Extracellular Vesicle-Based Bronchoalveolar Lavage Fluid Liquid Biopsy for EGFR Mutation Testing in Advanced Non-Squamous NSCLC
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Sample Collection
2.2. BALF Processing and EV Isolation
2.3. EV-Based BALF EGFR Genotyping
2.4. EGFR Genotyping of Tissue DNA and Plasma cfDNA
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Methods of Tissue Procurement for EGFR Genotyping
3.3. EGFR Mutation Detection Rate in Liquid Biopsy Using BALF and Plasma and Tissue Samples
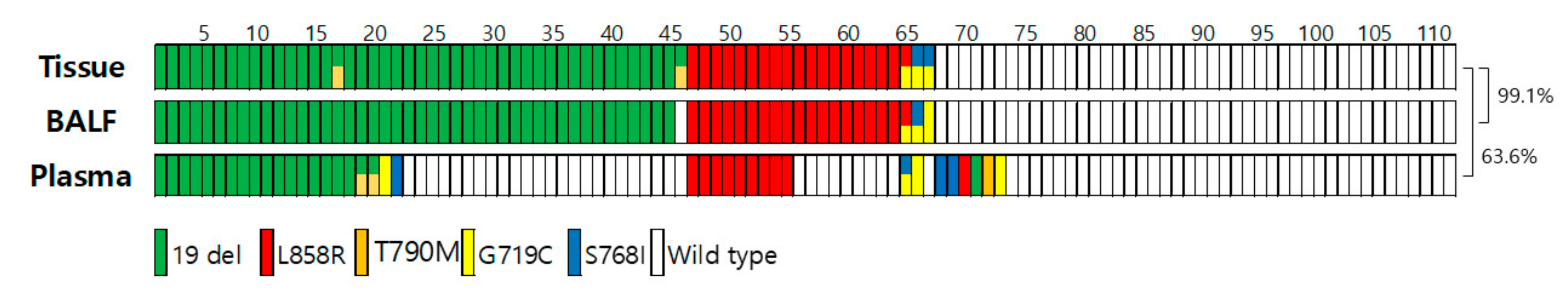
3.4. Concordance Rate for EGFR Genotyping among Tissue Biopsy, BALF, and Plasma Liquid Biopsy (n = 110)
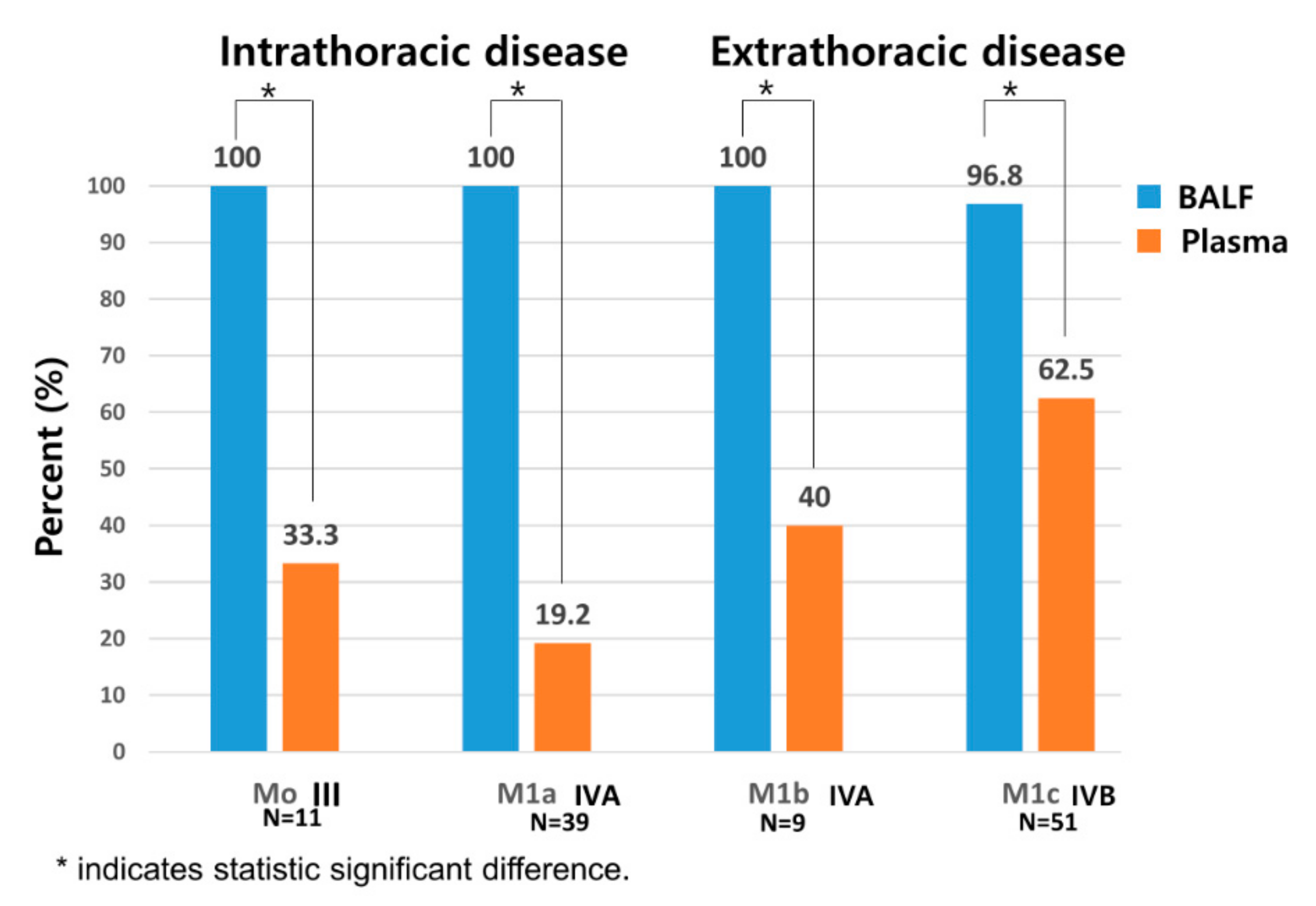
3.5. Comparison of Sensitivity between BALF and Plasma Liquid Biopsy Depending on Presence of Metastasis
3.6. Turnaround Time of EGFR Mutation Testing in BALF-Based and Tissue-Based Genotyping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mok, T.S.; Wu, Y.-L.; Thongprasert, S.; Yang, C.-H.; Chu, D.-T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Zhou, C.; Hu, C.-P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Lee, K.Y. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014, 15, 213–222. [Google Scholar] [CrossRef]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Loong, H.H.; Kwan, S.-c.S.; Mok, T.S.-k.; Lau, Y.-m. Therapeutic Strategies in EGFR Mutant Non-Small Cell Lung Cancer. Curr. Treat. Options Oncol. 2018, 19, 58. [Google Scholar] [CrossRef]
- Nour-Eldin, N.-E.A.; Alsubhi, M.; Emam, A.; Lehnert, T.; Beeres, M.; Jacobi, V.; Gruber-Rouh, T.; Scholtz, J.-E.; Vogl, T.J.; Naguib, N.N. Pneumothorax complicating coaxial and non-coaxial CT-guided lung biopsy: Comparative analysis of determining risk factors and management of pneumothorax in a retrospective review of 650 patients. Cardio Interv. Radiol. 2016, 39, 261–270. [Google Scholar] [CrossRef]
- Liu, H.E.; Vuppalapaty, M.; Wilkerson, C.; Renier, C.; Chiu, M.; Lemaire, C.; Che, J.; Matsumoto, M.; Carroll, J.; Crouse, S.; et al. Detection of EGFR Mutations in cfDNA and CTCs, and Comparison to Tumor Tissue in Non-Small-Cell-Lung-Cancer (NSCLC) Patients. Front. Oncol. 2020, 10, 572895. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579. [Google Scholar] [CrossRef]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Fu, S.; Piha-Paul, S.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. J. Clin. Oncol. 2018, 24, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-L.; Sequist, L.V.; Hu, C.-P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Schuler, M.; Mok, T.; et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: Analysis of LUX-Lung 3 and 6. Br. J. Cancer 2017, 116, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Ladrera, G.; Fuerte, F.; Margono, B.; Wu, L.; Tsai, J.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [PubMed] [Green Version]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating extracellular vesicles in human disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [PubMed]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rizaldos, E.; Grimm, D.G.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M.; et al. Exosome-Based Detection of EGFR T790M in Plasma from Non–Small Cell Lung Cancer Patients. Clin. Cancer Res. 2018, 24, 2944–2950. [Google Scholar]
- Wan, Y.; Liu, B.; Lei, H.; Zhang, B.; Wang, Y.; Huang, H.; Chen, S.; Feng, Y.; Zhu, L.; Gu, Y. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann. Oncol. 2018, 29, 2379–2383. [Google Scholar]
- Park, J.; Lee, C.; Eom, J.S.; Kim, M.-H.; Cho, Y.-K. Detection of EGFR Mutations Using Bronchial Washing-Derived Extracellular Vesicles in Patients with Non-Small-Cell Lung Carcinoma. Cancers 2020, 12, 2822. [Google Scholar]
- Purcell, E.; Owen, S.; Prantzalos, E.; Radomski, A.; Carman, N.; Lo, T.-W.; Zeinali, M.; Subramanian, C.; Ramnath, N.; Nagrath, S. Epidermal Growth Factor Receptor Mutations Carried in Extracellular Vesicle-Derived Cargo Mirror Disease Status in Metastatic Non-small Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 389. [Google Scholar] [CrossRef]
- Castellanos-Rizaldos, E.; Zhang, X.; Tadigotla, V.R.; Grimm, D.G.; Karlovich, C.; Raez, L.E.; Skog, J.K. Exosome-based detection of activating and resistance EGFR mutations from plasma of non-small cell lung cancer patients. Oncotarget 2019, 10, 2911. [Google Scholar]
- Krug, A.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.; Castellanos-Rizaldos, E. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 2018, 29, 700–706. [Google Scholar]
- Hur, J.Y.; Kim, H.J.; Lee, J.S.; Choi, C.-M.; Lee, J.C.; Jung, M.K.; Pack, C.G.; Lee, K.Y. Extracellular vesicle-derived DNA for performing EGFR genotyping of NSCLC patients. Mol. Cancer 2018, 17, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hur, J.Y.; Lee, J.S.; Kim, I.A.; Kim, H.J.; Kim, W.S.; Lee, K.Y. Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl. Lung Cancer Res. 2019, 8, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.-S.; Lim, S.-n.; An, J.Y.; Lee, K.-M.; Choe, K.H.; Le, K.H.; Kim, S.T.; Son, S.M.; Choi, S.Y.; Le, O.-J.; et al. Detection of EGFR Mutation Status in Lung Adenocarcinoma Specimens with Different Proportions of Tumor Cells Using Two Methods of Differential Sensitivity. J. Thorac. Oncol. 2012, 7, 355–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.-Y.; Choi, J.-J.; Kim, J.Y.; Han, Y.L.; Lee, G.K. PNA clamping-assisted fluorescence melting curve analysis for detecting EGFR and KRAS mutations in the circulating tumor DNA of patients with advanced non-small cell lung cancer. BMC Cancer 2016, 16, 627. [Google Scholar]
- Rantakokko-Jalava, K.; Jalava, J. Optimal DNA Isolation Method for Detection of Bacteria in Clinical Specimens by Broad-Range PCR. J. Clin. Mic. 2002, 40, 4211–4217. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.J.; Casal, R.F.; Lazarus, D.R.; Ost, D.E.; Eapen, G.A. Flexible bronchoscopy. Clin. Chest Med. 2018, 39, 1–16. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Kahlert, C. Liquid Biopsy: Is There an Advantage to Analyzing Circulating Exosomal DNA Compared to cfDNA or Are They the Same? Cancer Res. 2019, 79, 2462–2465. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, K.; Wang, Z.; Wang, Y.; Liu, J.; Lin, L.; Shao, Y.; Gao, L.; Yin, H.; Cui, C.; et al. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer 2016, 16, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hur, J.Y.; Lee, K.Y. Characteristics and Clinical Application of Extracellular Vesicle-Derived DNA. Cancers 2021, 13, 3827. [Google Scholar] [CrossRef] [PubMed]
- San Lucas, F.A.; Allenson, K.; Bernard, V.; Castillo, J.; Kim, D.U.; Ellis, K.; Ehli, E.A.; Davies, G.E.; Petersen, J.L.; Wistuba, I.; et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 2016, 27, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.E.; Park, H.Y.; Hur, J.Y.; Kim, H.J.; Kim, I.A.; Kim, W.S.; Lee, K.Y. Genomic profiling of extracellular vesicle-derived DNA from bronchoalveolar lavage fluid of patients with lung adenocarcinoma. Transl. Lung Cancer Res. 2021, 10, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Buttitta, F.; Felicioni, L.; Del Grammastro, M.; Filice, G.; Di Lorito, A.; Malatesta, S.; Viola, P.; Centi, I.; D’Antuono, T.; Zappacosta, R.; et al. Effective Assessment of egfr Mutation Status in Bronchoalveolar Lavage and Pleural Fluids by Next-Generation Sequencing. Clin. Cancer Res. 2013, 19, 691–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Cai, W.; Wang, Y.; Liao, M.; Tian, S. CT and clinical characteristics that predict risk of EGFR mutation in non-small cell lung cancer: A systematic review and meta-analysis. Int. J. Clin. Oncol. 2019, 24, 649–659. [Google Scholar] [CrossRef]
- Kim, N.; Cho, D.; Kim, H.; Kim, S.; Cha, Y.-J.; Greulich, H.; Bass, A.; Cho, H.-S.; Cho, J. Colorectal adenocarcinoma-derived EGFR mutants are oncogenic and sensitive to EGFR-targeted monoclonal antibodies, cetuximab and panitumumab. Int. J. Cancer 2020, 146, 2194–2200. [Google Scholar] [CrossRef]
- Kim, I.; Lee, J.S.; Kim, H.J.; Kim, W.S.; Lee, K.Y. Cumulative smoking dose affects the clinical outcomes of EGFR-mutated lung adenocarcinoma patients treated with EGFR-TKIs: A retrospective study. BMC Cancer 2018, 18, 1–10. [Google Scholar] [CrossRef]
| Characteristics | BALF, n (%) | Plasma, n (%) | p-Value |
|---|---|---|---|
| Number of patients | 224 | 110 | |
| Age (mean ± SD) | 67.7 ± 11.8 | 67.4 ± 10.1 | 0.82 |
| Sex | |||
| Male | 134 (59.8) | 53 (48.2) | 0.19 |
| Female | 90 (40.2) | 57 (51.8) | |
| Smoking history | |||
| Non-smoker | 106 (47.3) | 61 (55.5) | 0.38 |
| Ex-smoker | 58 (25.9) | 24 (21.8) | |
| Current smoker | 60 (26.8) | 25 (22.7) | |
| Histology | |||
| Adenocarcinoma | 187 (83.5) | 95 (86.4) | 0.52 |
| NSCLC | 37 (16.5) | 15 (13.6) | |
| Stage | |||
| III | 36 (16.1) | 11 (10) | 0.18 |
| IVA | 97 (43.3) | 48 (43.6) | |
| IVB | 91 (40.6) | 51 (46.4) | |
| Tissue EGFR mutation | |||
| Wild type | 131 (58.5) | 44 (40) | 0.12 |
| EGFR mutation | 93 (41.5) | 66 (60) | |
| Exon 19 del | 52 (55.9) | 43 (39.1) | |
| L858R | 33 (35.4) | 18 (16.4) | |
| Exon 19 del + T790M | 2 (2) | 2 (1.8) | |
| G719C + S768I | 2 (2) | 2 (1.8) | |
| L858R + G719C | 1 (1) | 1 (0.9) | |
| L858R + T790M | 1 (1) | 0 (0) | |
| G719C | 1 (1) | 0 (0) | |
| L861Q | 1 (1) | 0 (0) | |
| S768I | 0 (0) | 0 (0) | |
| T790M | 0 (0) | 0 (0) |
| Biopsy Methods | n | % |
|---|---|---|
| Endobronchial lesion (+) | 45 | 20.1 |
| Endobronchial biopsy | 29 | 12.9 |
| Endobronchial biopsy + EBUS | 15 | 6.7 |
| Negative endobronchial biopsy + PCNB | 1 | 0.4 |
| Endobronchial lesion (−) | 179 | 79.9 |
| EBUS-TBNA | 79 | 35.2 |
| CT-guided PCNB | 43 | 19.2 |
| TBLB | 22 | 9.8 |
| Surgical resection | 20 | 8.9 |
| Cytology * | 15 | 6.7 |
| EGFR Genotype | Tissue | BALF (n = 224) | Tissue | Plasma (n = 110) | ||
|---|---|---|---|---|---|---|
| Mutant | Wild Type | Mutant | Wild Type | |||
| Mutant type | 93 | 91 | 2 | 66 | 32 | 34 |
| Wild type | 131 | 3 | 128 | 44 | 6 | 38 |
| Sensitivity | 97.8% (91/93) | (95% CI, 92.4–99.7) | 48.5% (32/66) | (95% CI, 35.9–61.1) | ||
| Specificity | 97.7% (128/131) | (95% CI, 93.5–99.5) | 86.3% (38/44) | (95% CI, 72.6–94.8) | ||
| PPV | 96.8% (91/94) | (95% CI, 90.8–98.9) | 84.2% (32/38) | (95% CI, 70.8–92.1) | ||
| NPV | 98.4% (128/130) | (95% CI, 93.5–99.5) | 52.7% (38/72) | (95% CI, 46.2–72.6) | ||
| Concordance rate | 97.7% ((91 + 128)/224) (95% CI, 94.8–99.2) | 63.6% ((32 + 38)/110) (95% CI, 53.9–72.6) | ||||
| Sample Type | Mean (Days) | Median (Days) | p-Value |
|---|---|---|---|
| BALF | 2.6 ± 2.03 | 2 | <0.001 |
| Tissue | 13.9 ± 12.4 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.A.; Hur, J.Y.; Kim, H.J.; Kim, W.S.; Lee, K.Y. Extracellular Vesicle-Based Bronchoalveolar Lavage Fluid Liquid Biopsy for EGFR Mutation Testing in Advanced Non-Squamous NSCLC. Cancers 2022, 14, 2744. https://doi.org/10.3390/cancers14112744
Kim IA, Hur JY, Kim HJ, Kim WS, Lee KY. Extracellular Vesicle-Based Bronchoalveolar Lavage Fluid Liquid Biopsy for EGFR Mutation Testing in Advanced Non-Squamous NSCLC. Cancers. 2022; 14(11):2744. https://doi.org/10.3390/cancers14112744
Chicago/Turabian StyleKim, In Ae, Jae Young Hur, Hee Joung Kim, Wan Seop Kim, and Kye Young Lee. 2022. "Extracellular Vesicle-Based Bronchoalveolar Lavage Fluid Liquid Biopsy for EGFR Mutation Testing in Advanced Non-Squamous NSCLC" Cancers 14, no. 11: 2744. https://doi.org/10.3390/cancers14112744
APA StyleKim, I. A., Hur, J. Y., Kim, H. J., Kim, W. S., & Lee, K. Y. (2022). Extracellular Vesicle-Based Bronchoalveolar Lavage Fluid Liquid Biopsy for EGFR Mutation Testing in Advanced Non-Squamous NSCLC. Cancers, 14(11), 2744. https://doi.org/10.3390/cancers14112744






