Res. 2010, 70, 8127–8137. [Google Scholar] [CrossRef]
- Kato, C.; Miyazaki, K.; Nakagawa, A.; Ohira, M.; Nakamura, Y.; Ozaki, T.; Imai, T.; Nakagawara, A. Low Expression of Human Tubulin Tyrosine Ligase and Suppressed Tubulin Tyrosination/Detyrosination Cycle Are Associated with Impaired Neuronal Differentiation in Neuroblastomas with Poor Prognosis. Int. J. Cancer 2004, 112, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Mialhe, A.; Lafanechèere, L.; Treilleux, I.; Peloux, N.; Dumontet, C.; Brémond, A.; Panh, M.-H.; Payan, R.; Wehland, J.; Margolis, R.-L.; et al. Tubulin Detyrosination Is a Frequent Occurrence in Breast Cancers of Poor Prognosis. Cancer Res. 2001, 61, 5024–5027. [Google Scholar] [PubMed]
- Sato, Y. The Vasohibin Family: A Novel Family for Angiogenesis Regulation. J. Biochem. 2013, 153, 5. [Google Scholar] [CrossRef]
- Hara, H.; Ozawa, S.; Ninomiya, Y.; Yamamoto, M.; Ogimi, M.; Nabeshima, K.; Nakamura, K.; Kajiwara, H.; Nakamura, N.; Sato, Y. Prognostic Significance of Vasohibin-1 and Vasohibin-2 Immunohistochemical Expression in Gastric Cancer. Surg. Today 2020, 50, 1530–1543. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Ozawa, S.; Ninomiya, Y.; Koyanagi, K.; Oguma, J.; Kazuno, A.; Hara, H.; Yatabe, K.; Kajiwara, H.; Nakamura, N.; et al. Plasma Vasohibin-1 and Vasohibin-2 Are Useful Biomarkers in Patients with Esophageal Squamous Cell Carcinoma. Esophagus Off. J. Jpn. Esophageal Soc. 2020, 17, 289–297. [Google Scholar] [CrossRef]
- Ninomiya, Y.; Ozawa, S.; Oguma, J.; Kazuno, A.; Nitta, M.; Kajiwara, H.; Sato, Y. Expression of Vasohibin-1 and −2 Predicts Poor Prognosis among Patients with Squamous Cell Carcinoma of the Esophagus. Oncol. Lett. 2018, 16, 5265. [Google Scholar] [CrossRef]
- Sano, R.; Kanomata, N.; Suzuki, S.; Shimoya, K.; Sato, Y.; Moriya, T.; Shiota, M. Vasohibin-1 Is a Poor Prognostic Factor of Ovarian Carcinoma. Tohoku J. Exp. Med. 2017, 243, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, T.; Saga, Y.; Takahashi, Y.; Tamura, K.; Yoshiba, T.; Takahashi, S.; Taneichi, A.; Takei, Y.; Urabe, M.; Mizukami, H.; et al. Knockout of Vasohibin-2 Reduces Tubulin Carboxypeptidase Activity and Increases Paclitaxel Sensitivity in Ovarian Cancer. Cancer Med. 2021, 10, 2732–2739. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.; Schattschneider, J.; Blechner, C.; Krisp, C.; Schlüter, H.; Schweizer, M.; Nalaskowski, M.; Oliveira-Ferrer, L.; Windhorst, S. Tubulin Tyrosine Ligase Like 4 (TTLL4) Overexpression in Breast Cancer Cells Is Associated with Brain Metastasis and Alters Exosome Biogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 1–15. [Google Scholar] [CrossRef]
- Kashiwaya, K.; Nakagawa, H.; Hosokawa, M.; Mochizuki, Y.; Ueda, K.; Piao, L.; Chung, S.; Hamamoto, R.; Eguchi, H.; Ohigashi, H.; et al. Involvement of the Tubulin Tyrosine Ligase-Like Family Member 4 Polyglutamylase in PELP1 Polyglutamylation and Chromatin Remodeling in Pancreatic Cancer Cells. Cancer Res. 2010, 70, 4024–4033. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Xiao, Y.; Zhang, Y.; Yang, M.; Lin, Y.; Tang, J. Identification of a Novel Transcript Isoform of the TTLL12 Gene in Human Cancers. Oncol. Rep. 2016, 36, 3172–3180. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liang, Y.; Qian, H.; Li, Q. TTLL12 Expression in Ovarian Cancer Correlates with a Poor Outcome. Int. J. Clin. Exp. Pathol. 2020, 13, 239. [Google Scholar]
- He, W.; Wang, L. High Expression of AGBL2 Is a Novel Prognostic Factor of Adverse Outcome in Patients with Ovarian Carcinoma. Oncol. Lett. 2019, 18, 4900–4906. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Jin, X.H.; Cai, M.Y.; Li, H.G.; Chen, J.W.; Wang, F.W.; Wang, C.Y.; Hu, W.W.; Liu, F.; Xie, D. AGBL2 Promotes Cancer Cell Growth through IRGM-Regulated Autophagy and Enhanced Aurora A Activity in Hepatocellular Carcinoma. Cancer Lett. 2018, 414, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Peterfi, L.; Banyai, D.; Yusenko, M.V.; Bjercke, T.; Kovacs, G. Expression of RARRES1 and AGBL2 and Progression of Conventional Renal Cell Carcinoma. Br. J. Cancer 2020, 122, 1818. [Google Scholar] [CrossRef]
- Yang, Y.; Klionsky, D.J. Autophagy and Disease: Unanswered Questions. Cell Death Differ. 2020, 27, 858. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11. [Google Scholar] [CrossRef]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The Autophagosome: Origins Unknown, Biogenesis Complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Martens, S. Mechanisms of Selective Autophagy. J. Mol. Biol. 2016, 428, 1714. [Google Scholar] [CrossRef]
- Dossou, A.S.; Basu, A. The Emerging Roles of MTORC1 in Macromanaging Autophagy. Cancers 2019, 11, 1422. [Google Scholar] [CrossRef]
- Turco, E.; Witt, M.; Abert, C.; Bock-Bierbaum, T.; Su, M.-Y.; Trapannone, R.; Sztacho, M.; Danieli, A.; Shi, X.; Zaffagnini, G.; et al. FIP200 Claw Domain Binding to P62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol. Cell 2019, 74, 330. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Zhang, H. Formation and Maturation of Autophagosomes in Higher Eukaryotes: A Social Network. Curr. Opin. Cell Biol. 2018, 53, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Otomo, C.; Otomo, T. The Autophagic Membrane Tether ATG2A Transfers Lipids between Membranes. eLife 2019, 8, e45777. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (3rd Edition). Autophagy 2016, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; He, H.; Tang, Z.; Hattori, T.; Liu, Y.; Young, M.M.; Serfass, J.M.; Chen, L.; Gebru, M.; Chen, C.; et al. An Autophagy Assay Reveals the ESCRT-III Component CHMP2A as a Regulator of Phagophore Closure. Nat. Commun. 2018, 9, 2855. [Google Scholar] [CrossRef]
- Oyarzún, J.E.; Lagos, J.; Vázquez, M.C.; Valls, C.; de la Fuente, C.; Yuseff, M.I.; Alvarez, A.R.; Zanlungo, S. Lysosome Motility and Distribution: Relevance in Health and Disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 1076–1087. [Google Scholar] [CrossRef]
- Kaur, J.; Debnath, J. Autophagy at the Crossroads of Catabolism and Anabolism. Nat. Rev. Mol. Cell Biol. 2015, 16, 461–472. [Google Scholar] [CrossRef]
- Mackeh, R.; Perdiz, D.; Lorin, S.; Codogno, P.; Poüs, C. Autophagy and Microtubules—New Story, Old Players. J. Cell Sci. 2013, 126, 1071–1080. [Google Scholar] [CrossRef]
- Geeraert, C.; Ratier, A.; Pfisterer, S.G.; Perdiz, D.; Cantaloube, I.; Rouault, A.; Pattingre, S.; Proikas-Cezanne, T.; Codogno, P.; Poüs, C. Starvation-Induced Hyperacetylation of Tubulin Is Required for the Stimulation of Autophagy by Nutrient Deprivation. J. Biol. Chem. 2010, 285, 24184–24194. [Google Scholar] [CrossRef]
- Xie, R.; Nguyen, S.; McKeehan, K.; Wang, F.; McKeehan, W.L.; Liu, L. Microtubule-Associated Protein 1S (MAP1S) Bridges Autophagic Components with Microtubules and Mitochondria to Affect Autophagosomal Biogenesis and Degradation. J. Biol. Chem. 2011, 286, 10367. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, X.; Yang, F.; Yang, Y.; Wang, T.; Liu, W.; Zhou, H.; Zhao, W. RASSF1A Enhances Chemosensitivity of NSCLC Cells Through Activating Autophagy by Regulating MAP1S to Inactivate Keap1-Nrf2 Pathway. Drug Des. Dev. Ther. 2021, 15, 21. [Google Scholar] [CrossRef]
- Köchl, R.; Hu, X.W.; Chan, E.Y.W.; Tooze, S.A. Microtubules Facilitate Autophagosome Formation and Fusion of Autophagosomes with Endosomes. Traffic 2006, 7, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Pedersen, N.M.; Wang, L.; Torgersen, M.L.; Stenmark, H.; Raiborg, C. PtdIns3P Controls MTORC1 Signaling through Lysosomal Positioning. J. Cell Biol. 2017, 216, 4217. [Google Scholar] [CrossRef]
- Suzuki, R.; Gunarta, I.K.; Boldbaatar, J.; Erdenebaatar, P.; Odongoo, R.; Yoshioka, K. Functional Role of C-Jun NH2-Terminal Kinase-Associated Leucine Zipper Protein (JLP) in Lysosome Localization and Autophagy. Drug Discov. Ther. 2020, 14, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Saiki, S.; Lichtenberg, M.; Siddiqi, F.H.; Roberts, E.A.; Imarisio, S.; Jahreiss, L.; Sarkar, S.; Futter, M.; Menzies, F.M.; et al. Lysosomal Positioning Coordinates Cellular Nutrient Responses. Nat. Cell Biol. 2011, 13, 453–460. [Google Scholar] [CrossRef]
- Jia, R.; Bonifacino, J.S. Lysosome Positioning Influences MTORC2 and AKT Signaling. Mol. Cell 2019, 75, 26. [Google Scholar] [CrossRef]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as Dynamic Regulators of Cell and Organismal Homeostasis. Nat. Rev. Mol. Cell Biol. 2019, 21, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Acevedo-Arozena, A.; Imarisio, S.; Berger, Z.; Vacher, C.; O’Kane, C.J.; Brown, S.D.M.; Rubinsztein, D.C. Dynein Mutations Impair Autophagic Clearance of Aggregate-Prone Proteins. Nat. Genet. 2005, 37, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Noda, T.; Yoshimori, T. Dynein-Dependent Movement of Autophagosomes Mediates Efficient Encounters with Lysosomes. Cell Struct. Funct. 2008, 33, 109–122. [Google Scholar] [CrossRef]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The Itinerary of Autophagosomes: From Peripheral Formation to Kiss-and-Run Fusion with Lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.M.P.; Groth-Pedersen, L.; Høyer-Hansen, M.; Kirkegaard, T.; Corcelle, E.; Andersen, J.S.; Jäättelä, M.; Nylandsted, J. Depletion of Kinesin 5B Affects Lysosomal Distribution and Stability and Induces Peri-Nuclear Accumulation of Autophagosomes in Cancer Cells. PLoS ONE 2009, 4, e4424. [Google Scholar] [CrossRef]
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.-A.; Lamark, T.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. FYCO1 Is a Rab7 Effector That Binds to LC3 and PI3P to Mediate Microtubule plus End–Directed Vesicle Transport. J. Cell Biol. 2010, 188, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Kraft, L.J.; Manral, P.; Dowler, J.; Kenworthy, A.K. Nuclear LC3 Associates with Slowly Diffusing Complexes That Survey the Nucleolus. Traffic 2016, 17, 369. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, L.; Chen, H.; Xu, B.; Xu, W.; Sheng, Y.; Duan, Y. 2,3′,4,4′,5-Pentachlorobiphenyl Induced Autophagy of the Thyrocytes via DAPK2/PKD/VPS34 Pathway. Arch. Toxicol. 2019, 93, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Nowosad, A.; Creff, J.; Jeannot, P.; Culerrier, R.; Codogno, P.; Manenti, S.; Nguyen, L.; Besson, A. P27 Controls Autophagic Vesicle Trafficking in Glucose-Deprived Cells via the Regulation of ATAT1-Mediated Microtubule Acetylation. Cell Death Dis. 2021, 12, 481. [Google Scholar] [CrossRef]
- Xie, R.; Nguyen, S.; McKeehan, W.L.; Liu, L. Acetylated Microtubules Are Required for Fusion of Autophagosomes with Lysosomes. BMC Cell Biol. 2010, 11, 89. [Google Scholar] [CrossRef]
- Liu, K.P.; Zhou, D.; Ouyang, D.Y.; Xu, L.H.; Wang, Y.; Wang, L.X.; Pan, H.; He, X.H. LC3B-II Deacetylation by Histone Deacetylase 6 Is Involved in Serum-Starvation-Induced Autophagic Degradation. Biochem. Biophys. Res. Commun. 2013, 441, 970–975. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Koga, H.; Kawaguchi, Y.; Tang, W.; Wong, E.; Gao, Y.-S.; Pandey, U.B.; Kaushik, S.; Tresse, E.; Lu, J.; et al. HDAC6 Controls Autophagosome Maturation Essential for Ubiquitin-Selective Quality-Control Autophagy. EMBO J. 2010, 29, 969. [Google Scholar] [CrossRef]
- Liu, J.R.; Yu, C.W.; Hung, P.Y.; Hsin, L.W.; Chern, J.W. High-Selective HDAC6 Inhibitor Promotes HDAC6 Degradation Following Autophagy Modulation and Enhanced Antitumor Immunity in Glioblastoma. Biochem. Pharmacol. 2019, 163, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, Y.; Wang, H.; Li, H.; Zhang, M.; Zhou, L.; Meng, X.; Wu, Y.; Liu, P.; Liu, X.; et al. MicroRNA-221 Induces Autophagy through Suppressing HDAC6 Expression and Promoting Apoptosis in Pancreatic Cancer. Oncol. Lett. 2018, 16, 7295. [Google Scholar] [CrossRef]
- Yang, F.; Wang, F.; Liu, Y.; Wang, S.; Li, X.; Huang, Y.; Xia, Y.; Cao, C. Sulforaphane Induces Autophagy by Inhibition of HDAC6-Mediated PTEN Activation in Triple Negative Breast Cancer Cells. Life Sci. 2018, 213, 149–157. [Google Scholar] [CrossRef]
- Kaliszczak, M.; van Hechanova, E.; Li, Y.; Alsadah, H.; Parzych, K.; Auner, H.W.; Aboagye, E.O. The HDAC6 Inhibitor C1A Modulates Autophagy Substrates in Diverse Cancer Cells and Induces Cell Death. Br. J. Cancer 2018, 119, 1278. [Google Scholar] [CrossRef]
- Jung, K.H.; Noh, J.H.; Kim, J.K.; Eun, J.W.; Bae, H.J.; Chang, Y.G.; Kim, M.G.; Park, W.S.; Lee, J.Y.; Lee, S.-Y.; et al. Histone Deacetylase 6 Functions as a Tumor Suppressor by Activating C-Jun NH2-Terminal Kinase-Mediated Beclin 1-Dependent Autophagic Cell Death in Liver Cancer. Hepatology 2012, 56, 644–657. [Google Scholar] [CrossRef] [PubMed]
- Sharif, T.; Martell, E.; Dai, C.; Ghassemi-Rad, M.S.; Hanes, M.R.; Murphy, P.J.; Margam, N.N.; Parmar, H.B.; Giacomantonio, C.A.; Duncan, R.; et al. HDAC6 Differentially Regulates Autophagy in Stem-like versus Differentiated Cancer Cells. Autophagy 2018, 15, 686–706. [Google Scholar] [CrossRef]
- Inoue, T.; Nakayama, Y.; Li, Y.; Matsumori, H.; Takahashi, H.; Kojima, H.; Wanibuchi, H.; Katoh, M.; Oshimura, M. SIRT2 Knockdown Increases Basal Autophagy and Prevents Postslippage Death by Abnormally Prolonging the Mitotic Arrest That Is Induced by Microtubule Inhibitors. FEBS J. 2014, 281, 2623–2637. [Google Scholar] [CrossRef] [PubMed]
- Gal, J.; Bang, Y.; Choi, H.J. SIRT2 Interferes with Autophagy-Mediated Degradation of Protein Aggregates in Neuronal Cells under Proteasome Inhibition. Neurochem. Int. 2012, 61, 992–1000. [Google Scholar] [CrossRef]
- Esteves, A.R.; Palma, A.M.; Gomes, R.; Santos, D.; Silva, D.F.; Cardoso, S.M. Acetylation as a Major Determinant to Microtubule-Dependent Autophagy: Relevance to Alzheimer’s and Parkinson Disease Pathology. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 2008–2023. [Google Scholar] [CrossRef]
- Ragazzoni, Y.; Desideri, M.; Gabellini, C.; de Luca, T.; Carradori, S.; Secci, D.; Nescatelli, R.; Candiloro, A.; Condello, M.; Meschini, S.; et al. The Thiazole Derivative CPTH6 Impairs Autophagy. Cell Death Dis. 2013, 4, e524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kozako, T.; Mellini, P.; Ohsugi, T.; Aikawa, A.; Uchida, Y.; Honda, S.; Suzuki, T. Novel Small Molecule SIRT2 Inhibitors Induce Cell Death in Leukemic Cell Lines. BMC Cancer 2018, 18, 791. [Google Scholar] [CrossRef] [PubMed]
- D’Anneo, A.; Carlisi, D.; Lauricella, M.; Puleio, R.; Martinez, R.; di Bella, S.; di Marco, P.; Emanuele, S.; di Fiore, R.; Guercio, A.; et al. Parthenolide Generates Reactive Oxygen Species and Autophagy in MDA-MB231 Cells. A Soluble Parthenolide Analogue Inhibits Tumour Growth and Metastasis in a Xenograft Model of Breast Cancer. Cell Death Dis. 2013, 4, e891. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X.; Sun, J.; Yang, Y.; Li, W.; Song, J. Parthenolide Suppresses Pancreatic Cell Growth by Autophagy-Mediated Apoptosis. OncoTargets Ther. 2017, 10, 453–461. [Google Scholar] [CrossRef]
- Mohan, N.; Sorokina, E.M.; Verdeny, I.V.; Alvarez, A.S.; Lakadamyali, M. Detyrosinated Microtubules Spatially Constrain Lysosomes Facilitating Lysosome–Autophagosome Fusion. J. Cell Biol. 2019, 218, 632. [Google Scholar] [CrossRef]
- Veldhoen, R.A.; Banman, S.L.; Hemmerling, D.R.; Odsen, R.; Simmen, T.; Simmonds, A.J.; Underhill, D.A.; Goping, I.S. The Chemotherapeutic Agent Paclitaxel Inhibits Autophagy through Two Distinct Mechanisms That Regulate Apoptosis. Oncogene 2012, 32, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Yagdi Efe, E.; Mazumder, A.; Lee, J.Y.; Gaigneaux, A.; Radogna, F.; Nasim, M.J.; Christov, C.; Jacob, C.; Kim, K.W.; Dicato, M.; et al. Tubulin-Binding Anticancer Polysulfides Induce Cell Death via Mitotic Arrest and Autophagic Interference in Colorectal Cancer. Cancer Lett. 2017, 410, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Hasanain, M.; Sahai, R.; Pandey, P.; Maheshwari, M.; Choyal, K.; Gandhi, D.; Singh, A.; Singh, K.; Mitra, K.; Datta, D.; et al. Microtubule Disrupting Agent-mediated Inhibition of Cancer Cell Growth Is Associated with Blockade of Autophagic Flux and Simultaneous Induction of Apoptosis. Cell Prolif. 2020, 53, e12749. [Google Scholar] [CrossRef] [PubMed]
- Mercier, A.E.; Prudent, R.; Pepper, M.S.; De Koning, L.; Nolte, E.; Peronne, L.; Nel, M.; Lafanechère, L.; Joubert, A.M. Characterization of Signalling Pathways That Link Apoptosis and Autophagy to Cell Death Induced by Estrone Analogues Which Reversibly Depolymerize Microtubules. Molecules 2021, 26, 706. [Google Scholar] [CrossRef]
- das Mukherjee, D.; Kumar, N.M.; Tantak, M.P.; Datta, S.; Ghosh Dastidar, D.; Kumar, D.; Chakrabarti, G. NMK-BH2, a Novel Microtubule-Depolymerising Bis (Indolyl)-Hydrazide-Hydrazone, Induces Apoptotic and Autophagic Cell Death in Cervical Cancer Cells by Binding to Tubulin at Colchicine—Site. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2020, 1867, 118762. [Google Scholar] [CrossRef]
- Button, R.W.; Roberts, S.L.; Willis, T.L.; Oliver Hanemann, C.; Luo, S. Accumulation of Autophagosomes Confers Cytotoxicity. J. Biol. Chem. 2017, 292, 13599–13614. [Google Scholar] [CrossRef]
- Wei, R.J.; Lin, S.S.; Wu, W.R.; Chen, L.R.; Li, C.F.; Chen, H.D.; Chou, C.T.; Chen, Y.C.; Liang, S.S.; Chien, S.T.; et al. A Microtubule Inhibitor, ABT-751, Induces Autophagy and Delays Apoptosis in Huh-7 Cells. Toxicol. Appl. Pharmacol. 2016, 311, 88–98. [Google Scholar] [CrossRef]
- Hasanpourghadi, M.; Majid, N.A.; Mustafa, M.R. Activation of Autophagy by Stress-Activated Signals as a Cellular Self-Defense Mechanism against the Cytotoxic Effects of MBIC in Human Breast Cancer Cells in Vitro. Biochem. Pharmacol. 2018, 152, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, J.; Tian, H.; Fu, G.H. Mitochondrial ROS Accumulation Inhibiting JAK2/STAT3 Pathway Is a Critical Modulator of CYT997-Induced Autophagy and Apoptosis in Gastric Cancer. J. Exp. Clin. Cancer Res. 2020, 39, 119. [Google Scholar] [CrossRef] [PubMed]
- Dent, P.; Booth, L.; Poklepovic, A.; Hancock, J.F. Signaling Alterations Caused by Drugs and Autophagy. Cell. Signal. 2019, 64, 109416. [Google Scholar] [CrossRef] [PubMed]
- Hattori, K.; Takano, N.; Kazama, H.; Moriya, S.; Miyake, K.; Hiramoto, M.; Tsukahara, K.; Miyazawa, K. Induction of Synergistic Non-Apoptotic Cell Death by Simultaneously Targeting Proteasomes with Bortezomib and Histone Deacetylase 6 with Ricolinostat in Head and Neck Tumor Cells. Oncol. Lett. 2021, 22, 680. [Google Scholar] [CrossRef]
- Liu, P.; Xiao, J.; Wang, Y.; Song, X.; Huang, L.; Ren, Z.; Kitazato, K.; Wang, Y. Posttranslational Modification and beyond: Interplay between Histone Deacetylase 6 and Heat-Shock Protein 90. Mol. Med. 2021, 27, 1–12. [Google Scholar] [CrossRef] [PubMed]

| Tubulin PTM | Modification Sites | Enzyme | Impact on MTs |
|---|---|---|---|
| Acetylation | TubA Lys40 | ATAT1 | Resistance to mechanical bending |
| Deacetylation | TubA Lys40 | SIRT2, HDAC6 | Sensitivity to mechanical bending |
| Acetylation | TubB Lys252 | SAN acetyltransferase | MT depolymerization |
| Tyrosination | C-terminal Tyr residue | TTL | Binding of specific MAPs (e.g., MCAK121, CLIP170, dynein/dynactin/BICD2 complex) |
| Detyrosination | C-terminal Tyr residue | VASH1/2 | Associated with MT longevity and binding of specific MAPs (e.g., CENPE, kinesin-2) |
| Glutamylation/Polyglutamylation | C-terminal Glu residues | Monoglutamylases (TTLL4, -5, 7); Poliglutamylases (TTL-1, -6, -11, -13) | Fine-tuning of MT–MAP interactions |
| Deglutamylation/Polydeglutamylation | C-terminal Glu residues | CCP -1, -2, -3, -4, -5, -6 | Fine-tuning of MT–MAP interactions |
| Enzyme | Cancer Type | Experimental Approach | Impact on Cancer Cell Properties | References |
|---|---|---|---|---|
| Lysine AcetylTransferase | ||||
| ATAT1 | Lung | Overexpression | Attenuated cell migration, invasion, and metastasis | [79] |
| Lung | Overexpression | Drug resistance | [44] | |
| Lung | Downregulation | Mitotic catastrophe | [80] | |
| Breast | Downregulation | Attenuated tumor growth | [68] | |
| Colon | Downregulation | Attenuated cell invasion | [78] | |
| Lysine Deacetylase | ||||
| HDAC6 | Glioblastoma | Downregulation | Proliferation, clonogenicity and cell migration | [73,74] |
| Breast | Overexpression | Cell migration | [72] | |
| SIRT2 | Lung | Overexpression | Cell cycle arrest and apoptosis induction | [77] |
| Endometrial, renal cell carcinoma | Overexpression | Proliferation and stemness | [75,76] | |
| Tubulin-specific carboxypeptidase | ||||
| VASH2 | Ovarian | Downregulation | Drug sensitivity | [89] |
| AGBL2 | Breast, ovarian, prostate and hepatocellular carcinoma | Overexpression | Tumorigenesis and cancer progression | [94,95,96] |
| Tubulin monoglutamylase | ||||
| TTLL4 | Breast | Overexpression | Increased metastasis | [90] |
| Pancreatic | Downregulation | Increased cell proliferation | [91] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trisciuoglio, D.; Degrassi, F. The Tubulin Code and Tubulin-Modifying Enzymes in Autophagy and Cancer. Cancers 2022, 14, 6. https://doi.org/10.3390/cancers14010006
Trisciuoglio D, Degrassi F. The Tubulin Code and Tubulin-Modifying Enzymes in Autophagy and Cancer. Cancers. 2022; 14(1):6. https://doi.org/10.3390/cancers14010006
Chicago/Turabian StyleTrisciuoglio, Daniela, and Francesca Degrassi. 2022. "The Tubulin Code and Tubulin-Modifying Enzymes in Autophagy and Cancer" Cancers 14, no. 1: 6. https://doi.org/10.3390/cancers14010006
APA StyleTrisciuoglio, D., & Degrassi, F. (2022). The Tubulin Code and Tubulin-Modifying Enzymes in Autophagy and Cancer. Cancers, 14(1), 6. https://doi.org/10.3390/cancers14010006







