Added Value of Viscoelasticity for MRI-Based Prediction of Ki-67 Expression of Hepatocellular Carcinoma Using a Deep Learning Combined Radiomics (DLCR) Model
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Conventional MRI (cMRI)
2.3. Tomoelastography
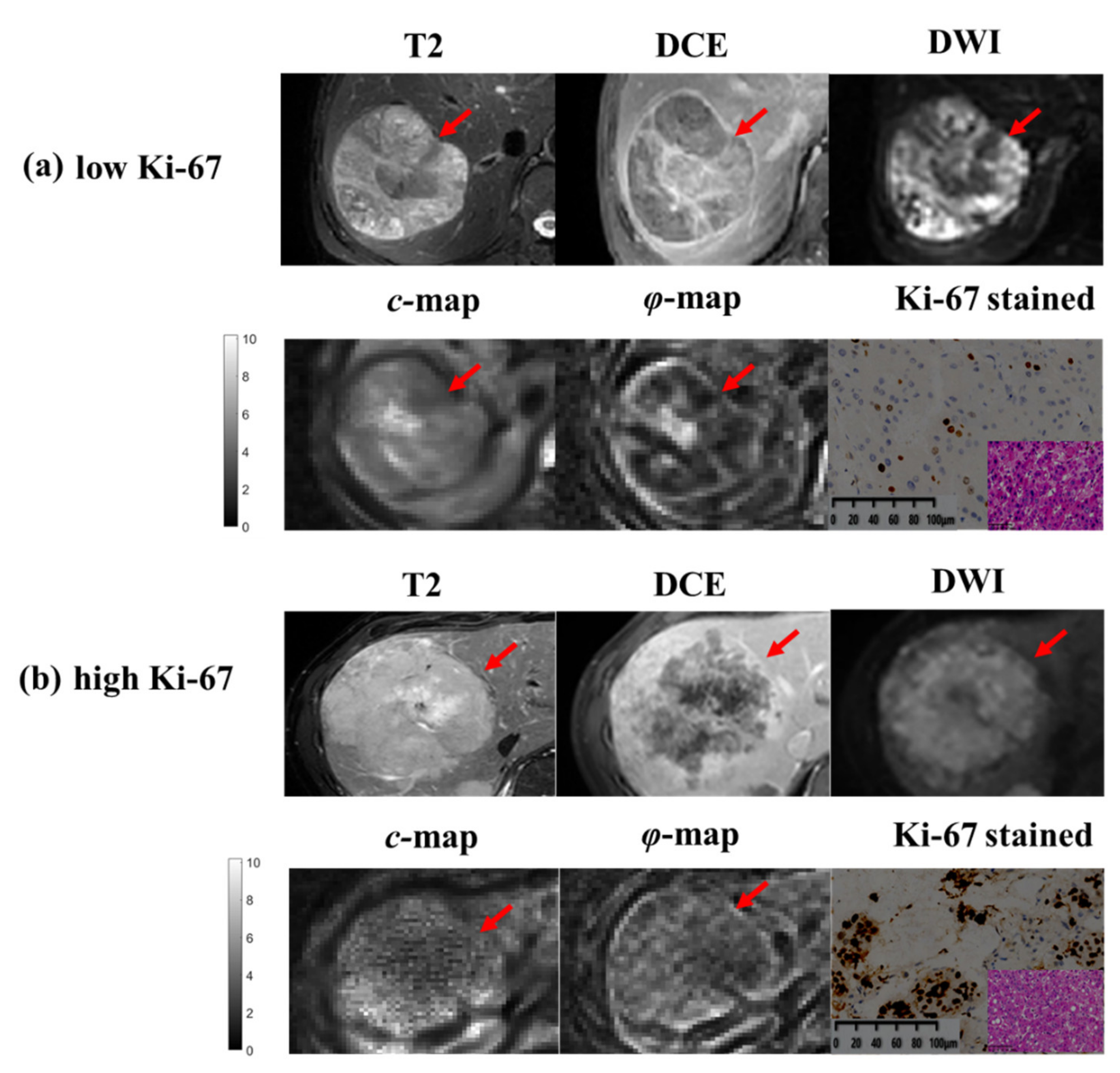
2.4. Histopathological Analysis
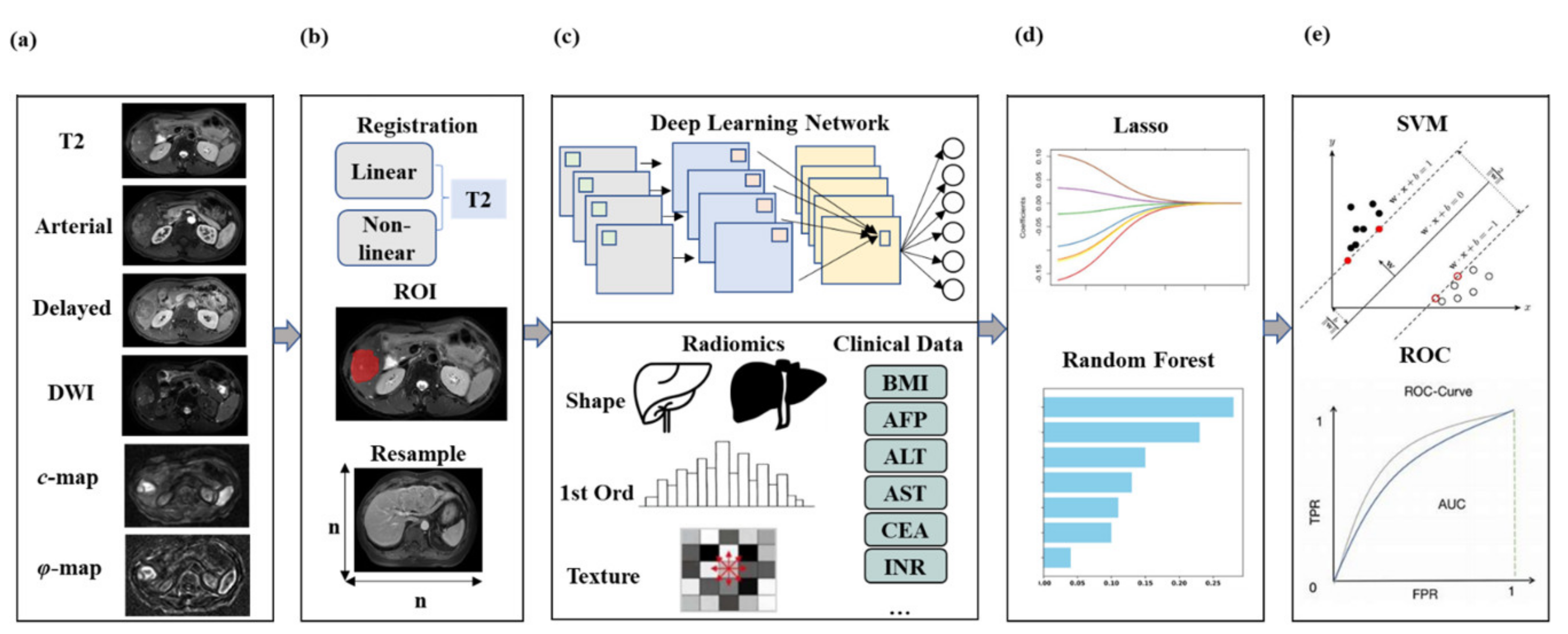
2.5. Image Preprocessing
2.6. cMRI-Based DLCR Model
2.7. cMRI-Based DLCR Model with Tomoelastography
2.8. Statistical Analysis
3. Results
3.1. Demographics and Clinical Characteristics
3.2. Optimization of cMRI-Based DLCR Models
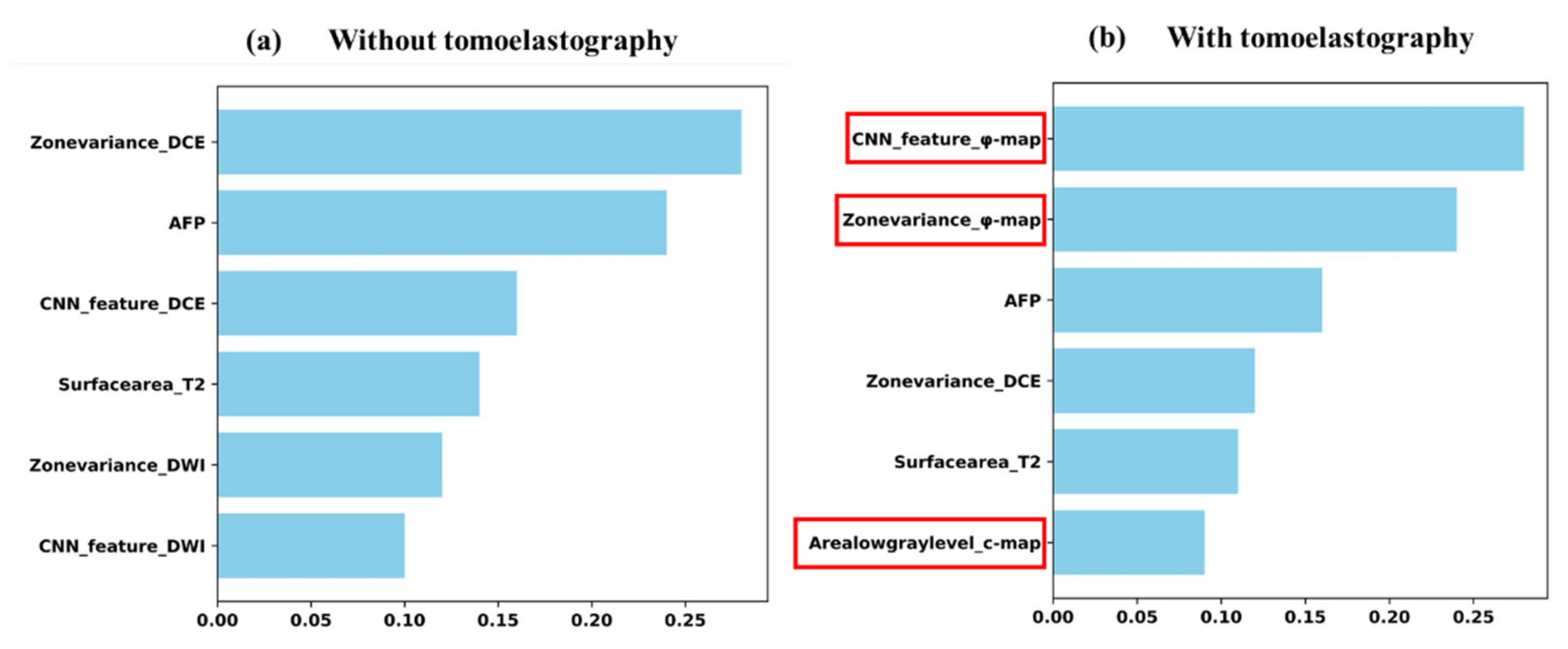
3.3. Comparison of DLCR Models with/without Tomoelastography
3.4. Contribution of Predictive Efficacy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktas, S.; Karakayali, H.; Moray, G.; Ozdemir, H.; Haberal, M. Effects of Risk Factors and Ki-67 on Rates of Recurrence on Patients Who Have Undergone Liver Transplant for Hepatocellular Carcinoma. Transplant. Proc. 2011, 43, 3807–3812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, R.; Zou, R.-H.; Shen, J.-X.; Wei, W.; Li, S.-H.; Ouyang, H.-Y.; Zhu, H.-B.; Xu, L.; Lao, X.-M.; et al. Efficacy and safety of preoperative chemoembolization for resectable hepatocellular carcinoma with portal vein invasion: A prospective comparative study. Eur. Radiol. 2016, 26, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, J.; Ding, M.; Xu, K.; Li, L.; Mao, L.; Zheng, J. Ki67 targeted strategies for cancer therapy. Clin. Transl. Oncol. 2018, 20, 570–575. [Google Scholar] [CrossRef]
- Sung, Y.S.; Park, B.; Park, H.J.; Lee, S.S. Radiomics and deep learning in liver diseases. J. Gastroenterol. Hepatol. 2021, 36, 561–568. [Google Scholar] [CrossRef]
- Hu, X.X.; Yang, Z.X.; Liang, H.Y.; Ding, Y.; Grimm, R.; Fu, C.X.; Liu, H.; Yan, X.; Ji, Y.; Zeng, M.S.; et al. Whole-tumor MRI histogram analyses of hepatocellular carcinoma: Correlations with Ki-67 labeling index. J. Magn. Reson. Imaging 2017, 46, 383–392. [Google Scholar] [CrossRef]
- Fan, Y.; Yu, Y.; Wang, X.; Hu, M.; Hu, C. Radiomic analysis of Gd-EOB-DTPA-enhanced MRI predicts Ki-67 expression in hepatocellular carcinoma. BMC Med. Imaging 2021, 21, 100. [Google Scholar] [CrossRef]
- Li, Y.; Yan, C.; Weng, S.; Shi, Z.; Sun, H.; Chen, J.; Xu, X.; Ye, R.; Hong, J. Texture analysis of multi-phase MRI images to detect expression of Ki67 in hepatocellular carcinoma. Clin. Radiol. 2019, 74, 813.e19–813.e27. [Google Scholar] [CrossRef]
- Sack, I.; Beierbach, B.; Hamhaber, U.; Klatt, D.; Braun, J. Non-invasive measurement of brain viscoelasticity using magnetic resonance elastography. NMR Biomed. 2008, 21, 265–271. [Google Scholar] [CrossRef]
- Sack, I.; Schaeffter, T. Quantification of Biophysical Parameters in Medical Imaging; Springer: Berlin, Germany, 2018; pp. 5–8. [Google Scholar]
- Shi, Y.; Qi, Y.-F.; Lan, G.-Y.; Wu, Q.; Ma, B.; Zhang, X.-Y.; Ji, R.-Y.; Ma, Y.-J.; Hong, Y. Three-Dimensional MR Elastography Depicts Liver Inflammation, Fibrosis, and Portal Hypertension in Chronic Hepatitis B or C. Radiology 2021, 301, 154–162. [Google Scholar] [CrossRef]
- Shahryari, M.; Tzschätzsch, H.; Guo, J.; Marticorena Garcia, S.R.; Böning, G.; Fehrenbach, U.; Stencel, L.; Asbach, P.; Hamm, B.; Käs, J.A.; et al. Tomoelastography Distinguishes Noninvasively between Benign and Malignant Liver Lesions. Cancer Res. 2019, 79, 5704–5710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manduca, A.; Bayly, P.J.; Ehman, R.L.; Kolipaka, A.; Royston, T.J.; Sack, I.; Sinkus, R.; Van Beers, B.E. MR elastography: Principles, guidelines, and terminology. Magn. Reson. Med. 2021, 85, 2377–2390. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; White, R.D.; Kolipaka, A. Advances and Future Direction of Magnetic Resonance Elastography. Top. Magn. Reson. Imaging 2018, 27, 363–384. [Google Scholar] [CrossRef]
- Li, M.; Guo, J.; Hu, P.; Jiang, H.; Chen, J.; Hu, J.; Asbach, P.; Sack, I.; Li, W. Tomoelastography Based on Multifrequency MR Elastography for Prostate Cancer Detection: Comparison with Multiparametric MRI. Radiology 2021, 299, E259. [Google Scholar] [CrossRef] [PubMed]
- Marticorena Garcia, S.R.; Zhu, L.; Gültekin, E.; Schmuck, R.; Burkhardt, C.; Bahra, M.; Geisel, D.; Shahryari, M.; Braun, J.; Hamm, B.; et al. Tomoelastography for Measurement of Tumor Volume Related to Tissue Stiffness in Pancreatic Ductal Adenocarcinomas. Investig. Radiol. 2020, 55, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zormpas-Petridis, K.; Boult, J.K.R.; Reeves, E.L.; Heindl, A.; Vinci, M.; Lopes, F.; Cummings, C.; Springer, C.J.; Chesler, L.; et al. Investigating the Contribution of Collagen to the Tumor Biomechanical Phenotype with Noninvasive Magnetic Resonance Elastography. Cancer Res. 2019, 79, 5874–5883. [Google Scholar] [CrossRef] [Green Version]
- Gültekin, E.; Wetz, C.; Braun, J.; Geisel, D.; Furth, C.; Hamm, B.; Sack, I.; Garcia, S.R.M. Added Value of Tomoelastography for Characterization of Pancreatic Neuroendocrine Tumor Aggressiveness Based on Stiffness. Cancers 2021, 13, 5185. [Google Scholar] [CrossRef]
- Zhu, L.; Guo, J.; Jin, Z.; Xue, H.; Dai, M.; Zhang, W.; Sun, Z.; Xu, J.; Garcia, S.R.M.; Asbach, P.; et al. Distinguishing pancreatic cancer and autoimmune pancreatitis with in vivo tomoelastography. Eur. Radiol. 2021, 31, 3366–3374. [Google Scholar] [CrossRef]
- Tustison, N.J.; Cook, P.A.; Holbrook, A.J.; Johnson, H.J.; Muschelli, J.; Devenyi, G.A.; Duda, J.T.; Das, S.R.; Cullen, N.C.; Gillen, D.L.; et al. The ANTsX ecosystem for quantitative biological and medical imaging. Sci. Rep. 2021, 11, 9068. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-ResNet and the impact of residual connections on learning of Presentation. In Proceedings of the AAAI Conference on AI, Phoenix, AZ, USA, 12–17 February 2016. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Eurhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions of Presentation. In Proceedings of the IEEE Computer Society, Beijing, China, 23–25 October 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition of Presentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition of Presentation. In Proceedings of the Computer Science, Berkeley, CA, USA, 4 September 2014. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions of Presentation. In Proceedings of the CVPR, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streitberger, K.-J.; Lilaj, L.; Schrank, F.; Braun, J.; Hoffmann, K.-T.; Reiss-Zimmermann, M.; Käs, J.A.; Sack, I. How tissue fluidity influences brain tumor progression. Proc. Natl. Acad. Sci. USA 2020, 117, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.J.; Youk, J.H.; Gil Kim, B.; Jung, W.H.; Cho, N.H. Lymphangiogenesis in Breast Cancer Correlates with Matrix Stiffness on Shear-Wave Elastography. Yonsei Med. J. 2016, 57, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.J.; Kim, H.H.; Cha, J.H.; Shin, H.J.; Kim, H.; Chae, E.Y.; Hong, M.J. Predicting Prognostic Factors of Breast Cancer Using Shear Wave Elastography. Ultrasound Med. Biol. 2014, 40, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.M.; Wang, J.; Chandan, V.S.; Glaser, K.J.; Roberts, L.R.; Ehman, R.L.; Venkatesh, S.K. MR elastography of hepatocellular carcinoma: Correlation of tumor stiffness with histopathology features—Preliminary findings. Magn. Reson. Imaging 2017, 37, 41–45. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Yin, Z.; Hu, B.; Guo, N.; Zhang, L.; Zhang, L.; Zhu, J.; Chen, W.; Yin, M.; Chen, J.; et al. MR Elastography-Based Shear Strain Mapping for Assessment of Microvascular Invasion in Hepatocellular Carcinoma. Eur. Radiol. 2022. [Google Scholar] [CrossRef]
- Harkos, C.; Svensson, S.F.; Emblem, K.E.; Stylianopoulos, T. Inducing Biomechanical Heterogeneity in Brain Tumor Modeling by MR Elastography: Effects on Tumor Growth, Vascular Density and Delivery of Therapeutics. Cancers 2022, 14, 884. [Google Scholar] [CrossRef]
- Pagé, G.; Tardieu, M.; Gennisson, J.-L.; Besret, L.; Garteiser, P.; Van Beers, B.E. Tumor Solid Stress: Assessment with MR Elastography under Compression of Patient-Derived Hepatocellular Carcinomas and Cholangiocarcinomas Xenografted in Mice. Cancers 2021, 13, 1891. [Google Scholar] [CrossRef]
- Zhao, Y.-F.; Xiong, X.; Chen, K.; Tang, W.; Yang, X.; Shi, Z.-R. Evaluation of the Therapeutic Effect of Adjuvant Transcatheter Arterial Chemoembolization Based on Ki67 After Hepatocellular Carcinoma Surgery. Front. Oncol. 2021, 11, 605234. [Google Scholar] [CrossRef]
- Murakami, K.; Kasajima, A.; Kawagishi, N.; Ohuchi, N.; Sasano, H. Microvessel density in hepatocellular carcinoma: Prognostic significance and review of the previous published work. Hepatol. Res. 2015, 45, 1185–1194. [Google Scholar] [CrossRef]
- Centonze, L.; De Carlis, R.; Vella, I.; Carbonaro, L.; Incarbone, N.; Palmieri, L.; Sgrazzutti, C.; Ficarelli, A.; Valsecchi, M.G.; Iacono, U.D.; et al. From LI-RADS Classification to HCC Pathology: A Retrospective Single-Institution Analysis of Clinico-Pathological Features Affecting Oncological Outcomes after Curative Surgery. Diagnostics 2022, 12, 160. [Google Scholar] [CrossRef] [PubMed]

| Variable | Total (n = 108) | Training (n = 87) | Testing (n = 21) | p-Value |
|---|---|---|---|---|
| Age (years) | 59.57 ± 10.97 | 59.38 ± 11.13 | 60.38 ± 10.20 | 0.19 |
| Sex, n (%) | 93 (87.03%) | 76 (88.51%) | 17 (80.95%) | 0.83 |
| BMI (kg/m2) | 23.81 ± 3.01 | 23.63 ± 2.94 | 24.59 ± 3.15 | 0.45 |
| Etiology, No. | -- | - | - | <0.05 |
| Hepatitis B virus | 83 | 64 | 18 | - |
| Hepatitis C virus | 4 | 3 | 1 | - |
| Others | 21 | 19 | 2 | - |
| AFP level (mg/mL) | - | - | - | <0.05 |
| <20 | 50 | 40 | 10 | - |
| ≥20 | 58 | 47 | 11 | - |
| Platelet count (×109/L) | 143.06 ± 64.93 | 141.81 ± 65.41 | 148.40 ± 62.56 | 0.39 |
| Prealbumin level (mg/L) | 195.99 ± 60.55 | 195.74 ± 62.60 | 197.05 ± 50.78 | 0.31 |
| ALT level (IU/L) | 41.08 ± 60.21 | 41.23 ± 65.27 | 40.45 ± 29.96 | 0.27 |
| AST level (IU/L) | 45.25 ± 64.87 | 46.37 ± 71.14 | 40.45 ± 22.68 | 0.25 |
| Total bilirubin (μmol/L) | 18.95 ± 12.09 | 18.52 ± 8.02 | 20.77 ± 22.23 | 0.16 |
| Direct bilirubin (μmol/L) | 3.89 ± 2.96 | 3.90 ± 2.72 | 3.83 ± 3.84 | 0.35 |
| Albumin level (g/L) | 34.77 ± 11.81 | 39.86 ± 5.87 | 40.30 ± 7.70 | 0.27 |
| Prothrombin time (s) | 12.54 ± 1.13 | 12.14 ± 1.36 | 12.89 ± 0.91 | 0.44 |
| INR | 1.04 ± 0.12 | 1.03 ± 0.12 | 1.10 ± 0.08 | 0.57 |
| Ki-67(%) | 27.28 ± 20.47 | 27.55 ± 19.52 | 26.14 ± 23.98 | 0.26 |
| Variable | Training (n = 87) | p Value | Validation (n = 21) | p Value | Testing (n = 43) | p Value | |||
|---|---|---|---|---|---|---|---|---|---|
| High Ki-67 (n = 40) | Low Ki-67 (n = 47) | High Ki-67 (n = 9) | Low Ki-67 (n = 12) | High Ki-67 (n = 17) | Low Ki-67 (n = 26) | ||||
| Age (years) | 56.8 ± 11.5 | 65.0 ± 7.9 | 0.07 | 61.8 ± 9.5 | 59.3 ± 10.6 | 0.09 | 59.4 ± 11.7 | 60.7 ± 10.5 | 0.17 |
| Sex, n (%) | 35 (87.50%) | 41 (87.23%) | 0.78 | 7 (77.78%) | 10 (83.33%) | 0.81 | 15 (88.24%) | 21 (80.77%) | 0.38 |
| BMI (kg/m2) | 23.30 ± 2.83 | 24.32 ± 3.05 | 0.57 | 26.02 ± 2.46 | 23.51 ± 3.19 | 0.67 | 23.18 ± 2.34 | 25.35 ± 4.06 | 0.27 |
| Etiology, No. | |||||||||
| Hepatitis B virus | 27 (67.50%) | 39 (82.98%) | 8 (88.89%) | 10 (83.33%) | 11 (64.71%) | 18 (69.23%) | |||
| Hepatitis C virus | 3 (7.50%) | 1 (2.13%) | 1 (11.11%) | 1 (8.33%) | 2 (11.76%) | 1 (3.85%) | |||
| Others | 10 (25.00%) | 7 (14.89%) | 0 (0%) | 1 (8.33%) | 4 (23.53%) | 7 (26.92%) | |||
| AFP level (mg/mL) | 0.03 | 0.02 | 0.04 | ||||||
| <20 | 6 (15.00%) | 38 (80.85%) | 3 (33.33%) | 8 (66.67%) | 4 (23.53%) | 15 (57.69%) | |||
| ≥20 | 34 (87.50%) | 9 (19.15%) | 6 (66.67%) | 4 (33.33%) | 13 (76.47%) | 11 (42.31%) | |||
| Platelet count (×109/L) | 142.32 ± 70.08 | 140.70 ± 53.78 | 0.35 | 133.50 ± 40.53 | 158.33 ± 71.99 | 0.34 | 156.00 ± 89.57 | 152.32 ± 73.11 | 0.36 |
| Prealbumin level (mg/L) | 187.86 ± 54.95 | 212.96 ± 73.84 | 0.24 | 194.13 ± 45.34 | 199.00 ± 54.01 | 0.31 | 177.18 ± 58.22 | 178.44 ± 53.98 | 0.27 |
| ALT level (IU/L) | 45.32 ± 77.39 | 32.30 ± 19.06 | 0.35 | 53.25 ± 36.98 | 31.92 ± 20.05 | 0.34 | 35.24 ± 22.25 | 33.40 ± 17.74 | 0.45 |
| AST level (IU/L) | 50.76 ± 84.47 | 36.78 ± 19.83 | 0.15 | 46.63 ± 27.50 | 36.33 ± 17.63 | 0.12 | 49.53 ± 41.14 | 36.80 ± 13.22 | 0.12 |
| Total bilirubin (μmol/L) | 18.23 ± 7.51 | 19.18 ± 9.00 | 0.36 | 15.35 ± 4.38 | 24.38 ± 27.90 | 0.17 | 17.09 ± 5.09 | 16.54 ± 7.70 | 0.24 |
| Direct bilirubin (μmol/L) | 3.99 ± 2.90 | 3.70 ± 2.26 | 0.26 | 3.00 ± 1.04 | 4.38 ± 4.81 | 0.28 | 3.79 ± 2.18 | 3.53 ± 2.24 | 0.23 |
| Albumin level (g/L) | 39.58 ± 4.68 | 40.48 ± 7.83 | 0.39 | 37.75 ± 2.90 | 42.00 ± 9.27 | 0.41 | 38.65 ± 4.73 | 38.92 ± 4.07 | 0.72 |
| Prothrombin time (s) | 12.21 ± 1.33 | 12.01 ± 1.39 | 0.81 | 12.66 ± 0.52 | 13.04 ± 1.07 | 0.89 | 12.47 ± 0.76 | 12.56 ± 1.11 | 0.82 |
| INR | 1.04 ± 0.12 | 1.02 ± 0.12 | 0.67 | 1.08 ± 0.05 | 1.11 ± 0.10 | 0.57 | 1.06 ± 0.07 | 1.07 ± 0.10 | 0.67 |
| c (rad) | 2.45 ± 0.65 | 2.26 ± 0.66 | 0.17 | 2.38 ± 0.85 | 2.23 ± 0.97 | 0.05 | 2.07 ± 0.58 | 2.11 ± 0.61 | 0.11 |
| φ (m/s) | 1.14 ± 0.25 | 1.05 ± 0.24 | 0.09 | 1.20 ± 0.24 | 0.99 ± 0.20 | 0.71 | 1.03 ± 0.22 | 1.02 ± 0.25 | 0.20 |
| Model | Inception-Resnet | Xception | Inception | Resnet | VGG16 | VGG19 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | 0.71 ± 0.04 | 0.61 ± 0.03 | 0.80 ± 0.03 | 0.71 ± 0.02 | 0.65 ± 0.03 | 0.56 ± 0.03 | 0.70 ± 0.04 | 0.62 ± 0.03 | 0.62 ± 0.03 | 0.53 ± 0.03 | 0.65 ± 0.03 | 0.55 ± 0.05 |
| (0.70–0.72) | (0.60–0.62) | (0.79–0.81) | (0.70–0.72) | (0.64–0.66) | (0.55–0.57) | (0.69–0.71) | (0.61–0.63) | (0.61–0.63) | (0.52–0.54) | (0.64–0.66) | (0.54–0.57) | |
| Accuracy | 0.71 ± 0.05 | 0.61 ± 0.04 | 0.77 ± 0.04 | 0.68 ± 0.03 | 0.66 ± 0.05 | 0.57 ± 0.04 | 0.70 ± 0.04 | 0.61 ± 0.04 | 0.62 ± 0.04 | 0.53 ± 0.03 | 0.64 ± 0.04 | 0.55 ± 0.03 |
| (0.70–0.72) | (0.60–0.62) | (0.76–0.78) | (0.67–0.69) | (0.65–0.67) | (0.56–0.58) | (0.69–0.71) | (0.60–0.62) | (0.61–0.63) | (0.52–0.54) | (0.63–0.65) | (0.54–0.56) | |
| Sensitivity | 0.68 ± 0.05 | 0.60 ± 0.03 | 0.76 ± 0.06 | 0.67 ± 0.04 | 0.65 ± 0.04 | 0.57 ± 0.05 | 0.67 ± 0.05 | 0.59 ± 0.03 | 0.59 ± 0.03 | 0.53 ± 0.04 | 0.66 ± 0.04 | 0.57 ± 0.02 |
| (0.67–0.69) | (0.59–0.61) | (0.75–0.77) | (0.66–0.68) | (0.65–0.67) | (0.55–0.58) | (0.66–0.68) | (0.58–0.60) | (0.58–0.60) | (0.52–0.54) | (0.65–0.67) | (0.56–0.58) | |
| Specificity | 0.72 ± 0.04 | 0.63 ± 0.02 | 0.78 ± 0.06 | 0.68 ± 0.04 | 0.67 ± 0.05 | 0.58 ± 0.04 | 0.72 ± 0.03 | 0.58 ± 0.04 | 0.64 ± 0.04 | 0.55 ± 0.03 | 0.62 ± 0.04 | 0.52 ± 0.03 |
| (0.71–0.73) | (0.62–0.64) | (0.77–0.79) | (0.67–0.69) | (0.66–0.68) | (0.57–0.59) | (0.71–0.73) | (0.57–0.59) | (0.63–0.65) | (0.54–0.56) | (0.61–0.63) | (0.51–0.53) | |
| PPV | 0.69 ± 0.03 | 0.63 ± 0.02 | 0.76 ± 0.03 | 0.65 ± 0.04 | 0.64 ± 0.02 | 0.55 ± 0.04 | 0.67 ± 0.02 | 0.58 ± 0.02 | 0.59 ± 0.04 | 0.52 ± 0.03 | 0.65 ± 0.03 | 0.56 ± 0.04 |
| (0.68–0.70) | (0.62–0.64) | (0.75–0.77) | (0.64–0.66) | (0.64–0.65) | (0.54–0.56) | (0.67–0.68) | (0.57–0.59) | (0.58–0.60) | (0.51–0.53) | (0.64–0.66) | (0.55–0.57) | |
| NPV | 0.71 ± 0.02 | 0.62 ± 0.03 | 0.77 ± 0.04 | 0.68 ± 0.03 | 0.68 ± 0.03 | 0.59 ± 0.04 | 0.72 ± 0.01 | 0.55 ± 0.03 | 0.64 ± 0.03 | 0.55 ± 0.02 | 0.63 ± 0.02 | 0.54 ± 0.03 |
| (0.71–0.72) | (0.62–0.64) | (0.76–0.78) | (0.67–0.69) | (0.67–0.69) | (0.58–0.60) | (0.72–0.73) | (0.54–0.56) | (0.63–0.65) | (0.54–0.56) | (0.63–0.64) | (0.53–0.55) | |
| Cohort | Parameter Combinations | Evaluation | |||||
|---|---|---|---|---|---|---|---|
| AUC | Accuracy | Sensitivity | Specificity | PPV | NPV | ||
| Internal validation cohort | cMRI + AFP | 0.84 ± 0.03 | 0.81 ± 0.04 | 0.80 ± 0.06 | 0.82 ± 0.06 | 0.78 ± 0.06 | 0.80 ± 0.03 |
| (0.83–0.85) | (0.80–0.82) | (0.79–0.81) | (0.81–0.83) | (0.77–0.79) | (0.79–0.81) | ||
| cMRI + AFP + MRE | 0.90 ± 0.03 | 0.87 ± 0.05 | 0.86 ± 0.04 | 0.93 ± 0.02 | 0.84 ± 0.03 | 0.87 ± 0.02 | |
| (0.89–0.91) | (0.86–0.88) | (0.85–0.87) | (0.93–0.94) | (0.83–0.85) | (0.87–0.88) | ||
| Independent testing cohort | cMRI + AFP | 0.74 ± 0.02 | 0.72 ± 0.03 | 0.72 ± 0.05 | 0.72 ± 0.04 | 0.68 ± 0.05 | 0.71 ± 0.03 |
| (0.73–0.75) | (0.71–0.73) | (0.71–0.74) | (0.71–0.73) | (0.67–0.70) | (0.70–0.72) | ||
| cMRI + AFP + MRE | 0.83 ± 0.03 | 0.83 ± 0.02 | 0.80 ± 0.03 | 0.86 ± 0.01 | 0.78 ± 0.02 | 0.80 ± 0.03 | |
| (0.82–0.84) | (0.82–0.84) | (0.79–0.81) | (0.86–0.87) | (0.77–0.79) | (0.79–0.81) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Zhou, J.; Li, Y.; Wang, Y.; Guo, J.; Sack, I.; Chen, W.; Yan, F.; Li, R.; Wang, C. Added Value of Viscoelasticity for MRI-Based Prediction of Ki-67 Expression of Hepatocellular Carcinoma Using a Deep Learning Combined Radiomics (DLCR) Model. Cancers 2022, 14, 2575. https://doi.org/10.3390/cancers14112575
Hu X, Zhou J, Li Y, Wang Y, Guo J, Sack I, Chen W, Yan F, Li R, Wang C. Added Value of Viscoelasticity for MRI-Based Prediction of Ki-67 Expression of Hepatocellular Carcinoma Using a Deep Learning Combined Radiomics (DLCR) Model. Cancers. 2022; 14(11):2575. https://doi.org/10.3390/cancers14112575
Chicago/Turabian StyleHu, Xumei, Jiahao Zhou, Yan Li, Yikun Wang, Jing Guo, Ingolf Sack, Weibo Chen, Fuhua Yan, Ruokun Li, and Chengyan Wang. 2022. "Added Value of Viscoelasticity for MRI-Based Prediction of Ki-67 Expression of Hepatocellular Carcinoma Using a Deep Learning Combined Radiomics (DLCR) Model" Cancers 14, no. 11: 2575. https://doi.org/10.3390/cancers14112575
APA StyleHu, X., Zhou, J., Li, Y., Wang, Y., Guo, J., Sack, I., Chen, W., Yan, F., Li, R., & Wang, C. (2022). Added Value of Viscoelasticity for MRI-Based Prediction of Ki-67 Expression of Hepatocellular Carcinoma Using a Deep Learning Combined Radiomics (DLCR) Model. Cancers, 14(11), 2575. https://doi.org/10.3390/cancers14112575






