Sensitization of Resistant Breast Cancer Cells with a Jumonji Family Histone Demethylase Inhibitor
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Drugs
2.2. Western Blotting
2.3. JIB-04 Treatment of Resistant Cancer Cells
2.4. Assay of Relative Resistance of Cells to Paclitaxel and Doxorubicin
3. Results
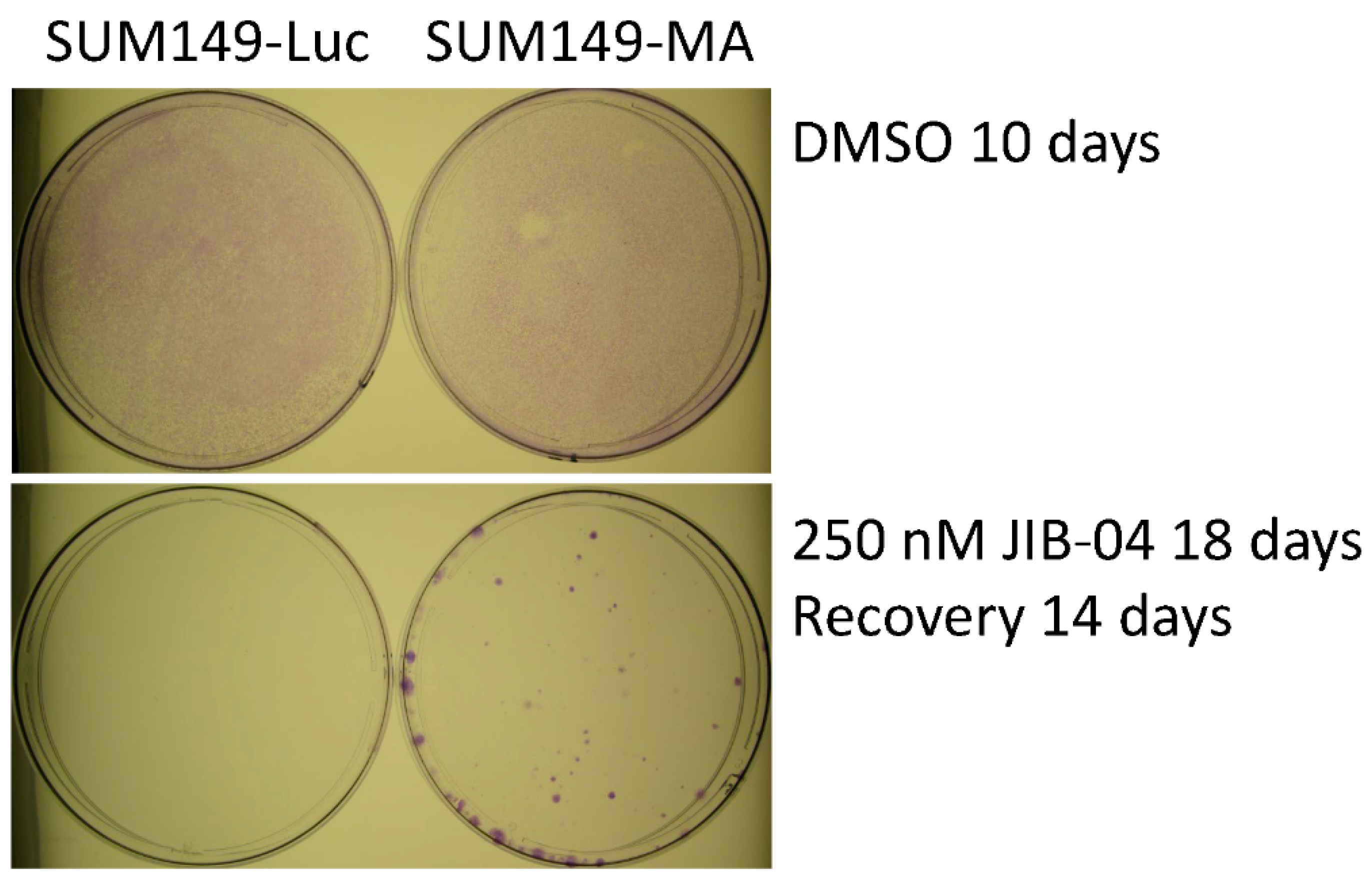
3.1. JIB-04 Resistance of SUM149-MA Cells
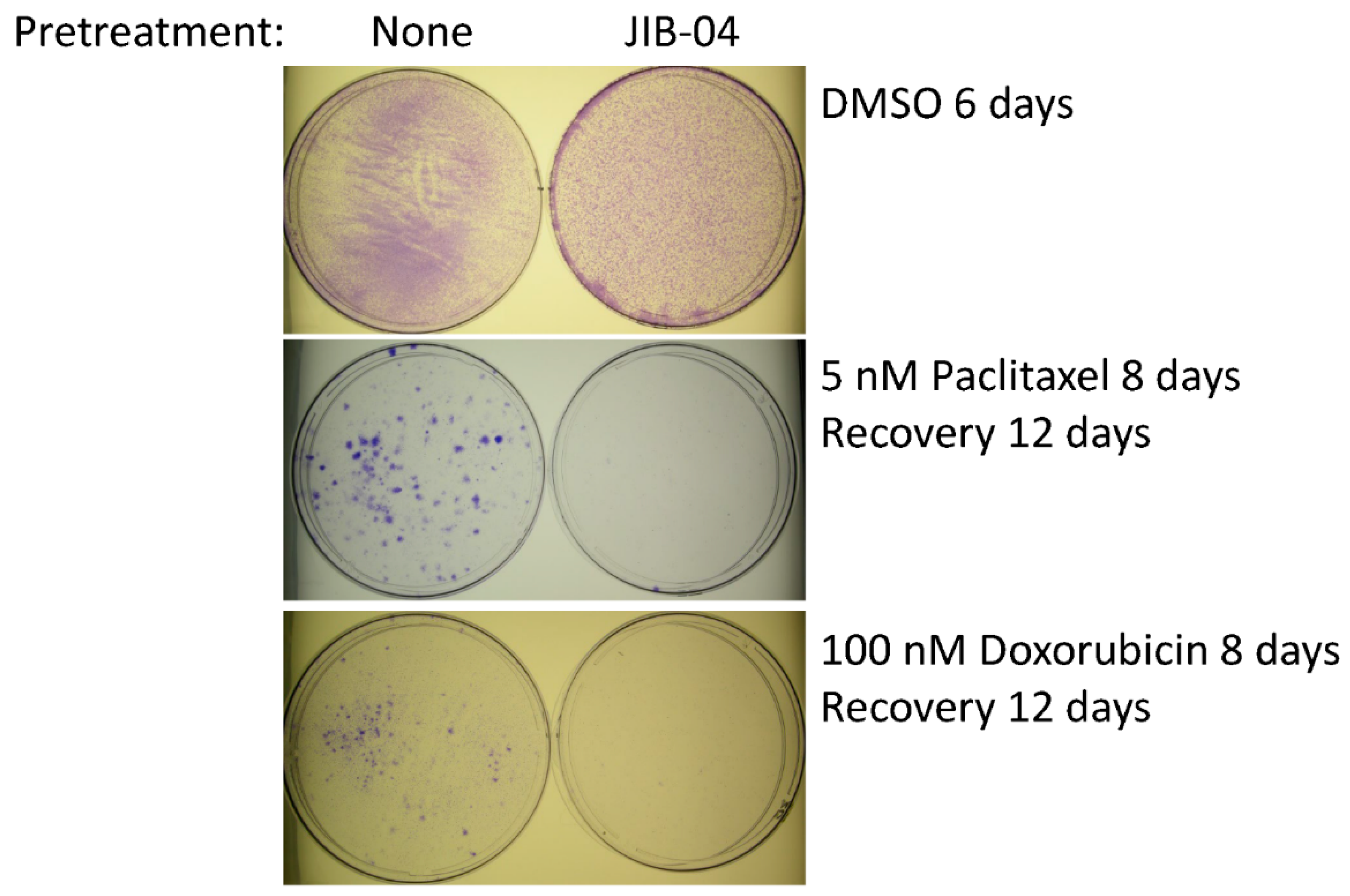
3.2. Sensitization of SUM149-MA Cells to Chemotherapeutic Drugs by Treatment with JIB-04
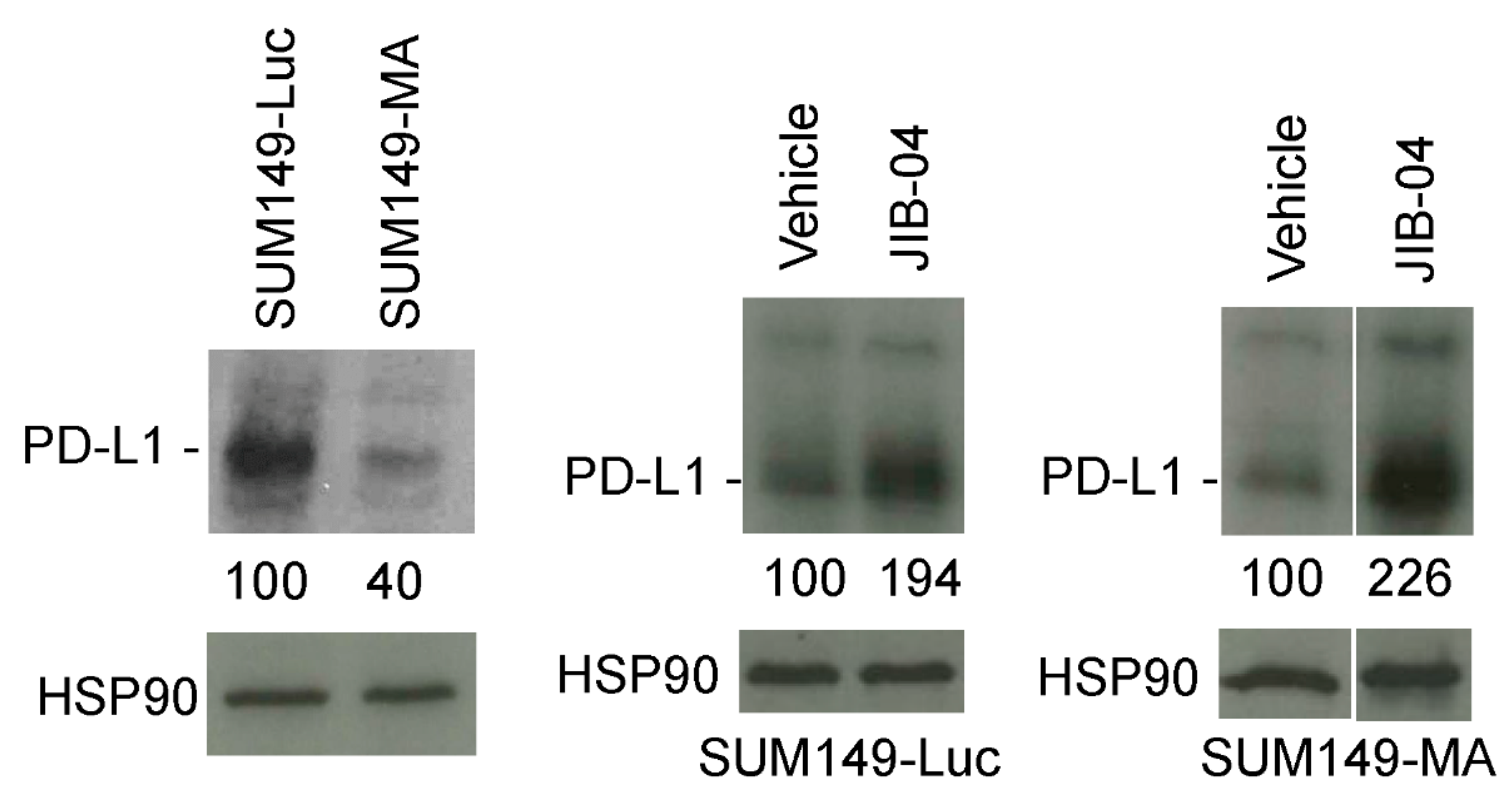
3.3. Increased PD-L1 Expression in SUM149-MA Cells upon Treatment with JIB-04
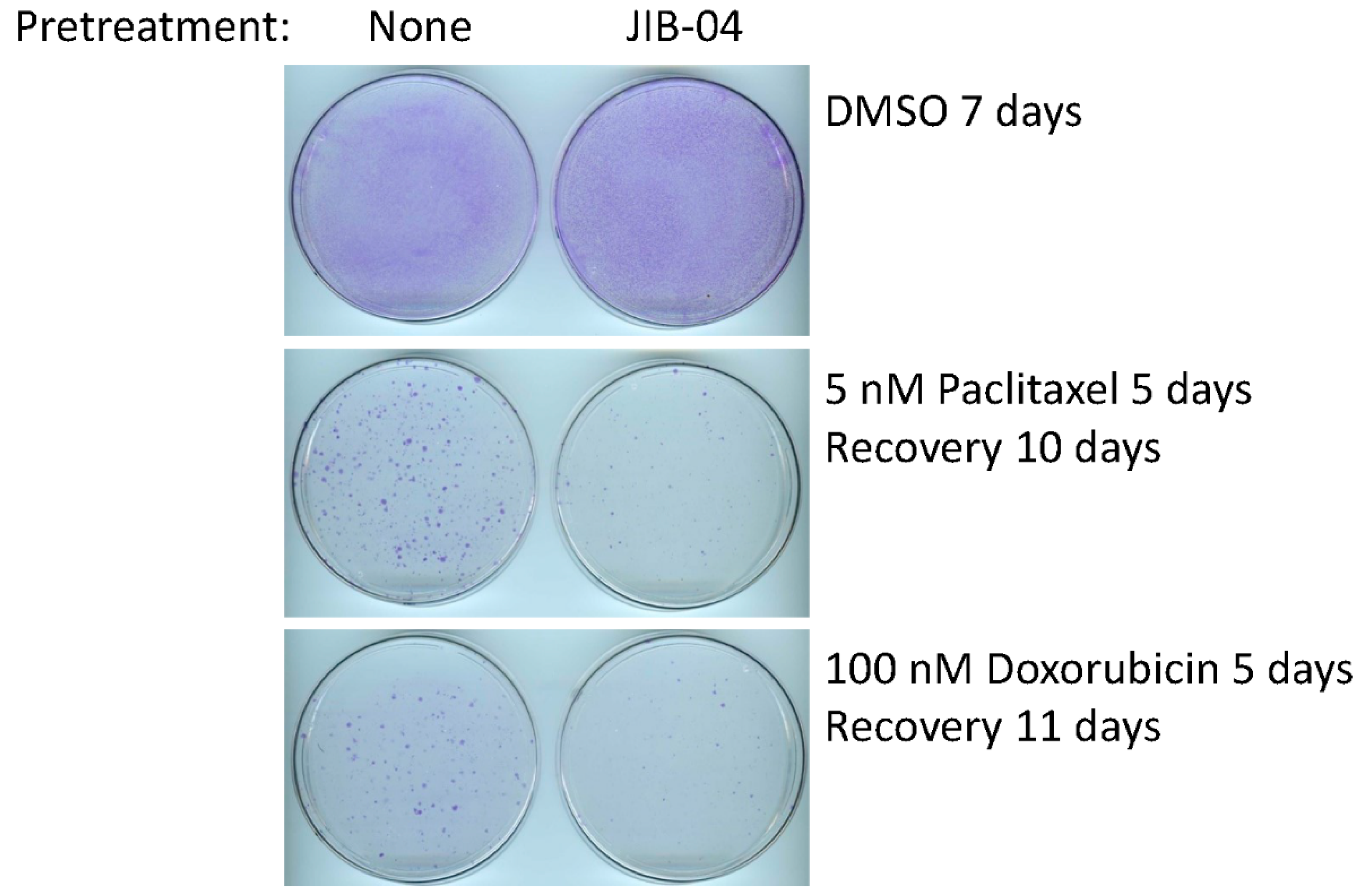
3.4. Sensitivity of FC-IBC02-MA Cells to Treatment with JIB-04
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Baldominos, P.; Barbera-Mourelle, A.; Barreiro, O.; Huang, Y.; Wight, A.; Cho, J.W.; Zhao, X.; Estivill, G.; Adam, I.; Sanchez, X.; et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell 2022, 185, 1694–1708.e19. [Google Scholar] [CrossRef] [PubMed]
- Mavrommati, I.; Johnson, F.; Echeverria, G.V.; Natrajan, R. Subclonal heterogeneity and evolution in breast cancer. NPJ Breast Cancer 2021, 7, 155. [Google Scholar] [CrossRef]
- Minussi, D.C.; Nicholson, M.D.; Ye, H.; Davis, A.; Wang, K.; Baker, T.; Tarabichi, M.; Sei, E.; Du, H.; Rabbani, M.; et al. Breast tumours maintain a reservoir of subclonal diversity during expansion. Nature 2021, 592, 302–308. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Somarelli, J.A.; Gardner, H.; Cannataro, V.L.; Gunady, E.F.; Boddy, A.M.; Johnson, N.A.; Fisk, J.N.; Gaffney, S.G.; Chuang, J.H.; Li, S.; et al. Molecular Biology and Evolution of Cancer: From Discovery to Action. Mol. Biol. Evol. 2020, 37, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Talmadge, J.E.; Wolman, S.R.; Fidler, I.J. Evidence for the clonal origin of spontaneous metastases. Science 1982, 217, 361–363. [Google Scholar] [CrossRef]
- Braun, S.; Vogl, F.D.; Naume, B.; Janni, W.; Osborne, M.P.; Coombes, R.C.; Schlimok, G.; Diel, I.J.; Gerber, B.; Gebauer, G.; et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N. Engl. J. Med. 2005, 353, 793–802. [Google Scholar] [CrossRef]
- Giancotti, F.G. Mechanisms governing metastatic dormancy and reactivation. Cell 2013, 155, 750–764. [Google Scholar] [CrossRef] [Green Version]
- Janni, W.J.; Rack, B.; Terstappen, L.W.; Pierga, J.Y.; Taran, F.A.; Fehm, T.; Hall, C.; de Groot, M.R.; Bidard, F.C.; Friedl, T.W.; et al. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin. Cancer Res. 2016, 22, 2583–2593. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Tai, K.; Madan, S.; Raythatha, M.R.; Cady, A.M.; Braunlin, M.; Irving, L.R.; Bajaj, A.; Lucci, A. Selection of metastatic breast cancer cells based on adaptability of their metabolic state. PLoS ONE 2012, 7, e36510. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Sarli, V.N.; Washburn, L.J.; Raythatha, M.R.; Lucci, A. A usable model of “decathlon winner” cancer cells in triple-negative breast cancer: Survival of resistant cancer cells in quiescence. Oncotarget 2018, 9, 11071–11082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Shamsnia, A.; Raythatha, M.R.; Milligan, R.D.; Cady, A.M.; Madan, S.; Lucci, A. Highly adaptable triple-negative breast cancer cells as a functional model for testing anticancer agents. PLoS ONE 2014, 9, e109487. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Sarli, V.N.; Lucci, A. Inhibition of resistant triple-negative breast cancer cells with low-dose 6-mercaptopurine and 5-azacitidine. Oncotarget 2021, 12, 626–637. [Google Scholar] [CrossRef]
- Singh, B.; Sarli, V.N.; Kinne, H.E.; Shamsnia, A.; Lucci, A. Evaluation of 6-mercaptopurine in a cell culture model of adaptable triple-negative breast cancer with metastatic potential. Oncotarget 2019, 10, 3681–3693. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Kinne, H.E.; Milligan, R.D.; Washburn, L.J.; Olsen, M.; Lucci, A. Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS ONE 2016, 11, e0159072. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 2013, 153, 56–69. [Google Scholar] [CrossRef] [Green Version]
- McKnight, S.L. Back to the future: Molecular biology meets metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 403–411. [Google Scholar] [CrossRef]
- Li, J.; Gonzalez-Angulo, A.M.; Allen, P.K.; Yu, T.K.; Woodward, W.A.; Ueno, N.T.; Lucci, A.; Krishnamurthy, S.; Gong, Y.; Bondy, M.L.; et al. Triple-negative subtype predicts poor overall survival and high locoregional relapse in inflammatory breast cancer. Oncologist 2011, 16, 1675–1683. [Google Scholar] [CrossRef] [Green Version]
- Menta, A.; Fouad, T.M.; Lucci, A.; Le-Petross, H.; Stauder, M.C.; Woodward, W.A.; Ueno, N.T.; Lim, B. Inflammatory Breast Cancer: What to Know About This Unique, Aggressive Breast Cancer. Surg. Clin. N. Am. 2018, 98, 787–800. [Google Scholar] [CrossRef]
- Wang, L.; Chang, J.; Varghese, D.; Dellinger, M.; Kumar, S.; Best, A.M.; Ruiz, J.; Bruick, R.; Pena-Llopis, S.; Xu, J.; et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 2013, 4, 2035. [Google Scholar] [CrossRef]
- Banelli, B.; Daga, A.; Forlani, A.; Allemanni, G.; Marubbi, D.; Pistillo, M.P.; Profumo, A.; Romani, M. Small molecules targeting histone demethylase genes (KDMs) inhibit growth of temozolomide-resistant glioblastoma cells. Oncotarget 2017, 8, 34896–34910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhan, A.; Ansari, K.I.; Chen, M.Y.; Jandial, R. Inhibition of Jumonji Histone Demethylases Selectively Suppresses HER2(+) Breast Leptomeningeal Carcinomatosis Growth via Inhibition of GMCSF Expression. Cancer Res. 2021, 81, 3200–3214. [Google Scholar] [CrossRef] [PubMed]
- Dalvi, M.P.; Wang, L.; Zhong, R.; Kollipara, R.K.; Park, H.; Bayo, J.; Yenerall, P.; Zhou, Y.; Timmons, B.C.; Rodriguez-Canales, J.; et al. Taxane-Platin-Resistant Lung Cancers Co-develop Hypersensitivity to JumonjiC Demethylase Inhibitors. Cell Rep. 2017, 19, 1669–1684. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Cho, H.I.; Yoon, H.J.; Ahn, Y.H.; Park, E.J.; Jin, Y.H.; Jang, Y.K. JIB-04, A Small Molecule Histone Demethylase Inhibitor, Selectively Targets Colorectal Cancer Stem Cells by Inhibiting the Wnt/beta-Catenin Signaling Pathway. Sci. Rep. 2018, 8, 6611. [Google Scholar] [CrossRef] [PubMed]
- Parrish, J.K.; McCann, T.S.; Sechler, M.; Sobral, L.M.; Ren, W.; Jones, K.L.; Tan, A.C.; Jedlicka, P. The Jumonji-domain histone demethylase inhibitor JIB-04 deregulates oncogenic programs and increases DNA damage in Ewing Sarcoma, resulting in impaired cell proliferation and survival, and reduced tumor growth. Oncotarget 2018, 9, 33110–33123. [Google Scholar] [CrossRef] [Green Version]
- Poulard, C.; Baulu, E.; Lee, B.H.; Pufall, M.A.; Stallcup, M.R. Increasing G9a automethylation sensitizes B acute lymphoblastic leukemia cells to glucocorticoid-induced death. Cell. Death Dis. 2018, 9, 1038. [Google Scholar] [CrossRef]
- Sobral, L.M.; Sechler, M.; Parrish, J.K.; McCann, T.S.; Jones, K.L.; Black, J.C.; Jedlicka, P. KDM3A/Ets1/MCAM axis promotes growth and metastatic properties in Rhabdomyosarcoma. Genes Cancer 2020, 11, 53–65. [Google Scholar] [CrossRef]
- Singh, B.; Cook, K.R.; Martin, C.; Huang, E.H.; Mosalpuria, K.; Krishnamurthy, S.; Cristofanilli, M.; Lucci, A. Evaluation of a CXCR4 antagonist in a xenograft mouse model of inflammatory breast cancer. Clin. Exp. Metastasis 2010, 27, 233–240. [Google Scholar] [CrossRef]
- Fernandez, S.V.; Robertson, F.M.; Pei, J.; Aburto-Chumpitaz, L.; Mu, Z.; Chu, K.; Alpaugh, R.K.; Huang, Y.; Cao, Y.; Ye, Z.; et al. Inflammatory breast cancer (IBC): Clues for targeted therapies. Breast Cancer Res. Treat 2013, 140, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Berry, J.A.; Shoher, A.; Ayers, G.D.; Wei, C.; Lucci, A. COX-2 involvement in breast cancer metastasis to bone. Oncogene 2007, 26, 3789–3796. [Google Scholar] [CrossRef] [Green Version]
- Gianni, L.; Huang, C.S.; Egle, D.; Bermejo, B.; Zamagni, C.; Thill, M.; Anton, A.; Zambelli, S.; Bianchini, G.; Russo, S.; et al. Pathologic complete response (pCR) to neoadjuvant treatment with or without atezolizumab in triple-negative, early high-risk and locally advanced breast cancer: NeoTRIP Michelangelo randomized study. Ann. Oncol. 2022, 33, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Winer, E.P.; Lipatov, O.; Im, S.A.; Goncalves, A.; Munoz-Couselo, E.; Lee, K.S.; Schmid, P.; Tamura, K.; Testa, L.; Witzel, I.; et al. Pembrolizumab versus investigator-choice chemotherapy for metastatic triple-negative breast cancer (KEYNOTE-119): A randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 499–511. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kummel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, B.; Sarli, V.N.; Lucci, A. Sensitization of Resistant Breast Cancer Cells with a Jumonji Family Histone Demethylase Inhibitor. Cancers 2022, 14, 2631. https://doi.org/10.3390/cancers14112631
Singh B, Sarli VN, Lucci A. Sensitization of Resistant Breast Cancer Cells with a Jumonji Family Histone Demethylase Inhibitor. Cancers. 2022; 14(11):2631. https://doi.org/10.3390/cancers14112631
Chicago/Turabian StyleSingh, Balraj, Vanessa N. Sarli, and Anthony Lucci. 2022. "Sensitization of Resistant Breast Cancer Cells with a Jumonji Family Histone Demethylase Inhibitor" Cancers 14, no. 11: 2631. https://doi.org/10.3390/cancers14112631
APA StyleSingh, B., Sarli, V. N., & Lucci, A. (2022). Sensitization of Resistant Breast Cancer Cells with a Jumonji Family Histone Demethylase Inhibitor. Cancers, 14(11), 2631. https://doi.org/10.3390/cancers14112631






