SAR131675, a VEGRF3 Inhibitor, Modulates the Immune Response and Reduces the Growth of Colorectal Cancer Liver Metastasis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and CR-LM Induction
2.2. Treatment Protocol
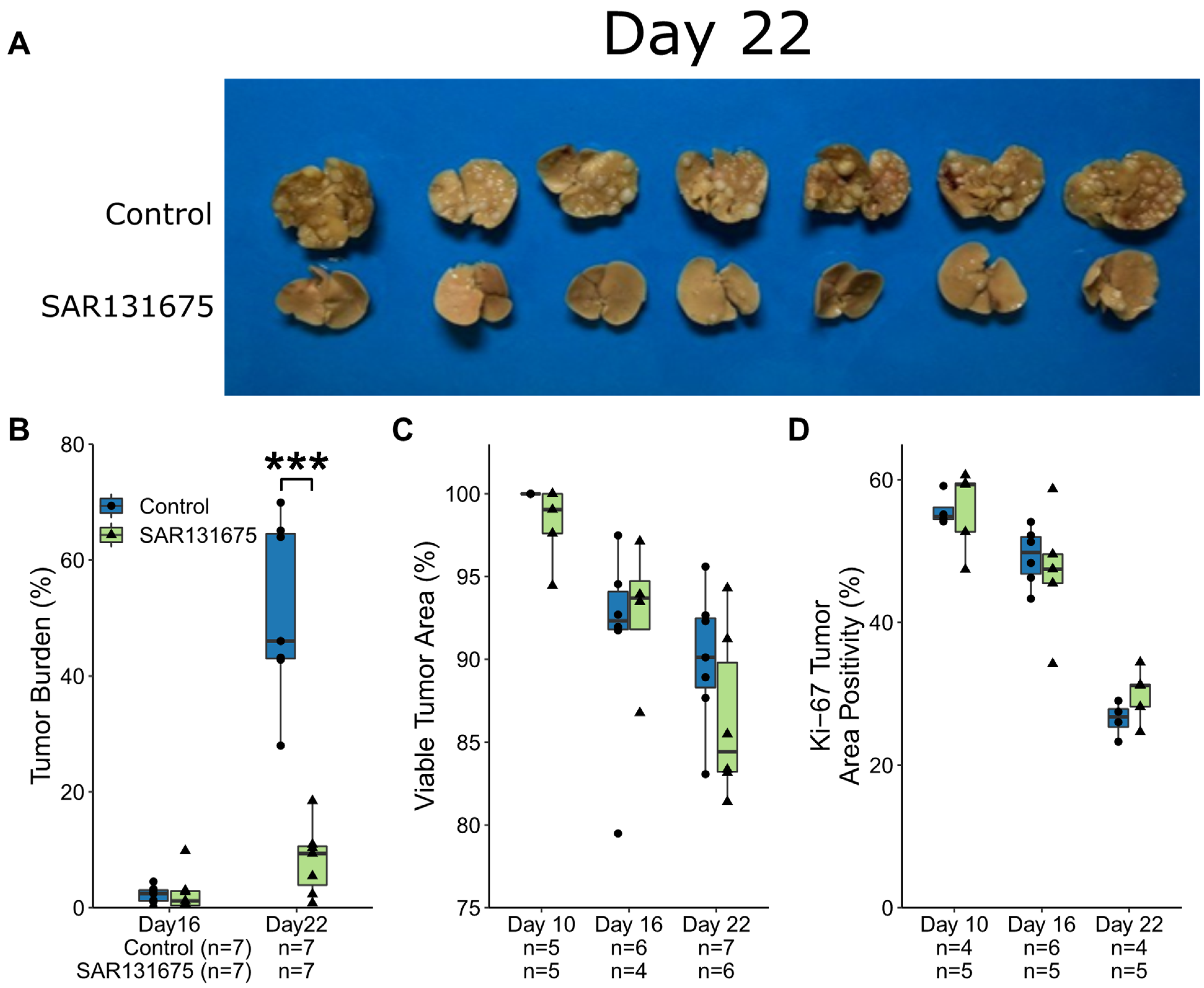
2.3. Tumor Burden and IHC Assessment
2.4. Image Caption and Analysis Algorithm
2.5. Flow Cytometry
2.6. Statistical Analysis
3. Results
3.1. CRLM Growth Kinetics Following SAR131675 Treatment
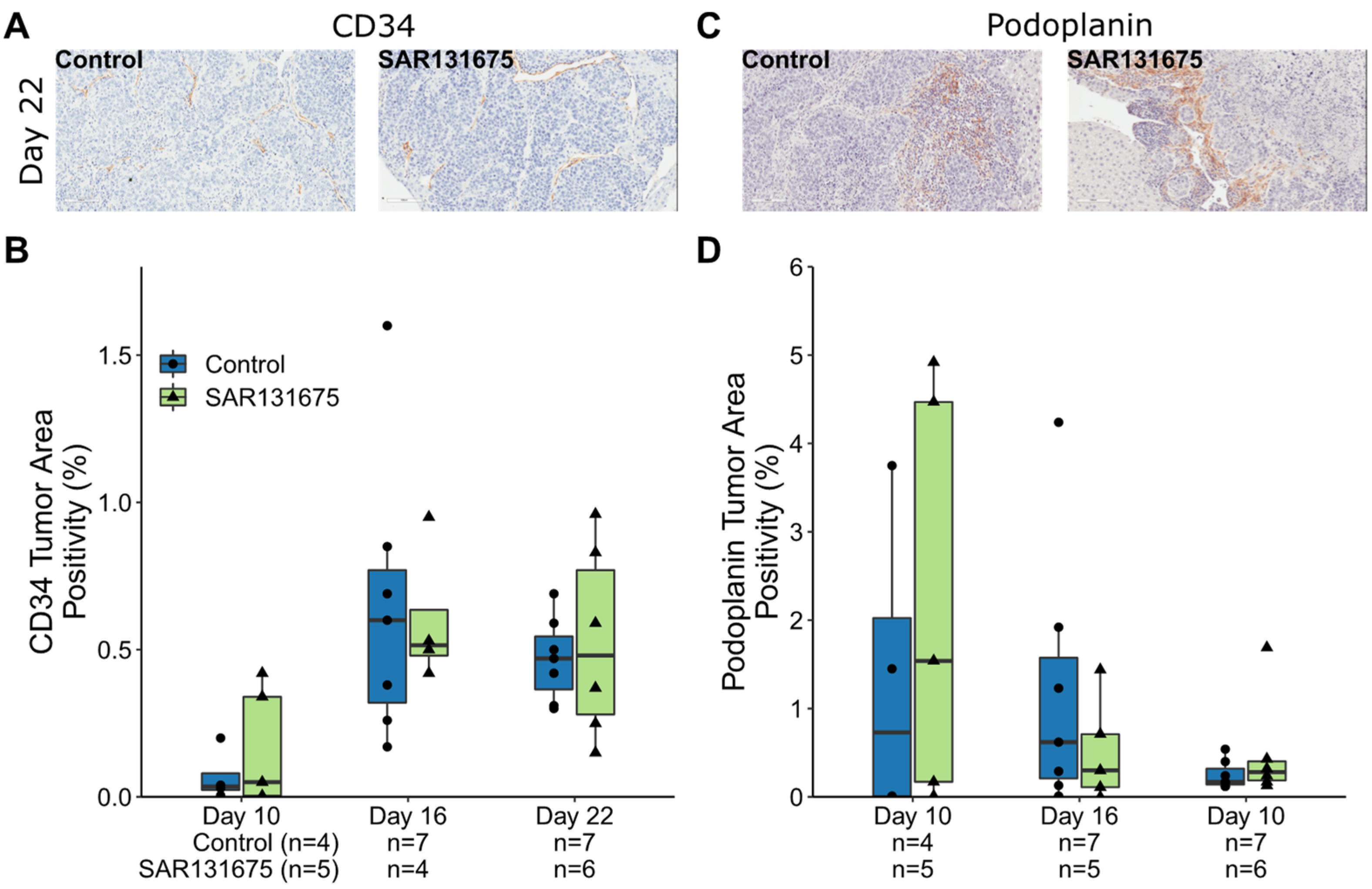
3.2. SAR131675 Treatment Did Not Affect Tumor Vascularization and Lymphatic Density
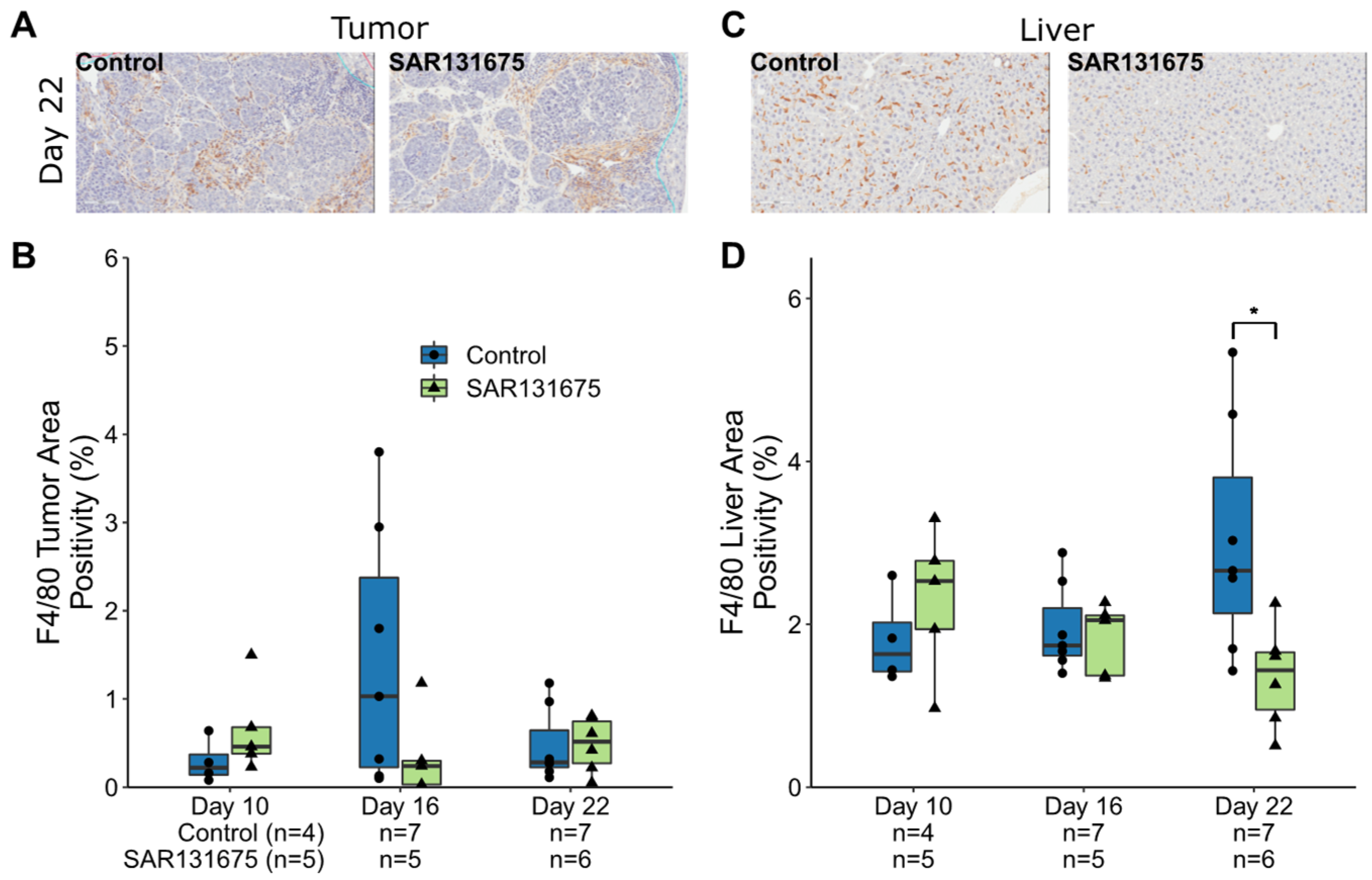
3.3. Macrophages Are Reduced within the Liver and CRLM by SAR131675 Treatment
3.4. SAR131675 Expression of VEGFR3 in Tumor and Liver Parenchyma
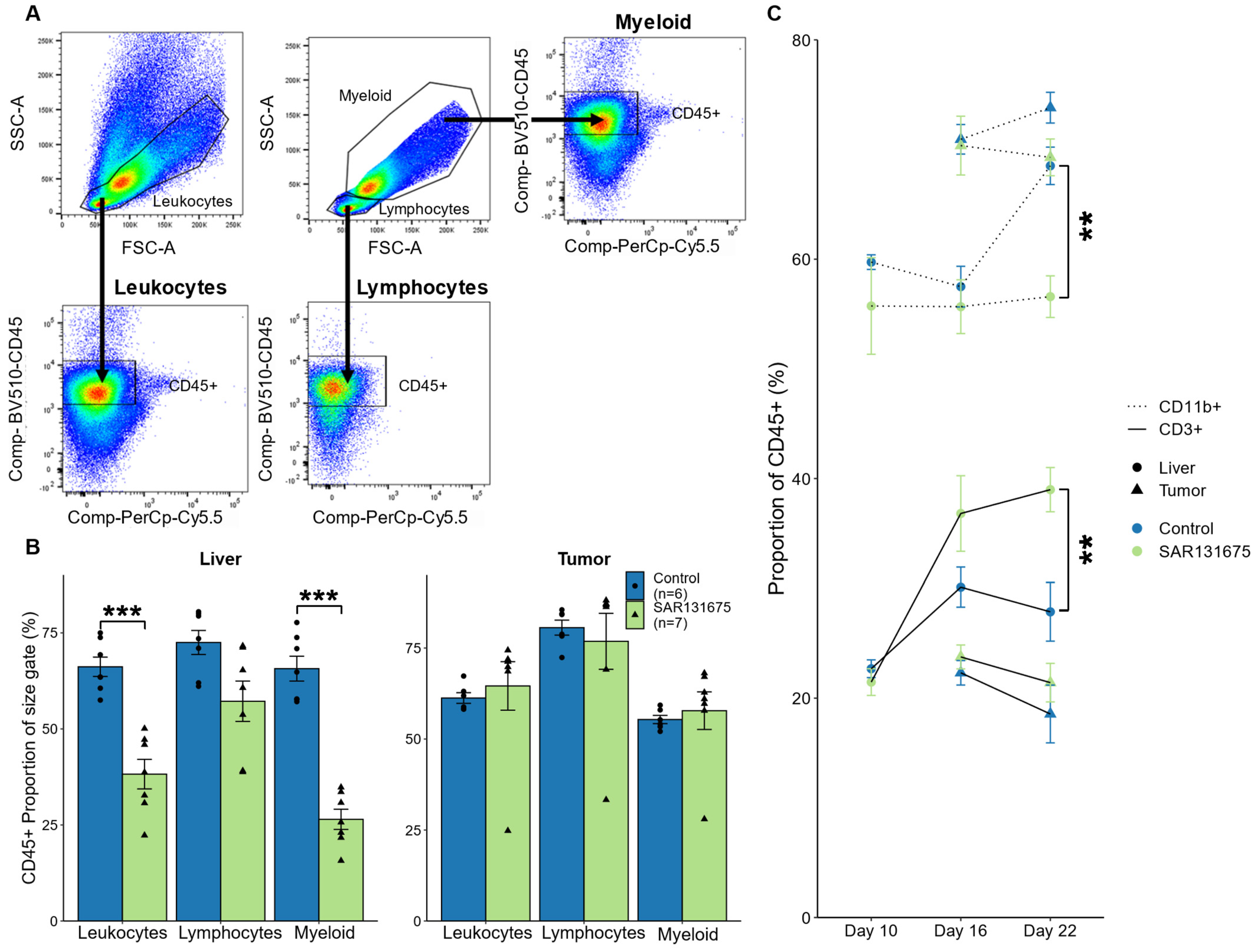
3.5. SAR131675 Treatment Significantly Alters the Balance of Lymphocytes and Myeloid Cells
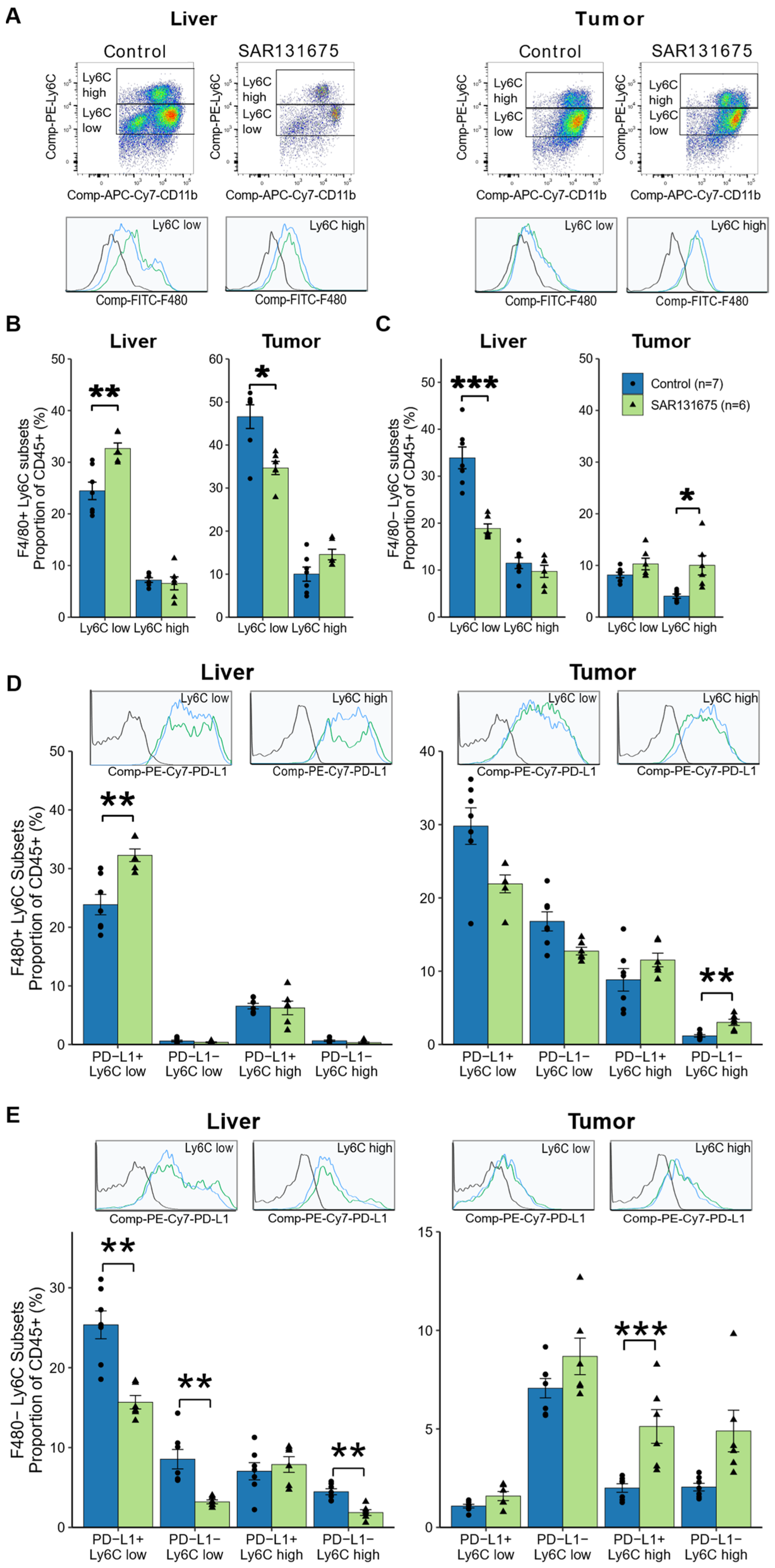
3.6. SAR131675 Treatment Differentially Modulates the Infiltration of Myeloid Subsets
3.7. SAR131675 Treatment Modulates the T Lymphocyte Responses to CRLM
3.8. Ratios of Myeloid Subsets/T Cell Subsets Are Significantly Altered by SAR131765 Treatment
Correlation Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pasetto, L.M.; Jirillo, A.; Iadicicco, G.; Rossi, E.; Paris, M.K.; Monfardini, S. FOLFOX versus FOLFIRI: A comparison of regimens in the treatment of colorectal cancer metastases. Anticancer Res. 2005, 25, 563–576. [Google Scholar] [PubMed]
- Lee, J.C.; Chow, N.H.; Wang, S.T.; Huang, S.M. Prognostic value of vascular endothelial growth factor expression in colorectal cancer patients. Eur. J. Cancer 2000, 36, 748–753. [Google Scholar] [CrossRef]
- Mazeda, I.; Martins, S.F.; Garcia, E.A.; Rodrigues, M.; Longatto, A. VEGF Expression in Colorectal Cancer Metastatic Lymph Nodes: Clinicopathological Correlation and Prognostic Significance. Gastrointest. Disord. 2020, 2, 267–280. [Google Scholar] [CrossRef]
- Regan, E.; Sibley, R.C.; Cenik, B.K.; Silva, A.; Girard, L.; Minna, J.D.; Dellinger, M.T. Identification of Gene Expression Differences between Lymphangiogenic and Non-Lymphangiogenic Non-Small Cell Lung Cancer Cell Lines. PLoS ONE 2016, 11, e0150963. [Google Scholar] [CrossRef] [PubMed]
- Vasudev, N.S.; Reynolds, A.R. Anti-angiogenic therapy for cancer: Current progress, unresolved questions and future directions. Angiogenesis 2014, 17, 471–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes-Coelho, F.; Martins, F.; Pereira, S.A.; Serpa, J. Anti-Angiogenic Therapy: Current Challenges and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 3765. [Google Scholar] [CrossRef]
- Gardner, V.; Madu, C.O.; Lu, Y. Anti-VEGF therapy in cancer: A double-edged sword. In Physiologic and Pathologic Angiogenesis-Signaling Mechanisms and Targeted Therapy; Simionescu, D., Simionescu, A., Eds.; IntechOpen: London, UK, 2017; pp. 385–410. [Google Scholar]
- Tacconi, C.; Ungaro, F.; Correale, C.; Arena, V.; Massimino, L. Activation of the VEGFC/VEGFR3 Pathway Induces Tumor Immune Escape in Colorectal Cancer. Cancer Res. 2019, 79, 4196–4210. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, H.; Feng, N.; Xin, X.; Xu, Y.; Huo, P.; Wang, X.; Zhang, N. Construction and antitumor effects of antitumor micelles with cyclic RGD-modified anlotinib. Nanomedicine 2020, 28, 102224. [Google Scholar] [CrossRef]
- Alam, A.; Blanc, I.; Gueguen-Dorbes, G.; Duclos, O.; Bonnin, J.; Barron, P.; Laplace, M.-C.; Morin, G.; Gaujarengues, F.; Dol, F.; et al. SAR131675, a potent and selective VEGFR-3-TK inhibitor with antilymphangiogenic, antitumoral, and antimetastatic activities. Mol. Cancer Ther. 2012, 11, 1637–1649. [Google Scholar] [CrossRef] [Green Version]
- Espagnolle, N.; Barron, P.; Mandron, M.; Blanc, I.; Bonnin, J.; Agnel, M.; Kerbelec, E.; Herault, J.P.; Savi, P.; Bono, F.; et al. Specific Inhibition of the VEGFR-3 Tyrosine Kinase by SAR131675 Reduces Peripheral and Tumor Associated Immunosuppressive Myeloid Cells. Cancers 2014, 6, 472–490. [Google Scholar] [CrossRef] [Green Version]
- Kuruppu, D.; Christophi, C.; Bertram, J.F.; O’Brien, P.E. Characterization of an animal model of hepatic metastasis. J. Gastroenterol. Hepatol. 1996, 11, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Vallejo Ardila, D.L.; Walsh, K.A.; Fifis, T.; Paolini, R.; Kastrappis, G.; Christophi, C.; Perini, M.V. Immunomodulatory effects of renin-angiotensin system inhibitors on T lymphocytes in mice with colorectal liver metastases. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Kuruppu, D.; Christophi, C.; O’Brien, P.E. Microvascular architecture of hepatic metastases in a mouse model. HPB Surg. 1997, 10, 149–157; discussion 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.; Fifis, T.; Christophi, C. Vascular disruptive agent OXi4503 and anti-angiogenic agent Sunitinib combination treatment prolong survival of mice with CRC liver metastasis. BMC Cancer 2016, 16, 533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loken, M.R.; Brosnan, J.M.; Bach, B.A.; Ault, K.A. Establishing optimal lymphocyte gates for immunophenotyping by flow cytometry. Cytometry 1990, 11, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Fifis, T.; Malcontenti-Wilson, C.; Chan, L.S.; Costa, P.N.; Nikfarjam, M.; Muralidharan, V.; Christophi, C. Spatial morphological and molecular differences within solid tumors may contribute to the failure of vascular disruptive agent treatments. BMC Cancer 2012, 12, 522. [Google Scholar] [CrossRef] [Green Version]
- Galdiero, M.R.; Garlanda, C.; Jaillon, S.; Marone, G.; Mantovani, A. Tumor associated macrophages and neutrophils in tumor progression. J. Cell Physiol. 2013, 228, 1404–1412. [Google Scholar] [CrossRef]
- Liu, Y.; Zugazagoitia, J.; Ahmed, F.S.; Henick, B.S.; Gettinger, S.N.; Herbst, R.S.; Schalper, K.A.; Rimm, D.L. Immune Cell PD-L1 Colocalizes with Macrophages and Is Associated with Outcome in PD-1 Pathway Blockade Therapy. Clin. Cancer Res. 2020, 26, 970–977. [Google Scholar] [CrossRef] [Green Version]
- Fifis, T.; Nguyen, L.; Malcontenti-Wilson, C.; Chan, L.S.; Nunes Costa, P.L.; Daruwalla, J.; Nikfarjam, M.; Muralidharan, V.; Waltham, M.; Thompson, E.W.; et al. Treatment with the vascular disruptive agent OXi4503 induces an immediate and widespread epithelial to mesenchymal transition in the surviving tumor. Cancer Med. 2013, 2, 595–610. [Google Scholar] [CrossRef] [Green Version]
- Webb, J.R.; Milne, K.; Nelson, B.H. Location, location, location: CD103 demarcates intraepithelial, prognostically favorable CD8(+) tumor-infiltrating lymphocytes in ovarian cancer. Oncoimmunology 2014, 3, e27668. [Google Scholar] [CrossRef] [Green Version]
- Molodtsov, A.; Turk, M.J. Tissue Resident CD8 Memory T Cell Responses in Cancer and Autoimmunity. Front. Immunol. 2018, 9, 2810. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Wei, M.; Wang, S.; Dong, J.; Wei, J. Pan-Cancer Analysis of Immune Cell Infiltration Identifies a Prognostic Immune-Cell Characteristic Score (ICCS) in Lung Adenocarcinoma. Front. Immunol. 2020, 11, 1218. [Google Scholar] [CrossRef] [PubMed]
- Awad, R.M.; De Vlaeminck, Y.; Maebe, J.; Goyvaerts, C.; Breckpot, K. Turn Back the TIMe: Targeting Tumor Infiltrating Myeloid Cells to Revert Cancer Progression. Front. Immunol. 2018, 9, 1977. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Kepp, O.; Kroemer, G. Immune parameters affecting the efficacy of chemotherapeutic regimens. Nat. Rev. Clin. Oncol. 2011, 8, 151–160. [Google Scholar] [CrossRef]
- García-Martínez, E.; Gil, G.L.; Benito, A.C.; González-Billalabeitia, E.; Conesa, M.A.; García García, T.; García-Garre, E.; Vicente, V.; Ayala de la Peña, F. Tumor-infiltrating immune cell profiles and their change after neoadjuvant chemotherapy predict response and prognosis of breast cancer. Breast Cancer Res. 2014, 16, 488. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Li, C.; Liu, T.; Dai, X.; Bazhin, A.V. Myeloid-Derived Suppressor Cells in Tumors: From Mechanisms to Antigen Specificity and Microenvironmental Regulation. Front. Immunol. 2020, 11, 1371. [Google Scholar] [CrossRef]
- Weston, C.J.; Zimmermann, H.W.; Adams, D.H. The Role of Myeloid-Derived Cells in the Progression of Liver Disease. Front. Immunol. 2019, 10, 893. [Google Scholar] [CrossRef]
- Kramer, E.D.; Abrams, S.I. Granulocytic Myeloid-Derived Suppressor Cells as Negative Regulators of Anticancer Immunity. Front. Immunol. 2020, 11, 1963. [Google Scholar] [CrossRef]
- Witte, D.; Thomas, A.; Ali, N.; Carlson, N.; Younes, M. Expression of the vascular endothelial growth factor receptor-3 (VEGFR-3) and its ligand VEGF-C in human colorectal adenocarcinoma. Anticancer Res. 2002, 22, 1463–1466. [Google Scholar]
- Kodama, M.; Kitadai, Y.; Tanaka, M.; Kuwai, T.; Tanaka, S.; Oue, N.; Yasui, W.; Chayama, K. Vascular endothelial growth factor C stimulates progression of human gastric cancer via both autocrine and paracrine mechanisms. Clin. Cancer Res. 2008, 14, 7205–7214. [Google Scholar] [CrossRef] [Green Version]
- Ran, S.; Wilber, A. Novel role of immature myeloid cells in formation of new lymphatic vessels associated with inflammation and tumors. J. Leukoc. Biol. 2017, 102, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Vetsika, E.-K.; Koukos, A.; Kotsakis, A. Myeloid-Derived Suppressor Cells: Major Figures that Shape the Immunosuppressive and Angiogenic Network in Cancer. Cells 2019, 8, 1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groth, C.; Hu, X.; Weber, R.; Fleming, V.; Altevogt, P.; Utikal, J.; Umansky, V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br. J. Cancer 2019, 120, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, S.J.; Hashimoto, M.; Gerner, M.Y.; Lee, J.; Kissick, H.T.; Burger, M.C.; Shan, Q.; Hale, J.S.; Lee, J.; Nasti, T.H.; et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016, 537, 417–421. [Google Scholar] [CrossRef]
- Tay, R.E.; Richardson, E.K.; Toh, H.C. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2020, 28, 5–17. [Google Scholar] [CrossRef]
- Oh, D.Y.; Fong, L. Cytotoxic CD4(+) T cells in cancer: Expanding the immune effector toolbox. Immunity 2021, 54, 2701–2711. [Google Scholar] [CrossRef]
- Spitzer, M.H.; Carmi, Y.; Reticker-Flynn, N.E.; Kwek, S.S.; Madhireddy, D.; Martins, M.M.; Gherardini, P.F.; Prestwood, T.R.; Chabon, J.; Bendall, S.C.; et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 2017, 168, 487–502.e15. [Google Scholar] [CrossRef] [Green Version]
- Oh, D.Y.; Kwek, S.S.; Raju, S.S.; Li, T.; McCarthy, E.; Chow, E.; Aran, D.; Ilano, A.; Pai, C.S.; Rancan, C.; et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 2020, 181, 1612–1625.e13. [Google Scholar] [CrossRef]
- Alishekevitz, D.; Gingis-Velitski, S.; Kaidar-Person, O.; Gutter-Kapon, L.; Scherer, S.D.; Raviv, Z.; Merquiol, E.; Ben-Nun, Y.; Miller, V.; Rachman-Tzemah, C.; et al. Macrophage-Induced Lymphangiogenesis and Metastasis following Paclitaxel Chemotherapy Is Regulated by VEGFR3. Cell Rep. 2016, 17, 1344–1356. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.A.; Patel, V.G. The role of PD-L1 expression as a predictive biomarker: An analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.; Cheung, J.; Navarro, A.; Lianoglou, S.; Haley, B.; Totpal, K.; Sanders, L.; Koeppen, H.; Caplazi, P.; McBride, J.; et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat. Commun. 2017, 8, 14572. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, C.; Zografos, G.; Aronis, K.; Troupis, T.G.; Gorgoulis, V.G.; Sechas, M.N.; Skalkeas, G. The effect of various types of splenectomy on the development of B-16 melanoma in mice. Anticancer Res. 1999, 19, 4235–4239. [Google Scholar] [PubMed]
- Long, X.; Wang, J.; Zhao, J.P.; Liang, H.F.; Zhu, P.; Cheng, Q.; Chen, Q.; Wu, Y.H.; Zhang, Z.G.; Zhang, B.X.; et al. Splenectomy suppresses growth and metastasis of hepatocellular carcinoma through decreasing myeloid-derived suppressor cells in vivo. J. Huazhong Univ. Sci. Technol. Med. Sci. 2016, 36, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.T.; Struk, A.; Malcontenti-Wilson, C.; Christophi, C.; Lynch, G.S. Physiological characterization of a mouse model of cachexia in colorectal liver metastases. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R854–R864. [Google Scholar] [CrossRef] [Green Version]


| Liver | Tumor | |
| Control mean ± SD | 4.73 ± 1.88 | 5.87 ± 1.94 |
| SAR131675 mean ± SD | 2.6 ± 0.40 | 3.09 ± 0.77 |
| p value | 0.024 | 0.015 |
| Liver | Tumor | |||
| Subset | Total | PD1+ | Total | PD1+ |
| Control mean ± SD | 0.75 ± 0.20 | 0.65 ± 0.28 | 0.39 ± 0.08 | 0.18 ± 0.07 |
| SAR131675 mean ± SD | 1.01 ± 0.28 | 1.28 ± 0.82 | 0.30 ± 0.03 | 0.20 ± 0.04 |
| p value | 0.071 | 0.094 | 0.0167 | 0.6777 |
| Liver | Tumor | |||||||
| Macrophage | MDSC | Macrophage | MDSC | |||||
| PD-L1+ | PD-L1− | PD-L1+ | PD-L1− | PD-L1+ | PD-L1− | PD-L1+ | PD-L1− | |
| Control mean ± SD | 3.69 ± 0.69 | 0.91 ± 0.28 | 4.23 ± 2.03 | 1.90 ± 0.47 | 3.81 ± 1.28 | 16.10 ± 6.78 | 0.57 ± 0.17 | 3.64 ± 1.11 |
| SAR131675 mean ± SD | 6.60 ± 4.23 | 1.73 ± 0.78 | 2.15 ± 0.69 | 2.12 ± 1.26 | 1.97 ± 0.48 | 4.65 ± 1.58 | 0.35 ± 0.15 | 2.09 ± 0.87 |
| p value | 0.15 | 0.048 | 0.036 | 0.70 | 0.008 | 0.004 | 0.033 | 0.017 |
| Liver | Tumor | |||||||
|---|---|---|---|---|---|---|---|---|
| Subset | Ly6Clo PD-L1+ | Ly6Clo PD-L1− | Ly6Chi PD-L1+ | Ly6Chi PD-L1− | Ly6Clo PD-L1+ | Ly6Clo PD-L1− | Ly6Chi PD-L1+ | Ly6Chi PD-L1− |
| MAC (F4/80+) | ||||||||
| Control | - | −0.849 * | - | - | - | - | 0.931 ** | - |
| SAR131675 | - | - | - | - | - | −0.908 * | - | - |
| MDSC (F4/80−) | ||||||||
| Control | - | −0.835 * | 0.825 * | - | - | - | 0.839 * | 0.845 * |
| SAR131675 | - | - | - | - | - | - | - | - |
| Liver | Tumor | |||||||
|---|---|---|---|---|---|---|---|---|
| Subset | Ly6Clo PD-L1+ | Ly6Clo PD-L1− | Ly6Chi PD-L1+ | Ly6Chi PD-L1− | Ly6Clo PD-L1+ | Ly6Clo PD-L1− | Ly6Chi PD-L1+ | Ly6Chi PD-L1− |
| MAC (F4/80+) | ||||||||
| Control | - | −0.803 * | - | −0.755 * | - | - | 0.962 *** | - |
| SAR131675 | - | - | - | - | - | - | - | - |
| MDSC (F4/80−) | ||||||||
| Control | - | - | - | - | - | - | 0.819 * | 0.893 ** |
| SAR131675 | - | - | - | - | - | - | 0.878 * | 0.911 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walsh, K.A.; Kastrappis, G.; Fifis, T.; Paolini, R.; Christophi, C.; Perini, M.V. SAR131675, a VEGRF3 Inhibitor, Modulates the Immune Response and Reduces the Growth of Colorectal Cancer Liver Metastasis. Cancers 2022, 14, 2715. https://doi.org/10.3390/cancers14112715
Walsh KA, Kastrappis G, Fifis T, Paolini R, Christophi C, Perini MV. SAR131675, a VEGRF3 Inhibitor, Modulates the Immune Response and Reduces the Growth of Colorectal Cancer Liver Metastasis. Cancers. 2022; 14(11):2715. https://doi.org/10.3390/cancers14112715
Chicago/Turabian StyleWalsh, Katrina A., Georgios Kastrappis, Theodora Fifis, Rita Paolini, Christopher Christophi, and Marcos V. Perini. 2022. "SAR131675, a VEGRF3 Inhibitor, Modulates the Immune Response and Reduces the Growth of Colorectal Cancer Liver Metastasis" Cancers 14, no. 11: 2715. https://doi.org/10.3390/cancers14112715
APA StyleWalsh, K. A., Kastrappis, G., Fifis, T., Paolini, R., Christophi, C., & Perini, M. V. (2022). SAR131675, a VEGRF3 Inhibitor, Modulates the Immune Response and Reduces the Growth of Colorectal Cancer Liver Metastasis. Cancers, 14(11), 2715. https://doi.org/10.3390/cancers14112715






