Prognostic and Predictive Effects of Tumor and Plasma miR-200c-3p in Locally Advanced and Metastatic Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Clinical Diagnosis and Treatment of Breast Cancer Patients
2.3. Plasma Levels of miR-200c-3p
2.4. Pathology
2.5. miRNA and mRNA Isolation and RT-qPCR Analysis
2.6. Statistical Methods
3. Results
3.1. Patients and Tumor Characteristics
3.2. Plasma Levels of miR-200c-3p in Breast Cancer Patients and Controls
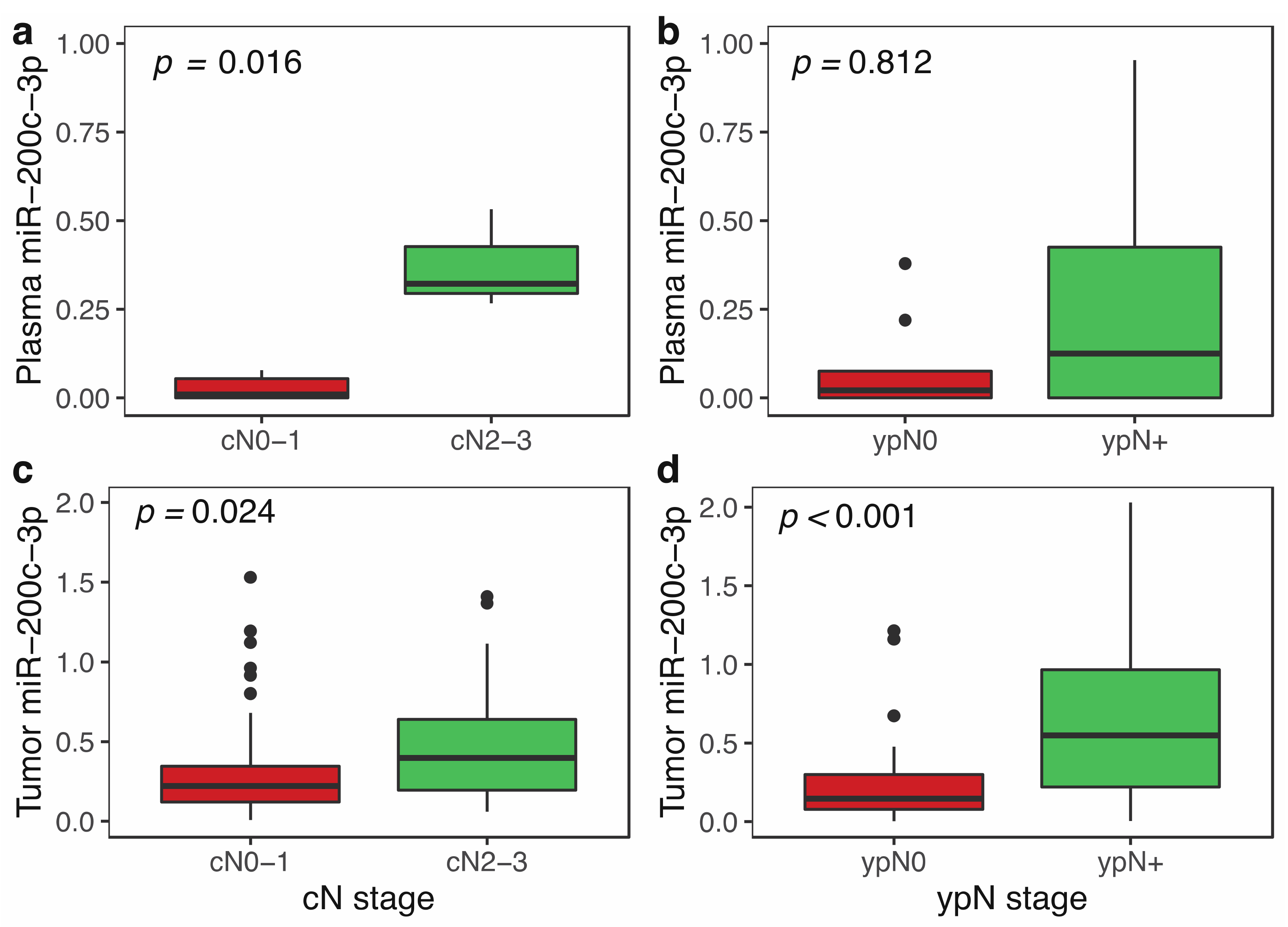
3.3. miR-200c-3p Plasma Levels and Clinicopathological Features in Locally Advanced Breast Cancer
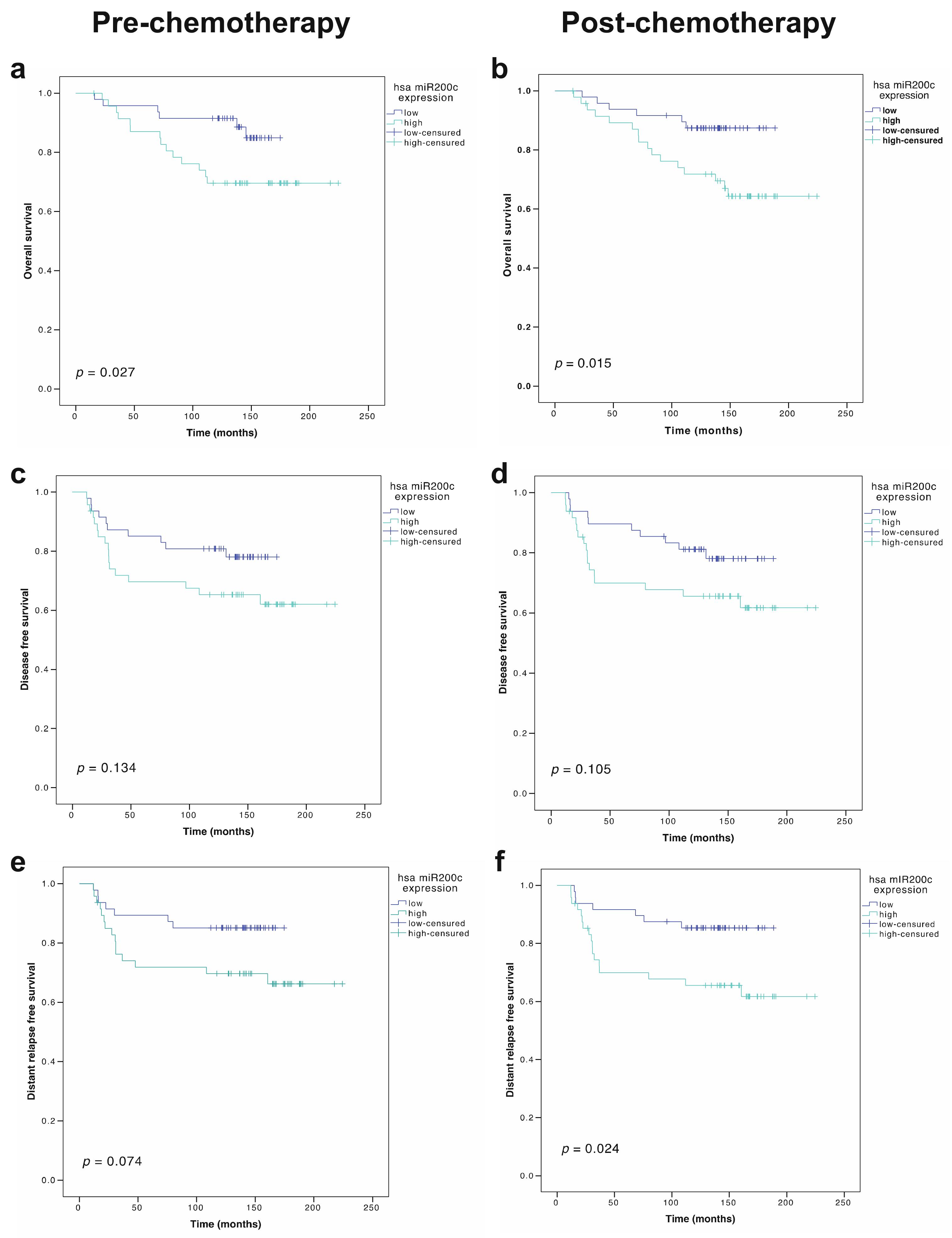
3.4. Prognostic Role of miR-200c-3p Plasma Levels
3.5. miR-200c-3p Expression in Tumor and Clinicopathological Features in Locally Advanced Breast Cancer Patients
3.6. Prognostic Role of miR-200c-3p Tumor Expression in Locally Advanced Breast Cancer Patients
3.7. Correlation between Plasma and Tumor Expression of miR-200c-3p in Locally Advanced Breast Cancer Patients
3.8. Changes in miR-200c-3p-Dependent Proliferation and EMT Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garcia-Murillas, I.; Chopra, N.; Comino-Méndez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Yu, Z.; Song, M.; Chouchane, L.; Ma, X. Functional Genomic Analysis of Breast Cancer Metastasis: Implications for Diagnosis and Therapy. Cancers 2021, 13, 3276. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Vickers, K.C. Intercellular transport of MicroRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 186–192. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Li, L.; Zhu, D.; Huang, L.; Zhang, J.; Bian, Z.; Chen, X.; Liu, Y.; Zhang, C.Y.; Zen, K. Argonaute 2 Complexes Selectively Protect the Circulating MicroRNAs in Cell-Secreted Microvesicles. PLoS ONE 2012, 7, e46957. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Ell, B.J.; Buffa, F.M.; Ibrahim, T.; Blanco, M.A.; Celià-Terrassa, T.; Mercatali, L.; Khan, Z.; Goodarzi, H.; Hua, Y.; et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011, 17, 1101–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlmann, S.; Zhang, J.D.; Schwäger, A.; Mannsperger, H.; Riazalhosseini, Y.; Burmester, S.; Ward, A.; Korf, U.; Wiemann, S.; Sahin, O. MiR-200bc/429 cluster targets PLCγ1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 2010, 29, 4297–4306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Du, R.; Zhou, L.; Xu, J.; Chen, S.; Chen, J.; Yang, X.; Liu, D.X.; Shao, Z.M.; Zhang, L.; et al. miR-200c/141 regulates breast cancer stem cell heterogeneity via targeting HIPK1/β-catenin axis. Theranostics 2018, 8, 5801–5813. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, M.; Sharifi-Zarchi, A.; Zarghami, N.; Geranpayeh, L.; Ebrahimi, M.; Alizadeh, E. Down-Regulation of miR-200c and Up-Regulation of miR-30cTarget both Stemness and Metastasis Genes in Breast Cancer. Cell J. 2020, 21, 467. [Google Scholar] [CrossRef]
- Jin, T.; Kim, H.S.; Choi, S.K.; Hwang, E.H.; Woo, J.; Ryu, H.S.; Kim, K.; Moon, A.; Moon, W.K. microRNA-200c/141 upregulates SerpinB2 to promote breast cancer cell metastasis and reduce patient survival. Oncotarget 2017, 8, 32769–32782. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; He, H.; Huang, W.; Lin, Y.; Luo, S.; Du, Q.; Duan, R. Decreased expression of MicroRNA-200 family in human breast cancer is associated with lymph node metastasis. Clin. Transl. Oncol. 2016, 18, 283–288. [Google Scholar] [CrossRef]
- Song, C.; Liu, L.; Pe, X.; Liu, X.; Yang, L.; Ye, F.; Xie, X.; Chen, J.; Tang, H.; Xie, X. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget 2015, 6, 34968–34978. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.P.; Huang, S.F.; Li, C.F.; Chien, H.T.; Chen, S.C. Differential microRNA expression in breast cancer with different onset age. PLoS ONE 2018, 13, e0191195. [Google Scholar] [CrossRef] [Green Version]
- Fontana, A.; Barbano, R.; Dama, E.; Pasculli, B.; Rendina, M.; Morritti, M.G.; Melocchi, V.; Castelvetere, M.; Valori, V.M.; Ravaioli, S.; et al. Combined analysis of miR-200 family and its significance for breast cancer. Sci. Rep. 2021, 11, 2980. [Google Scholar] [CrossRef]
- Madhavan, D.; Zucknick, M.; Wallwiener, M.; Cuk, K.; Modugno, C.; Scharpff, M.; Schott, S.; Heil, J.; Turchinovich, A.; Yang, R.; et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin. Cancer Res. 2012, 18, 5972–5982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadaki, C.; Stoupis, G.; Tsalikis, L.; Monastirioti, A.; Papadaki, M.; Maliotis, N.; Stratigos, M.; Mastrostamatis, G.; Mavroudis, D.; Agelaki, S. Circulating miRNAs as a marker of metastatic disease and prognostic factor in metastatic breast cancer. Oncotarget 2019, 10, 966–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhavan, D.; Peng, C.; Wallwiener, M.; Zucknick, M.; Nees, J.; Schott, S.; Rudolph, A.; Riethdorf, S.; Trumpp, A.; Pantel, K.; et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis 2016, 37, 461–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Sun, J.; Lu, Y.; Liu, Y.; Cao, H.; Zhang, H.; Calin, G. MiR-200 family and cancer: From a meta-analysis view. Mol. Asp. Med. 2019, 70, 57–71. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef]
- Wang, J.; Yang, M.; Li, Y.; Han, B. The Role of MicroRNAs in the Chemoresistance of Breast Cancer. Drug Dev. Res. 2015, 76, 368–374. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, Y.; Chen, L.; Xu, X.; Zhang, X.; Wang, B.; Min, L.; Liu, W. miRNA-200c increases the sensitivity of breast cancer cells to doxorubicin through the suppression of E-cadherin-mediated PTEN/Akt signaling. Mol. Med. Rep. 2013, 7, 1579–1584. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Chang, E.; Lee, E.; Lee, H.; Kang, H.; Chun, K.; Woo, Y.; Kong, H.; Ko, J.; Suzuki, H.; et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014, 74, 7573–7582. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.; Zheng, W.; Liu, J.; Tang, Q.; Wang, S.; Yang, X. MiR-34a-5p/PD-L1 axis regulates cisplatin chemoresistance of ovarian cancer cells. Neoplasma 2020, 67, 93–101. [Google Scholar] [CrossRef]
- Knezevic, J.; Pfefferle, A.D.; Petrovic, I.; Greene, S.B.; Perou, C.M.; Rosen, J.M. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene 2015, 34, 5997–6006. [Google Scholar] [CrossRef] [Green Version]
- Alam, F.; Mezhal, F.; El Hasasna, H.; Nair, V.; Aravind, S.; Saber Ayad, M.; El-Serafi, A.; Abdel-Rahman, W. The role of p53-microRNA 200-Moesin axis in invasion and drug resistance of breast cancer cells. Tumor Biol. 2017, 39, 1010428317714634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roncati, L.; Barbolini, G.; Gatti, A.; Pusiol, T.; Piscioli, F.; Maiorana, A. The Uncontrolled Sialylation is Related to Chemoresistant Metastatic Breast Cancer. Pathol. Oncol. Res. 2016, 22, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Chuthapisith, S.; Eremin, J.; El-Sheemey, M.; Eremin, O. Breast cancer chemoresistance: Emerging importance of cancer stem cells. Surg. Oncol. 2010, 19, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, W.; Liu, C.; Li, G. miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Sci. Rep. 2019, 9, 18844. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, M.; Sun, H.; Zhu, T.; Wang, X. Downregulation of LINC00894-002 Contributes to Tamoxifen Resistance by Enhancing the TGF-β Signaling Pathway. Biochemistry 2018, 83, 603–611. [Google Scholar] [CrossRef]
- Satyarsa, A.B.S.; Supadmanaba, I.; Adiputra, P. The role of microRNA-200c in chemoresistant breast cancer. Indones. J. Health Sci. 2020, 4, 74–83. [Google Scholar] [CrossRef]
- McGrogan, B.T.; Gilmartin, B.; Carney, D.N.; McCann, A. Taxanes, microtubules and chemoresistant breast cancer. Biochim. Biophys. Acta-Rev. Cancer 2008, 1785, 96–132. [Google Scholar] [CrossRef]
- Buzdar, A.U. Preoperative chemotherapy treatment of breast cancer—A review. Cancer 2007, 110, 2394–2407. [Google Scholar] [CrossRef]
- Schwartz, L.H.; Litière, S.; de Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Update and Clarification: From the RECIST Committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.S.; Taylor, J.; Valentine, H.R.; Irlam, J.J.; Eustace, A.; Hoskin, P.J.; Miller, C.J.; West, C.M.L. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br. J. Cancer 2012, 107, 684–694. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Zhang, W.; Li, B.; Stringer-Reasor, E.; Chu, C.; Sun, L.; Bae, S.; Chen, D.; Wei, S.; Jiao, K.; et al. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors. Genes Dev. 2009, 23, 1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, M.; Hamar, P.; Guo, C.; Basar, E.; Perdigão-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haakensen, V.; Nygaard, V.; Greger, L.; Aure, M.; Fromm, B.; Bukholm, I.; Lüders, T.; Chin, S.; Git, A.; Caldas, C.; et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int. J. Cancer 2016, 139, 1117–1128. [Google Scholar] [CrossRef]
- Damiano, V.; Brisotto, G.; Borgna, S.; di Gennaro, A.; Armellin, M.; Perin, T.; Guardascione, M.; Maestro, R.; Santarosa, M. Epigenetic silencing of miR-200c in breast cancer is associated with aggressiveness and is modulated by ZEB1. Genes Chromosom. Cancer 2017, 56, 147–158. [Google Scholar] [CrossRef]
- Piasecka, D.; Braun, M.; Kordek, R.; Sadej, R.; Romanska, H. MicroRNAs in regulation of triple-negative breast cancer progression. J. Cancer Res. Clin. Oncol. 2018, 144, 1401–1411. [Google Scholar] [CrossRef] [Green Version]
- Gulei, D.; Mehterov, N.; Ling, H.; Stanta, G.; Braicu, C.; Berindan-Neagoe, I. The “good-cop bad-cop” TGF-beta role in breast cancer modulated by non-coding RNAs. Biochim. Biophys. Acta-Gen. Subj. 2017, 1861, 1661–1675. [Google Scholar] [CrossRef]
- Perdigão-Henriques, R.; Petrocca, F.; Altschuler, G.; Thomas, M.; Le, M.; Tan, S.; Hide, W.; Lieberman, J. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 2016, 35, 158–172. [Google Scholar] [CrossRef]
- D’Ippolito, E.; Plantamura, I.; Bongiovanni, L.; Casalini, P.; Baroni, S.; Piovan, C.; Orlandi, R.; Gualeni, A.; Gloghini, A.; Rossini, A.; et al. miR-9 and miR-200 Regulate PDGFRβ-Mediated Endothelial Differentiation of Tumor Cells in Triple-Negative Breast Cancer. Cancer Res. 2016, 76, 5562–5572. [Google Scholar] [CrossRef] [Green Version]
- Noman, M.; Janji, B.; Abdou, A.; Hasmim, M.; Terry, S.; Tan, T.; Mami-Chouaib, F.; Thiery, J.; Chouaib, S. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 2017, 6, e1263412. [Google Scholar] [CrossRef]
- Gollavilli, P.N.; Parma, B.; Siddiqui, A.; Yang, H.; Ramesh, V.; Napoli, F.; Schwab, A.; Natesan, R.; Mielenz, D.; Asangani, I.A.; et al. The role of miR-200b/c in balancing EMT and proliferation revealed by an activity reporter. Oncogene 2021, 40, 2309–2322. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cid, L.; Pons, M.; Lozano, J.J.; Rubio, N.; Guerra-Rebollo, M.; Soriano, A.; Paris-Coderch, L.; Segura, M.F.; Fueyo, R.; Arguimbau, J.; et al. MicroRNA-200, associated with metastatic breast cancer, promotes traits of mammary luminal progenitor cells. Oncotarget 2017, 8, 83384–83406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala de la Peña, F.; Andrés, R.; Garcia-Sáenz, J.A.; Manso, L.; Margelí, M.; Dalmau, E.; Pernas, S.; Prat, A.; Servitja, S.; Ciruelos, E. SEOM clinical guidelines in early stage breast cancer (2018). Clin. Transl. Oncol. 2019, 21, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E.; ESMO Guidelines Committee. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef] [Green Version]
- Nami, B.; Wang, Z. Genetics and expression profile of the Tubulin gene superfamily in breast cancer subtypes and its relation to taxane resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Saura, C.; Tseng, L.-M.; Chan, S.; Chacko, R.T.; Campone, M.; Manikhas, A.; Nag, S.M.; Leichman, C.G.; Dasappa, L.; Fasching, P.A.; et al. Neoadjuvant Doxorubicin/Cyclophosphamide Followed by Ixabepilone or Paclitaxel in Early Stage Breast Cancer and Evaluation of βIII-Tubulin Expression as a Predictive Marker. Oncologist 2013, 18, 787–794. [Google Scholar] [CrossRef] [Green Version]
- Hatse, S.; Brouwers, B.; Dalmasso, B.; Laenen, A.; Kenis, C.; Schöffski, P.; Wildiers, H. Circulating MicroRNAs as easy-to-measure aging biomarkers in older breast cancer patients: Correlation with chronological age but not with fitness/frailty status. PLoS ONE 2014, 9, e110644. [Google Scholar] [CrossRef]


| Characteristics | Category | n (%) |
|---|---|---|
| Age, median, range | 56 (21–79) | |
| Histological type | Ductal | 164 (95.9) |
| Lobular | 5 (2.9) | |
| Other | 2 (1.2) | |
| Histological grade | GI | 8 (4.7) |
| GII | 64 (37.4) | |
| GIII | 84 (49.1) | |
| N/A | 15 (8.8) | |
| Tumor phenotype | HR+ HER2− | 78 (45.6) |
| HR+ HER2+ | 34 (19.9) | |
| HR− HER2+ | 16 (9.3) | |
| Triple negative | 39 (22.8) | |
| N/A | 4 (2.3) | |
| cT | cT1-2 | 84 (49.2) |
| cT3 | 72 (42.1) | |
| cT4 | 14 (8.2) | |
| cTx | 1 (0.6) | |
| cN | cN0 | 54 (31.6) |
| cN1 | 59 (34.5) | |
| cN2 | 34 (19.9) | |
| cN3 | 22 (12.9) | |
| cNx | 2 (1.2) | |
| Stage | IIA | 35 (20.5) |
| IIB | 48 (28.1) | |
| IIIA | 57 (33.3) | |
| IIIB | 9 (5.3) | |
| IIIC | 22 (12.9) | |
| Pathologic complete response (pCR) | pCR (ypT0/is ypN0) | 32 (18.9) |
| pCR breast (ypT0/ypTis) | 36 (21.1) |
| Characteristics | Category | n (%) |
|---|---|---|
| Age, median (range) | 59 (31–78) | |
| Histological Type | Ductal | 35 (83,3) |
| Lobular | 7 (16,7) | |
| Stage at diagnosis | M0 (recurrence) | 9 (21,4) |
| M1 (de novo metastatic disease) | 33 (78,6) | |
| Estrogen Receptor | Negative | 6 (13,4) |
| Positive | 36 (85,7) | |
| HER2-neu | Negative | 33 (78,6) |
| Positive | 9 (21,4) | |
| Immunohistochemical subtype | HR+ HER2− | 31 (73,8) |
| HR+ HER2+ | 6 (14,3) | |
| HR− HER2+ | 3 (7,1) | |
| TNBC | 2 (4,8) | |
| ECOG | 0 | 13 (30.9) |
| 1 | 22 (52.4) | |
| 2 | 4 (9.5) | |
| 3 | 2 (4,7) | |
| Metastatic site | Bone+/-Soft Tissues | 23 (54.8) |
| Visceral | 7 (16.7) | |
| Visceral+Bone+/-Soff Tissues | 12 (28.5) | |
| CNS | 1 (2.4) | |
| N/A | 1 (2.4) | |
| Visceral Metastases | No | 23 (54.8) |
| Yes | 19 (45.2) | |
| Bone Metastases | No | 7 (16.7) |
| Yes | 35 (83.3) | |
| Treatment | Endocrine-based therapy | 10 (23.8) |
| Chemotherapy | 18 (42.9) | |
| Chemotherapy + biological | 13 (30.9) | |
| No treatment | 1 (2.4) |
| DFS | DRFS | OS | ||||
|---|---|---|---|---|---|---|
| UNIVARIATE ANALYSIS | HR (95% CI) | p-Value * | HR (95% CI) | p-Value * | HR (95% CI) | p-Value * |
| Grade 3 | 1.58 (0.74–3.38) | 0.237 | 1.07 (0.52–2.19) | 0.849 | 1.98 (0.83–4.74) | 0.125 |
| Stage III (vs. stage II) | 1.59 (0.77–3.27) | 0.211 | 2.42 (1.13–5.21) | 0.024 | 1.82 (0.81–4.08) | 0.147 |
| cN2-3 | 2.04 (2.01–4.13) | 0.047 | 3.12 (1.55–6.27) | 0.001 | 2.54 (1.18–5.51) | 0.018 |
| Estrogen receptor positivity | 0.69 (0.34–1.41) | 0.311 | 1.27 (0.59–2.76) | 0.537 | 0.70 (0.32–1.53) | 0.376 |
| HER2 amplification | 0.99 (0.44–2.22) | 0.988 | 1.02 (0.46–2.27) | 0.965 | 0.91 (0.36–2.28) | 0.842 |
| Pathologic complete response (pCR) | 0.27 (0.06–1.15) | 0.076 | 0.14 (0.02–1.00) | 0.050 | 0.16 (0.08–1.52) | 0.163 |
| miR-200c-3p (high vs. low), tumor, pre-NCT | 1.82 (0.83–3.99) | 0.134 | 2.27 (0.92–5.60) | 0.074 | 2.76 (1.07–7.11) | 0.027 |
| miR-200c-3p (high vs. low), tumor, post-NCT | 1.914(0.87–4.19) | 0.105 | 2.77 (1.14–6.69) | 0.024 | 3.15 (1.25–7.97) | 0.015 |
| MULTIVARIATE MODEL (with pre-NCT miR-200c-3p tumor expression) | ||||||
| pCR | 0.21 (0.05–0.92) | 0.039 | 0.12 (0.01–0.89) | 0.038 | 0.06 (0.01–0.49) | 0.009 |
| ER positivity | ----- | ----- | ----- | ----- | 0.54 (0.21–1.40) | 0.206 |
| Grade 3 | ----- | ----- | ----- | ----- | 2.35 (0.85–6.54) | 0.101 |
| cN2-3 | 2.56 (1.19–5.52) | 0.017 | 3.43 (1.46–8.00) | 0.005 | 3.01 (1.21–7.52) | 0.018 |
| miR-200c-3p (high vs. low) pre-NCT | 1.73 (0.79–3.79) | 0.170 | 2.15 (0.87–5.29) | 0.097 | 3.98 (1.49–10.60) | 0.006 |
| MULTIVARIATE MODEL (with post-NCT miR-200c-3p tumor expression) | ||||||
| pCR | 0.30 (0.07–1.33) | 0.113 | 0.18 (0.02–1.39) | 0.101 | 0.46 (0.10–2.06) | 0.310 |
| cN2-3 | 2.50 (1.16–5.41) | 0.020 | 2.68 (1.18–6.07) | 0.018 | 2.44 (1.08–5.50) | 0.032 |
| miR-200c-3p (high vs. low) post-NCT | 1.52 (0.67–3.41) | 0.313 | 2.06 (0.83–5.11) | 0.118 | 2.63 (1.01–6.86) | 0.049 |
| Pre-Chemotherapy | Post-Chemotherapy | ||||
|---|---|---|---|---|---|
| Total LABC | HR+ HER2− | HER2+ | TNBC | Total LABC | |
| n = 105 | n = 52 | n = 28 | n = 25 | n = 77 | |
| MKI67 | |||||
| Rho | 0.49 | 0.44 | 0.41 | 0.66 | 0.38 |
| p | <0.001 | 0.0067 | 0.055 | 0.004 | 0.003 |
| MYBL2 | |||||
| Rho | 0.46 | 0.47 | 0.32 | 0.64 | 0.38 |
| p | <0.001 | 0.003 | 0.142 | 0.006 | 0.003 |
| CCNB1 | |||||
| Rho | 0.56 | 0.62 | 0.32 | 0.71 | 0.23 |
| p | <0.001 | <0.001 | 0.139 | 0.001 | 0.075 |
| CDH1 | |||||
| Rho | 0.20 | 0.41 | 0.10 | 0.21 | 0.32 |
| p | 0.053 | 0.007 | 0.635 | 0.341 | 0.006 |
| VIM | |||||
| Rho | −0.28 | −0.42 | −0.16 | −0.08 | −0.35 |
| p | 0.006 | 0.004 | 0.465 | 0.713 | 0.002 |
| ESR1 | |||||
| Rho | 0.14 | 0.31 | −0.01 | 0.10 | −0.03 |
| p | 0.185 | 0.008 | 0.953 | 0.689 | 0.792 |
| TUBB3 | |||||
| Rho | 0.01 | 0.16 | −0.44 | 0.19 | 0.03 |
| p | 0.937 | 0.370 | 0.047 | 0.446 | 0.884 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro-Manzano, E.; Luengo-Gil, G.; González-Conejero, R.; García-Garre, E.; García-Martínez, E.; García-Torralba, E.; Chaves-Benito, A.; Vicente, V.; Ayala de la Peña, F. Prognostic and Predictive Effects of Tumor and Plasma miR-200c-3p in Locally Advanced and Metastatic Breast Cancer. Cancers 2022, 14, 2390. https://doi.org/10.3390/cancers14102390
Navarro-Manzano E, Luengo-Gil G, González-Conejero R, García-Garre E, García-Martínez E, García-Torralba E, Chaves-Benito A, Vicente V, Ayala de la Peña F. Prognostic and Predictive Effects of Tumor and Plasma miR-200c-3p in Locally Advanced and Metastatic Breast Cancer. Cancers. 2022; 14(10):2390. https://doi.org/10.3390/cancers14102390
Chicago/Turabian StyleNavarro-Manzano, Esther, Ginés Luengo-Gil, Rocío González-Conejero, Elisa García-Garre, Elena García-Martínez, Esmeralda García-Torralba, Asunción Chaves-Benito, Vicente Vicente, and Francisco Ayala de la Peña. 2022. "Prognostic and Predictive Effects of Tumor and Plasma miR-200c-3p in Locally Advanced and Metastatic Breast Cancer" Cancers 14, no. 10: 2390. https://doi.org/10.3390/cancers14102390







