Simple Summary
To study various biomarkers, it is necessary to analyze multiple tissue sections through serial histological sections, which is challenging when only a small tissue sample is available. In this work we have developed a validated and objective method for combined biomarker immunostaining and its digital image analysis using open informatics tools, which is necessary for comprehensive understanding of the tumor microenvironment in rare cancers and in cases of limited samples with very significant clinical features.
Abstract
Penile, vulvar and anal neoplasms show an incidence lower than 0.5% of the population per year and therefore can be considered as rare cancers but with a dramatic impact on quality of life and survival. This work describes the experience of a Chilean cancer center using multiplexed immunofluorescence to study a case series of four penile cancers, two anal cancers and one vulvar cancer and simultaneous detection of CD8, CD68, PD-L1, Cytokeratin and Ki-67 in FFPE samples. Fluorescent image analyses were performed using open sources for automated tissue segmentation and cell phenotyping. Our results showed an objective and reliable counting of objects with a single or combined labeling or within a specific tissue compartment. The variability was below 10%, and the correlation between analytical events was 0.92–0.97. Critical cell phenotypes, such as TILs, PD-L1+ or proliferative tumor cells were detected in a supervised and unsupervised manner with a limit of detection of less than 1% of relative abundance. Finally, the observed diversity and abundance of the different cell phenotypes within the tumor microenvironment for the three studied tumor types confirmed that our methodology is useful and robust to be applicable for many other solid tumors.
1. Introduction
Anal, vulvar, and penile cancer are tumors of external genitalia considered as rare due to their low incidence among the global population, showing an incidence of 0.65, 1.2 and 0.92 cases per 100,000 habitants in 2020, respectively [1,2]. In Chile, these types of cancer are also rare, with between 100 and 140 new cases diagnosed per year. During 2020, the cases of anal, vulvar and penile cancer were 134, 128 and 93, respectively, with a prevalence at five years of 1.8, 3.5, and 2.6 per 100,000 Chileans [1].
All of these tumors have in common their development from a premalignant lesion related to the Human Papilloma Virus (HPV) infection, and in some cases arising through an HPV independent pathway [3,4].
HPV oncoproteins can induce genetic alterations affecting cell proliferation, the cell cycle, and genomic stability [5,6,7]. In addition, HPV has been proven to induce alterations in the immune microenvironment of infected tissue such as the suppression of immune recognition, promoting acute inflammation and the upregulation of immune checkpoint proteins [8,9,10].
The most common treatment for these rare cancers is chemo-radiotherapy and surgery; however, several other strategies are now in clinical trials. As with many other types of cancer, immunotherapy has also been evaluated as a treatment option [10,11,12,13], therefore a comprehensive characterization of tissue biomarkers within the tumor microenvironment of rare cancers is essential.
Tumor-infiltrating lymphocytes (TILs) have shown a crucial role in the anti-tumor immune response as have macrophages, and penile, vulvar, and anal cancer are no exception, with several groups describing the presence of these markers and their role in outcome and survival [9,14,15,16,17,18,19,20,21]. On the other hand, tumor cells can evade immune anti-tumor response using several mechanisms, such as the expression of negative immune checkpoint molecules and the modulation of the tumor microenvironment [22,23,24,25]. The expression of PD-L1 has been demonstrated in penile [26,27], vulvar [28,29] and anal [30,31] carcinomas, with different percentages of positive cells, and these previous reports about PD-L1 expression, including TILs presence, in rare cancer tissues had been evaluated using a classic immunohistochemistry (IHC) [26,27,28,29,30,31]. Even though the IHC is considered as a hallmark for cancer diagnosis, this tool has several limitations, including inter-observer variability, and the fact that it only allows the analysis of one to two biomarkers at a time [32]. Hence, in order to analyze several markers, several histological sections are required, which is challenging when only a small sample of tissue is available [33], or when the cancer type of interest is classified as rare and the tissue sample is limited by the frequency of appearance.
Several tools have been developed for multiplexing targets during histological studies [34]; however, the least expensive is based on multiplexed immunofluorescence (mIF), which advances those challenges by interrogating panels of multiple biomarkers with 5–6 being the most common and up to 9 depending on the instruments and methodology. Due to this advantage, the mIF analysis is a useful tool to characterize the tumor microenvironment, particularly in tumor types with low incidences such as penile, vulvar and anal cancer, and a limited amount of tissue for further studies.
In this study we describe the technical approach during the implementation of two panels of validated antibodies to evaluate the proliferation rate of tumor cells based on Ki67 signal, tumor infiltrating immune cells by CD8 signal, and by CD68 detection for macrophages, and the tissue localization of the immune checkpoint PD-L1, applying tissue segmentation and cell phenotyping to interrogate the cytokeratin positive and negative compartments in retrospective cases of rare cancer in Chilean patients.
2. Materials and Methods
2.1. Patients and Case Selection
This is a single-institution retrospective study at Instituto Oncológico Fundación Arturo López Pérez (Santiago, Chile), reviewed and approved by the institutional ethical committee (Prot. 007-RES-ANC-MUL). Deceased patients with histologically proven invasive squamous cell carcinoma of penis, vulva and anus diagnosed in the Institution during 2010–2014 who died because of these diseases between 2014 and 2017 were considered eligible. The respective historical FFPE block was collected from the internal biorepository after the Institutional Ethical Committee approval of the waiver of a signed consent letter. Twenty patient records were retrospectively found, however four cases of penile cancer, one case of vulvar cancer and two of anal cancer had complete clinical records and a suitable tissue sample (Table 1). Clinical and demographic data were retrieved from patient medical records and managed according to privacy regulations.

Table 1.
Clinical and demographic information of the case series of anal, vulvar and penile cancer included in this research.
2.2. Sample Processing and Multiplexed Immunofluorescence
For each tumor type, all available tissue cores were requested and examined by a pathologist to confirm the histology and the integrity of the material. Based on hematoxylin-eosin visual inspection, all cores with an area of less than 5% of the tumor were excluded. Immunostaining was carried out as published [35] and each primary cocktail (or panel) was developed using validated antibodies [35,36]. In panel 1: PD-L1 (clone E1L3N), Cytokeratin (clone AE1/AE3), CD68 (clone PG-M1) and CD8a (clone C8/144B). In panel 2: PD-1 (clone D4W2J), Cytokeratin (clone AE1/AE3) and Ki67 (clone MIB-1). Briefly, after antigen retrieval, each whole tissue slide was incubated overnight at 4 °C with the selected panel separately. Isotype-specific HRP-conjugated antibodies and tyramide-based amplification systems (Perkin Elmer, Waltham, MA, USA) were used for signal detection. After co-staining with DAPI, each slide was cover slipped in Prolong Gold mounting medium (Invitrogen, Carlsbad, CA, USA). An index TMA was stained in parallel as quality control. This array included human tissue as naturally positive and negative controls, such as skeletal muscle (negative/noise threshold), tonsil (positive for immune cells), and placenta (positive for PD-L1 immune checkpoint). The dynamic range of positivity for each biomarker was estimated from the ratio between the mean fluorescent signal from a positive control tissue and a negative control tissue. Pearson’s coefficients were calculated between independent staining runs or for single markers to verify concordances between experiments.
2.3. Cases and FFPE Cores Considerations
For some analysis considering average values in penile cancer, only cases with more than one FFPE core were considered, however for single case representation all cases were included. For the single case of vulvar cancer, the three FFPE cores available were processed and analyzed in some figures as average values or individual values. For anal cancer, each case and its respective FFPE core was analyzed separately.
2.4. Slide Scanning and Image Generation
All slides were scanned at 20× of magnification using an Aperio VERSA 200 microscope (Leica Biosystems, Vista, CA, USA). Exposure times were determined for each target using an index TMA including normal human tissue as positive and negative controls, such as skeletal muscle, tonsil and placenta. The optimal exposure time considered the optimal signal to noise ratio measured for each target and the supervised observation of the operator for the visual positive signal and the expected staining pattern. Later, for each antibody panel, an unsupervised template was designed for the automatic and simultaneous scanning of the fluorescent signal and applied to the tumor whole tissue section. The templates considered the following steps: an initial low-resolution scanning at 1.25× for automatic tissue detection, a second scanning at 5× for fluorescent signal detection (using DAPI); and a final scanning at 20× including sequentially FITC, Cy3, Cy5 and Cy7 filters. Images were saved in scn extension and size varied from 26,976 × 32,249 to 60,440 × 32,249 pixels. After acquisition, all images were revised and exported for further analysis.
2.5. Analysis for Single Objects
Whole-slide scans from the fluorescent-labeled sections were analyzed using QuPath v.0.3.2 software (University of Edinburgh. Edinburgh, Scotland, UK) [37]. DAPI channel was used for cell segmentation using the following settings: Pixel size: 0.5 μm; Background radius: 8.0 μm; Median filter radius: 0.0 μm; Minimum area: 10.0 μm2; Maximum area: 200 μm2; Threshold: 5.0.
After cell detection, cells were classified using the Object classifier module to determine the number of positive cells for each marker after applying the defined visual cutoff (fluorescent intensity ratio). At least 400 cells from different cores were used to train each object classifier. After every object classifier was trained, they were run for each image using the script editor for batch analysis. Data was exported and analyzed using Microsoft Office tools. The number of total positive cells for each biomarker was determined by the addition of the positive cells for individual biomarkers and the positive ones for more than one biomarker. Percentages were calculated based on the total cell detections.
2.6. Analysis for Segmentation and Masking
The pixel classifier from QuPath was trained under manual supervision to detect four different regions: Ignore, corresponding to the sections of the image that had no tissue (glass); Negative, corresponding to regions with auto-fluorescence, mostly red blood cells; Tumor region, defined by Cytokeratin and DAPI positive signal, and Stroma region, corresponding to areas without signal for Cytokeratin, but including DAPI positive objects. For the pixel classifier, 28 fields of view from different tissue images were generated and used for the image training, and the trained classifier was applied in batch or as a single run to each digitized image. The automatic tumor masking was verified visually based on nuclei morphology. Once Pixel classifiers were verified, the tumor and stromal areas were set as annotations and cells within each annotation (tumor or stroma) were classified using the Object Classifier plugin. Data was exported and analyzed in Microsoft Excel. Percentages were calculated based on the total cell detections.
3. Results
3.1. Immunostaining Verification
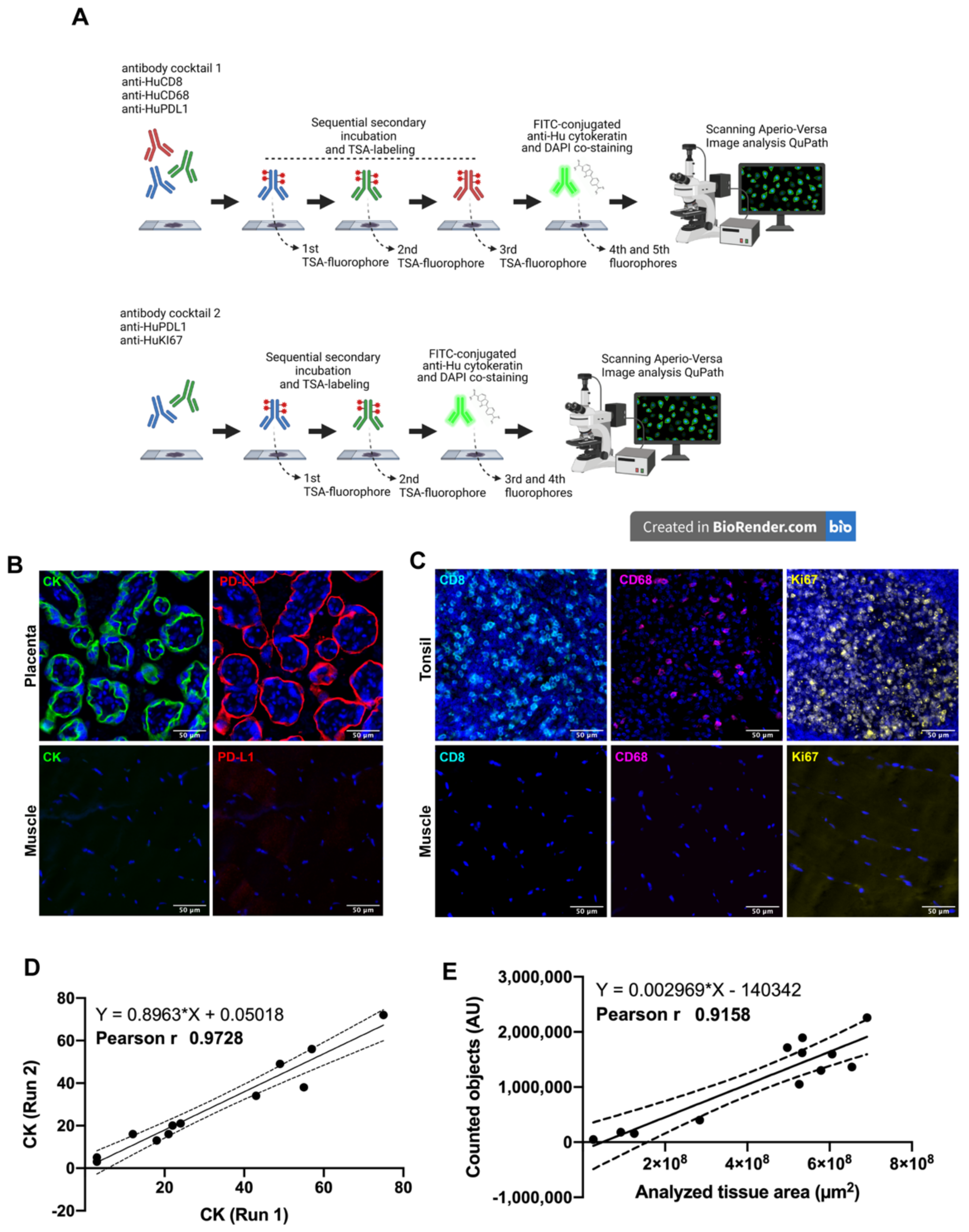
Two antibody panels using multiplexed immunofluorescence were designed for the simultaneous detection of CD8+ T-lymphocytes, CD68+ macrophages, Cytokeratin (CK) for tumor cells, PD-L1 immune checkpoint andKi67 as a proliferation marker (Figure 1A). Naturally positive and negative tissues were used as a procedure control: skeletal muscle was used as a negative control while placenta and tonsil tissues were used as positive controls, placenta for antibodies anti-PD-L1 and anti-CK, and tonsil for antibodies anti-Ki67, anti-CD8 and anti-CD68. As we expected, each control tissue showed a very distinctive staining pattern in agreement with previous validation and other reports [35,36]. Placenta showed a membranous and cytoplasmic distribution of PD-L1 and cytokeratin in the trophoblast cells (Figure 1B). Regarding the immune cells, CD8+ lymphocytes and CD68+ macrophages were detected mostly at the medullar and interfollicular area of human tonsils (Figure 1C), and most of the proliferative cells were located within the germinal center as observed through the Ki67 positive signal, which was very well defined and distributed in the nuclear compartment (DAPI co-staining), and only a few cases showed a granular signal at the periphery of the nuclei (Figure 1D). Skeletal muscle did not show fluorescent signal for any tested antibody (Figure 1C,D, lower inserts).
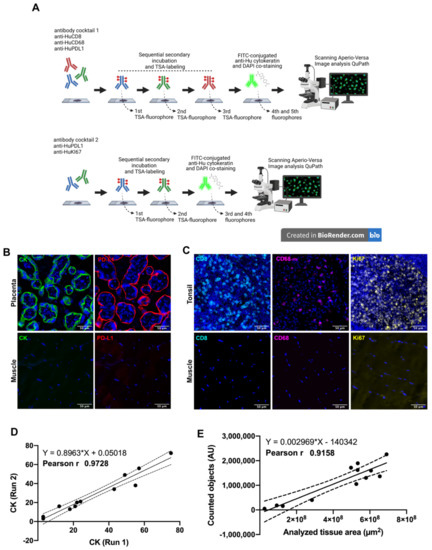
Figure 1.
Multiplex Immunofluorescence workflow. In (A) after antigen retrieval, each whole tissue slide was incubated with the indicated antibody panel. Sequential signal detection required isotype-specific HRP-conjugated antibodies, tyramide-based fluorophores and digitalization using Aperio VERSA 200 microscope (designed using Biorender). In (B,C), the staining pattern for positive control tissues are shown. Images are representative for cytokeratin (CK) and PD-L1 in human placenta (B). Immune markers CD8 and CD68, and proliferation as Ki67 were verified in human tonsil (C). Skeletal muscle was used as negative control. Correlation between two independent staining runs is shown for CK (D) as well for the correlation between the scanned area and counted objects (E). Scale bar represents 50 µm. Nuclei were stained with DAPI.
The CK intensity (signal) between two independent staining runs showed a Pearson’s coefficient of 0.97 (Figure 1D), as well as the amount of counted elements using DAPI classifier showing a positive correlation (r 0.91 and p-value 0.001) after analyzing several tissue sizes (Figure 1E). Both parameters were representative for other staining procedures.
3.2. Multiplexed Immunofluorescence in Rare Cancer Cases Series
Seven retrospective cases with a diagnosis of a rare cancer were selected to interrogate the immune composition and the characteristics of squamous tumor cells in anal, vulvar and penile cancer (Table 1).
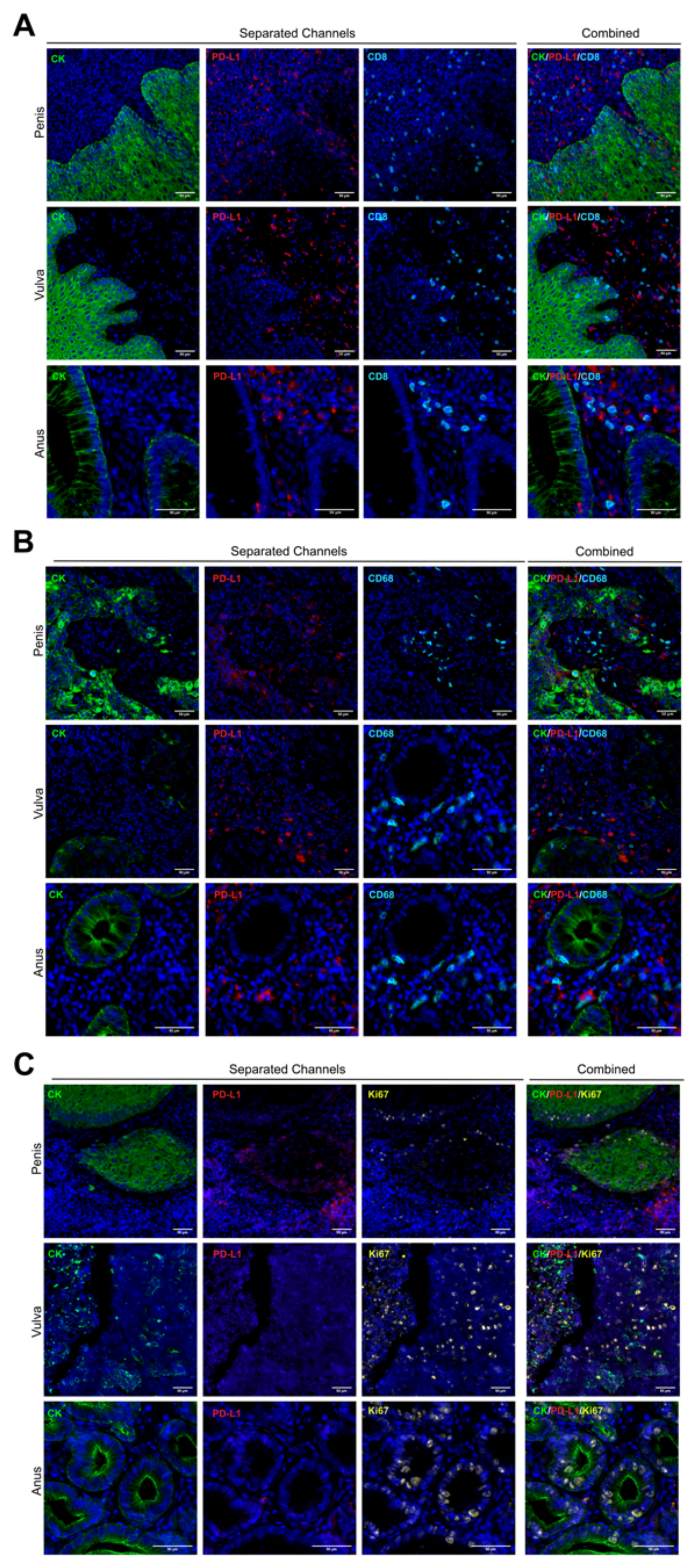
Based on a methodology previously described [35], we stained whole tissue sections of formalin-fixed paraffin-embedded (FFPE) samples from the chosen cases described above. No visual differences in staining patterns were observed for any tumor type (Figure 2). As we expected, tumor cores were reactive for the cytokeratin (CK) antibody clone AE1/AE3, and they showed a membranous-cytoplasmic staining pattern in all cases (Figure 2A–C).
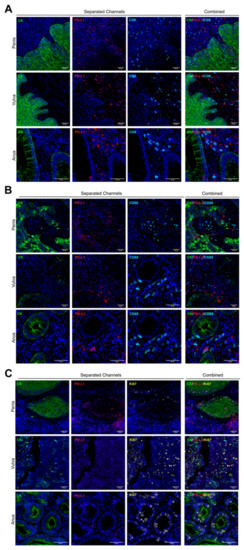
Figure 2.
Multiplexed biomarker staining patterns. Whole tissue sections were assessed by multiplexed immunofluorescence. Staining patterns for each biomarker are shown in separated and combined channels, and the images are representative for all cores and tumor types. The cytokeratin CK and PD-L1 area are pseudo-colored in green and red, respectively (A–C). CD8 (A) and CD68 (B) are presented in cyan, and Ki67 is was colored in yellow (C). 20× of magnification. Scale bar represents 50 µm.
PD-L1 protein was mostly positive in penile tumor cells but with very low detection in anal and vulvar squamous tumor cells (Figure 2). Several non-tumor cells (CK negative) within the stromal area showed a strong signal for PD-L1, but without a clear trend for any tumor types or analyzed core (Figure 2 and Figure 3E). Lymphocytes and macrophages were observed surrounding and infiltrating the tumor compartment for the three tumor types; however, TILs showed a preferential stromal distribution in vulvar cancer (Supplementary Figure S1). Based on the Ki67 signal, a broad range of proliferative cells were observed, without a significant mean trend for any cancer type (Figure 2C).
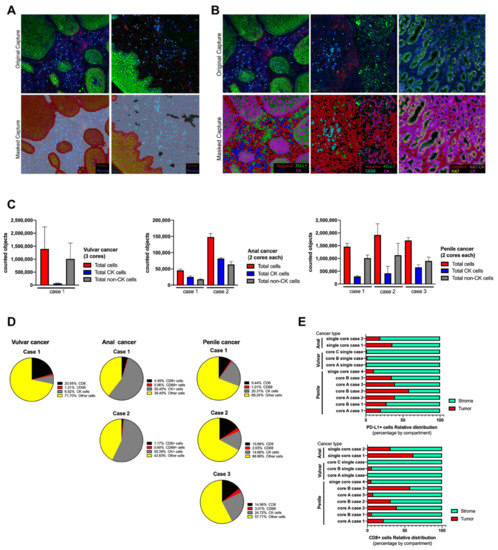
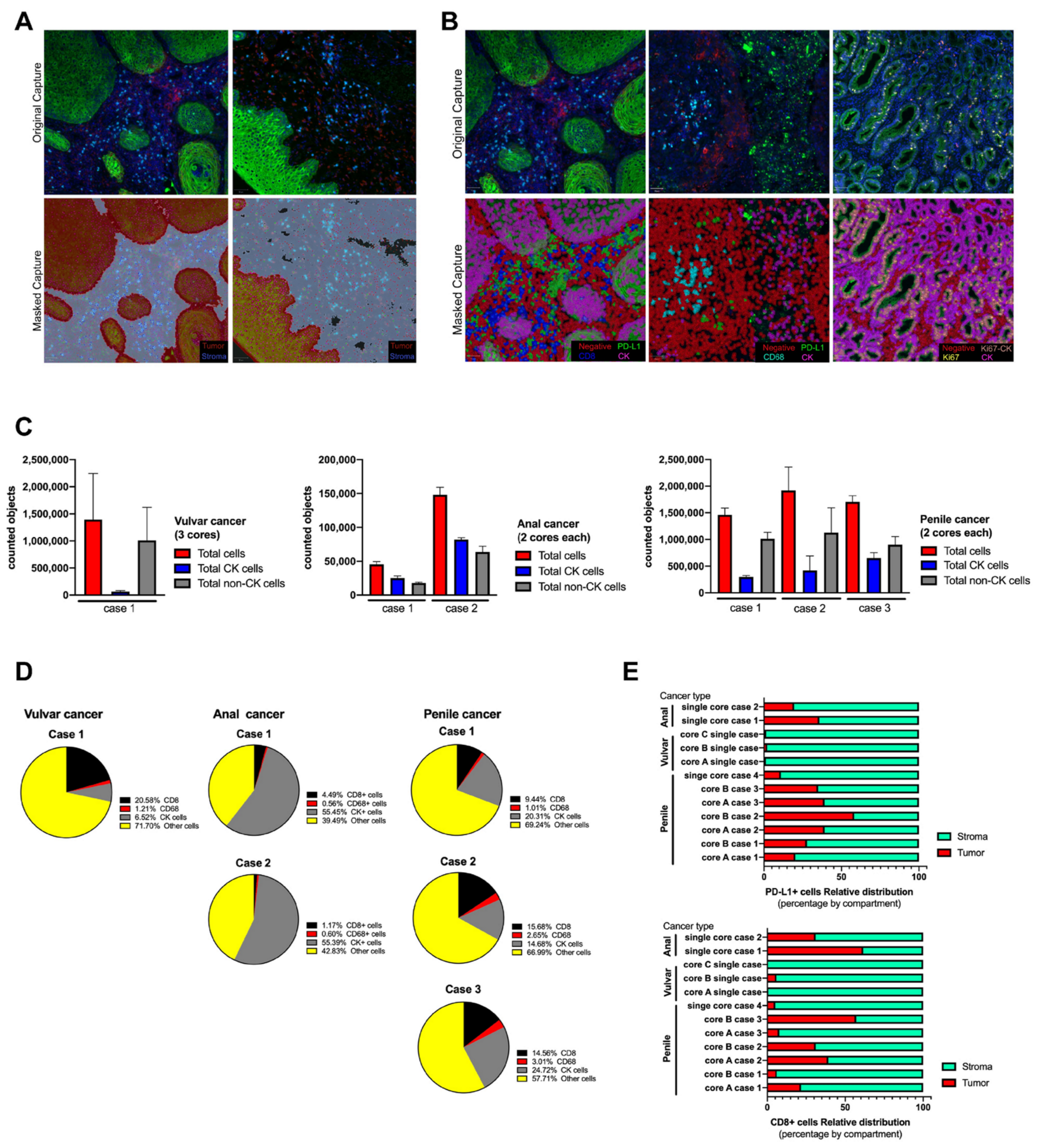
Figure 3.
Tissue segmentation and cell phenotyping in cases series of anal, vulvar and penile cancer. Based on QuPath algorithms, the scanned whole tissue sections were masked as tumor and stroma compartments (A) using cytokeratin (CK) signal and using single classifiers all cells were labeled and phenotyped (B). All counted objects were classified within the tissue section as tumor and non-tumor cells and represented as the average between cores per each case and tumor type (C). The relative abundance of infiltrating immune cells and tumor cells was expressed in percentage from the total counted cells (D) and calculated for each studied case. After tissue segmentation as tumor (CK positive area) and stroma (CK negative area), CD8 and PD-L1 signal was counted as individual and its distribution within both compartments was expressed as a percentage (E). To test variability, the anal cancer cases were processed twice, and for penile cancer only cases with more than one core were included.
3.3. Image Analysis by Tissue Segmentation and Cell Phenotyping
As a first approach, two antibody panels for mIF were designed and considered PD-L1 and cytokeratin as common targets to be used as reference to identify common elements and perform association between biomarkers detected in different slides.
For the cell segmentation analysis, all images were analyzed using the positive cell detection module of QuPath software and DAPI as reference for single counts [37]. No significant differences were observed when an area of 100µm2 was manually counted and compared with the counts using QuPath, and the estimated error for the automated single cell segmentation was 2–5% (data not shown). After cell segmentation by nuclei detection, QuPath object classifier module generated a single-color mask for each targeted biomarker. Positive cells were masked in color based on QuPath color code (Figure 3A,B). Tumor and stroma compartments were built using the positive and negative areas for CK signal, respectively, and masked as shown (Figure 3A). The accuracy increased with the intensity of the fluorescent signal and the depth of the selected pixels. A similar observation was verified with the cell classifier (Figure 3B), however, because the area per single cell was smaller, the accuracy depended on the initial DAPI detection. Each tissue section was analyzed twice, “one by one” and “in batch” mode, and the masking images were comparable with each other. For each biomarker, the estimated error of counted cells between cores was below 10%, and the highest variability was observed for the proportion of CK+ cells with respect to the total cells counted within the stained section per core (data not shown).
After defining the settings for an automated tissue segmentation and cell phenotyping, all images were processed and the abundance of each cell type was counted per scanned area and expressed as the average between cores for the same case (Figure 3C,D). All sections showed variability in the number of counted objects without affecting the trend for each tumor case (Supplementary Figure). Regarding immune cells, CD68 macrophages were the less abundant object for any analyzed tissue (Figure 3D) which was related to its biology per se and not to the intensity of signal as in cytokeratin. On the other hand, CD8 TILs were frequently detected and their distribution was preferentially within the stroma compartment (Figure 3E and Supplementary Materials). Very few cells showed reactivity for PD-L1, which limited the analysis, however this immune checkpoint did not show a predominant distribution or compartmentalization between stroma and tumor. Penile and anal cancer cases showed a clear expression of PD-L1 and the variability between tissue cores was smaller than the observed distribution of CD8 cells (Figure 3D). For a better understanding, the cell phenotyping analysis was applied in tumor cells using the combination of key biomarkers, such as PD-L1 and Ki67, and expressed as a percentage of relative abundance within a selected region (Table 2); the counted objects allowed to express it as an objective score and was able to count less than 1% of a specific cell phenotype within the total scanned area.

Table 2.
Tumor cells phenotyping and multiple biomarker labeling.
4. Discussion
The Society for Immunotherapy of Cancer (SITC) has provided the fundamentals of best practices for multiplex immunohistochemistry (mIHC) and immunofluorescence (mIF) staining and validation, and suggested that mIHC/mIF technologies are becoming standard tools for biomarker studies and are likely to enter routine clinical practice in the near future [38]. For that reason, a careful assay optimization and validation is required to ensure that outputs are robust and comparable across laboratories as well as potentially across mIHC/mIF platforms.
This study represents the beginning of the use of multiplexing and its validation process developed by a Chilean cancer center based on the recommendations of the SITC, and included an objective and reliable approach for the characterization of a tumor microenvironment in FPEE samples from invasive squamous cell carcinomas of the penis, vulva and anus. These types of tumors are classified as rare for their low incidence in the population compared to other types of malignancies [1,39,40]. Since the number of samples is always limited for this type of cancer, we have developed a multiplex immunofluorescence analysis that allowed us to obtain more information from a single histological section.
After reviewing three different case series of two anal cancers (single tissue core each), of one vulvar cancer (three tissue cores), and of four penile cancers (two cores and one single core), using a single biomarker for cell phenotyping may reduce the possibility for the researcher and the pathologist to detect the natural variability between cores and cases, and the combination of biomarkers may highlight the differences between patients and even between tissue cores from a same patient.
As far as we know, this is one of the first studies to analyze FFPE samples from rare cancers in Chilean patients by multiplexed immunofluorescence (mIF). Previously, studies based on image analysis from FFPE samples from these types of cancer were performed using classic IHC, which has a limitation regarding the number of biomarkers to analyze and a high inter-observer variability [32,33].
The mIF facilitates the study of these rare cancers because it enables us to analyze the co-expression of different biomarkers for a better understanding of the tumor microenvironment, identifying different cell phenotypes combining biomarkers and maintaining the spatial distribution for each detected element [41]. Using an open-source software such as QuPath allows anyone, including research laboratories with limited resources, to collect a large amount of data, from single counts, location and distribution and regional abundance using positive pixel training to identify the target of interest [42,43,44,45]. Based on the software’s plugins, we analyzed the number of positive cells for each biomarker in the whole tissue section and ran the analysis in batch, which reduced time and variability inter-reader [45,46]. After managing the single cell identifier, larger areas of tissue can be analyzed by specific signal masking, such as tumor nest and stroma. The QuPath plugin, through a supervised training, identifies pixels with a pan-cytokeratin antibody signal, segregating them into pixels with and without information to define an algorithm for the automatic tissue masking and the execution of a secondary analysis, such as counting single elements within the selected area as a regular machine learning operation [47,48].
The selection of biomarkers used in this work was based on their clinical relevance and role in tumor microenvironment adaptation. Tumor-infiltrating CD8 lymphocytes are considered as the major inflammatory component in almost all known solid tumors and are responsible for killing tumor cells by their cytotoxic activity [49,50] In addition, these TILs may become exhausted because they are exposed to a sustained immune suppressive environment, such as tumor cells and myeloid cells expressing PD-L1. As reported by our collaborators and other researchers, using immune checkpoint blockers may reinvigorate some cytotoxic CD8 T cells and restore their effector role against several tumor types [51,52]. In addition, radiotherapy is frequently used to treat these type of cancers, and recently an abscopal effect after irradiation regimens was described in preclinical models where CD8 T cells were shown to be crucial in improving the response to immunomodulatory monoclonal antibodies, such as anti-PD1 or anti-CD137 [52].
Different mechanisms of immune evasion have been studied in penile [11,21,53,54] vulvar [14,18,20,55] and anal cancers [17,19], and included immune checkpoint proteins and exhausted phenotypes for infiltrating-lymphocytes, suggesting that patients with these type of tumors can benefit from anti PD-L1 therapy [9,27,56,57,58,59]. Currently, there are several clinical trials ongoing for the use of immune therapy targeting PD-L1 (or its receptor PD-1) as treatment for penile, vulvar or anal cancer [60,61,62,63,64,65,66,67,68,69]. Therefore, multiplexed immunofluorescence and an objective analysis of the tumor microenvironment may contribute significantly to improve the understanding of different therapeutic targets when the access to biological material is reduced or limited.
Flow cytometry has been the most extended tool to characterize specific cell phenotypes based on the multiple detection of surface and intracellular biomarkers, in particular for non-tumor cells within the tumor microenvironment, such as infiltrating lymphocytes [70] and tumor-associated macrophages (TAM) [71]. We did not include specific phenotypes, such as M1 or M2 macrophages, because of the lack of validated antibodies for histochemistry or the complexity of combining some of them, and CD68 alone may be considered as an indicator of TAM-like cells. Other limitations of this study include the small number of analyzed cases which may affect the ability of the trained algorithms to recognize a variable related to a specific-tissue type, in particular in the only one case of vulvar cancer. In addition, we were aware of the absence of statistical power, which limited the analysis but it does not invalidate the histological findings and developed tools. Finally, we determined the localization of single objects but we did not carry out algorithms to measure distance between objects, which has also been described as being determinant in tumor microenvironment characterization [72].
5. Conclusions
We have described two multiplex antibody panels to identified biomarkers and developed a supervised machine learning analysis based on positive pixels that is able to do tissue segmentation, cell phenotyping and object counting for the objective and automated scoring of immune checkpoint proteins, tissue immune infiltration and proliferation in FFPE samples from solid tumors such as penile, vulvar, or anal cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14112815/s1, Figure S1: Counted elements per case and tissue core.
Author Contributions
Conceptualization, F.V.-E.; methodology, F.V.-E., R.G.-S. and K.C.; software, K.C., N.B. and R.J.; validation, F.V.-E. and K.C.; formal analysis, F.V.-E. and K.C.; investigation, F.V.-E. and K.C.; resources, F.V.-E., K.C. and R.G.-S.; data curation, N.B. and K.C.; writing—original draft preparation, K.C. and F.V.-E.; writing—review and editing, F.V.-E., R.G.-S., K.C. and R.J.; visualization, F.V.-E. and K.C.; supervision, F.V.-E.; project administration, F.V.-E. and R.G.-S.; funding acquisition, F.V.-E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Foundation Arturo Lopez Perez (FALP) internal grant number FALP-LMT-2020 (FVE), National Fund for Scientific and Technological Development, FONDECYT grant number 1221415 (FVE). The APC was funded by FALP.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Foundation Arturo Lopez Perez FALP (protocol 007-RES-ANC-MUL, 08/09/2020). A waiver of a signed consent letter was authorized for FFPE samples from deceased cases.
Informed Consent Statement
Patient consent was waived due to all cases included in this work were deceased by the time of ethical revision.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical restrictions.
Acknowledgments
We thank Jose Pablo Aguilera and Carolina Selman from the Anatomical Pathology Unit of FALP for helping with the FFPE sample rescue and Kathia D’Appollonio for the identification of relevant cases.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Globocan 2020 World Health Organization International Agency for Research on Cancer (IARC). GLOBOCAN 2020: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2020. Available online: https://gco.iarc.fr/ (accessed on 1 March 2020).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Douglawi, A.; Masterson, T.A. Updates on the Epidemiology and Risk Factors for Penile Cancer. Transl. Androl. Urol. 2017, 6, 785–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, P.A.; Anderson, L.; Eva, L.; Scurry, J. Clinical and Molecular Classification of Vulvar Squamous Pre-Cancers. Int. J. Gynecol. Cancer 2019, 29, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-Wide Analysis of HPV Integration in Human Cancers Reveals Recurrent, Focal Genomic Instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodelon, C.; Untereiner, M.E.; Machiela, M.J.; Vinokurova, S.; Wentzensen, N. Genomic Characterization of Viral Integration Sites in HPV-Related Cancers. Int. J. Cancer 2016, 139, 2001–2011. [Google Scholar] [CrossRef]
- Yang, W.; Liu, Y.; Dong, R.; Liu, J.; Lang, J.; Yang, J.; Wang, W.; Li, J.; Meng, B.; Tian, G. Accurate Detection of HPV Integration Sites in Cervical Cancer Samples Using the Nanopore MinION Sequencer Without Error Correction. Front. Genet. 2020, 11, 660. [Google Scholar] [CrossRef]
- Shamseddine, A.A.; Burman, B.; Lee, N.Y.; Zamarin, D.; Riaz, N. Tumor Immunity and Immunotherapy for HPV-Related Cancers. Cancer Discov. 2021, 11, 1896–1912. [Google Scholar] [CrossRef]
- Balermpas, P.; Martin, D.; Wieland, U.; Rave-Fränk, M.; Strebhardt, K.; Rödel, C.; Fokas, E.; Rödel, F. Human Papilloma Virus Load and PD-1/PD-L1, CD8+ and FOXP3 in Anal Cancer Patients Treated with Chemoradiotherapy: Rationale for Immunotherapy. Oncoimmunology 2017, 6, e1288331. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.Y.; Allen, C.T. Immunotherapy for HPV Malignancies. Semin. Radiat. Oncol. 2021, 31, 361–370. [Google Scholar] [CrossRef]
- de Vries, H.M.; Ottenhof, S.R.; Horenblas, S.; van der Heijden, M.S.; Jordanova, E.S. Defining the Tumor Microenvironment of Penile Cancer by Means of the Cancer Immunogram. Eur. Urol. Focus 2019, 5, 718–721. [Google Scholar] [CrossRef] [Green Version]
- Pessia, B.; Romano, L.; Giuliani, A.; Lazzarin, G.; Carlei, F.; Schietroma, M. Squamous Cell Anal Cancer: Management and Therapeutic Options. Ann. Med. Surg. 2020, 55, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Vanthoor, J.; Vos, G.; Albersen, M. Penile Cancer: Potential Target for Immunotherapy? World J. Urol. 2021, 39, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, Z.; Kortekaas, K.E.; De Vos Van Steenwijk, P.J.; Van Der Burg, S.H.; Van Poelgeest, M.I.E. The Immune Microenvironment in Vulvar (Pre)Cancer: Review of Literature and Implications for Immunotherapy. Expert Opin. Biol. Ther. 2018, 18, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Aydin, A.M.; Chahoud, J.; Adashek, J.J.; Azizi, M.; Magliocco, A.; Ross, J.S.; Necchi, A.; Spiess, P.E. Understanding Genomics and the Immune Environment of Penile Cancer to Improve Therapy. Nat. Rev. Urol. 2020, 17, 555–570. [Google Scholar] [CrossRef]
- Chu, C.; Yao, K.; Lu, J.; Zhang, Y.; Chen, K.; Lu, J.; Zhang, C.Z.; Cao, Y. Immunophenotypes Based on the Tumor Immune Microenvironment Allow for Unsupervised Penile Cancer Patient Stratification. Cancers 2020, 12, 1796. [Google Scholar] [CrossRef] [PubMed]
- Geltzeiler, C.B.; Xu, Y.; Carchman, E.; Ghouse, Y.; Beczkiewicz, J.; Son, J.; Voils, C.I.; Striker, R. CD4/CD8 Ratio as a Novel Marker for Increased Risk of High-Grade Anal Dysplasia and Anal Cancer in HIV+ Patients: A Retrospective Cohort Study. Dis. Colon Rectum 2020, 63, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Heeren, A.M.; Rotman, J.; Samuels, S.; Zijlmans, H.J.M.A.A.; Fons, G.; Van De Vijver, K.K.; Bleeker, M.C.G.; Kenter, G.G.; Jordanova, E.J.; De Gruijl, T.D. Immune Landscape in Vulvar Cancer-Draining Lymph Nodes Indicates Distinct Immune Escape Mechanisms in Support of Metastatic Spread and Growth. J. Immunother. Cancer 2021, 9, e003623. [Google Scholar] [CrossRef]
- Hu, W.H.; Miyai, K.; Cajas-Monson, L.C.; Luo, L.; Liu, L.; Ramamoorthy, S.L. Tumor-Infiltrating CD8+ T Lymphocytes Associated with Clinical Outcome in Anal Squamous Cell Carcinoma. J. Surg. Oncol. 2015, 112, 421–426. [Google Scholar] [CrossRef]
- Kortekaas, K.E.; Santegoets, S.J.; Abdulrahman, Z.; Van Ham, V.J.; Van Der Tol, M.; Ehsan, I.; Van Doorn, H.C.; Bosse, T.; Van Poelgeest, M.I.E.; Van Der Burg, S.H. High Numbers of Activated Helper T Cells Are Associated with Better Clinical Outcome in Early Stage Vulvar Cancer, Irrespective of HPV or P53 Status. J. Immunother. Cancer 2019, 7, 236. [Google Scholar] [CrossRef] [Green Version]
- Ottenhof, S.R.; Djajadiningrat, R.S.; Thygesen, H.H.; Jakobs, P.J.; Józwiak, K.; Heeren, A.M.; de Jong, J.; Sanders, J.; Horenblas, S.; Jordanova, E.S. The Prognostic Value of Immune Factors in the Tumor Microenvironment of Penile Squamous Cell Carcinoma. Front. Immunol. 2018, 9, 1253. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zhang, J.; Wang, J.; Fu, J.; Li, T.; Zheng, X.; Wang, B.; Gu, S.; Jiang, P.; Fan, J.; et al. Publisher Correction: Landscape of B Cell Immunity and Related Immune Evasion in Human Cancers. Nat. Genet. 2019, 51, 1068. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Zhang, S.; Gong, Z.; Li, X.; Xiang, B.; Deng, H.; Zhou, M.; Li, G.; Li, Y.; Xiong, W.; et al. Emerging Role of Metabolic Reprogramming in Tumor Immune Evasion and Immunotherapy. Sci. China Life Sci. 2021, 64, 534–547. [Google Scholar] [CrossRef] [PubMed]
- Daassi, D.; Mahoney, K.M.; Freeman, G.J. The Importance of Exosomal PDL1 in Tumour Immune Evasion. Nat. Rev. Immunol. 2020, 20, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Stanger, B.Z. How Tumor Cell Dedifferentiation Drives Immune Evasion and Resistance to Immunotherapy. Cancer Res. 2020, 80, 4037–4041. [Google Scholar] [CrossRef] [PubMed]
- Davidsson, S.; Carlsson, J.; Giunchi, F.; Harlow, A.; Kirrander, P.; Rider, J.; Fiorentino, M.; Andrén, O. PD-L1 Expression in Men with Penile Cancer and Its Association with Clinical Outcomes. Eur. Urol. Oncol. 2019, 2, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Udager, A.M.; Liu, T.Y.; Skala, S.L.; Magers, M.J.; McDaniel, A.S.; Spratt, D.E.; Feng, F.Y.; Siddiqui, J.; Cao, X.; Fields, K.L.; et al. Frequent PD-L1 Expression in Primary and Metastatic Penile Squamous Cell Carcinoma: Potential Opportunities for Immunotherapeutic Approaches. Ann. Oncol. 2016, 27, 1706–1712. [Google Scholar] [CrossRef] [Green Version]
- Choschzick, M.; Gut, A.; Fink, D. PD-L1 Receptor Expression in Vulvar Carcinomas Is HPV-Independent. Virchows Arch. 2018, 473, 513–516. [Google Scholar] [CrossRef]
- Thangarajah, F.; Morgenstern, B.; Pahmeyer, C.; Schiffmann, L.M.; Puppe, J.; Mallmann, P.; Hamacher, S.; Buettner, R.; Alidousty, C.; Holz, B.; et al. Clinical Impact of PD-L1 and PD-1 Expression in Squamous Cell Cancer of the Vulva. J. Cancer Res. Clin. Oncol. 2019, 145, 1651–1660. [Google Scholar] [CrossRef]
- Bucau, M.; Gault, N.; Sritharan, N.; Valette, E.; Charpentier, C.; Walker, F.; Couvelard, A.; Abramowitz, L. PD-1/PD-L1 Expression in Anal Squamous Intraepithelial Lesions. Oncotarget 2020, 11, 3582–3589. [Google Scholar] [CrossRef]
- Govindarajan, R.; Gujja, S.; Siegel, E.R.; Batra, A.; Saeed, A.; Lai, K.; James, J.D.; Fogel, B.J.; Williamson, S. Programmed Cell Death-Ligand 1 (PD-L1) Expression in Anal Cancer. Am. J. Clin. Oncol. Cancer Clin. Trials 2018, 41, 638–642. [Google Scholar] [CrossRef]
- Varga, Z.; Diebold, J.; Dommann-Scherrer, C.; Frick, H.; Kaup, D.; Noske, A.; Obermann, E.; Ohlschlegel, C.; Padberg, B.; Rakozy, C.; et al. How Reliable Is Ki-67 Immunohistochemistry in Grade 2 Breast Carcinomas? A QA Study of the Swiss Working Group of Breast- and Gynecopathologists. PLoS ONE 2012, 7, e37379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Herck, Y.; Antoranz, A.; Andhari, M.D.; Milli, G.; Bechter, O.; De Smet, F.; Bosisio, F.M. Multiplexed Immunohistochemistry and Digital Pathology as the Foundation for Next-Generation Pathology in Melanoma: Methodological Comparison and Future Clinical Applications. Front. Oncol. 2021, 11, 636681. [Google Scholar] [CrossRef] [PubMed]
- Cereceda, K.; Jorquera, R.; Villarroel-Espíndola, F. Advances in Mass Cytometry and Its Applicability to Digital Pathology in Clinical-Translational Cancer Research. Adv. Lab. Med. 2021, 3, 5–16. [Google Scholar] [CrossRef]
- Villarroel-Espindola, F.; Yu, X.; Datar, I.; Mani, N.; Sanmamed, M.; Velcheti, V.; Syrigos, K.; Toki, M.; Zhao, H.; Chen, L.; et al. Spatially Resolved and Quantitative Analysis of Vista/Pd-1h as a Novel Immunotherapy Target in Human Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 1562–1573. [Google Scholar] [CrossRef] [Green Version]
- Pelekanou, V.; Villarroel-espindola, F.; Schalper, K.A.; Pusztai, L.; Rimm, D.L. CD68, CD163, and matrix metalloproteinase 9 (MMP-9) Co-Localization in Breast Tumor Microenvironment Predicts Survival Differently in ER-Positive and -Negative Cancers. Breast Cancer Res. 2018, 20, 1–10. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open Source Software for Digital Pathology Image Analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Taube, J.M.; Akturk, G.; Angelo, M.; Engle, E.L.; Gnjatic, S.; Greenbaum, S.; Greenwald, N.F.; Hedvat, C.V.; Hollmann, T.J.; Juco, J.; et al. The Society for Immunotherapy in Cancer Statement on Best Practices for Multiplex Immunohistochemistry (IHC) and Immunofluorescence (IF) Staining and Validation. J. Immunother. Cancer 2020, 8, e000155. [Google Scholar] [CrossRef]
- Eslick, G.D. What Is a Rare Cancer? Hematol. Oncol. Clin. N. Am. 2012, 26, 1137–1141. [Google Scholar] [CrossRef]
- Bondei, I.T.; Suteu, O.; Todor, N.; Nagy, V.M. Rare Tumors: A Comprehensive Analysis of Cancer. J. BUON 2019, 24, 2173–2179. [Google Scholar]
- Wong, P.F.; Wei, W.; Smithy, J.W.; Acs, B.; Toki, M.I.; Blenman, K.R.M.; Zelterman, D.; Kluger, H.M.; Rimm, D.L. Multiplex Quantitative Analysis of Tumor-Infiltrating Lymphocytes and Immunotherapy Outcome in Metastatic Melanoma. Clin. Cancer Res. 2019, 25, 2442–2449. [Google Scholar] [CrossRef] [Green Version]
- Viratham Pulsawatdi, A.; Craig, S.G.; Bingham, V.; McCombe, K.; Humphries, M.P.; Senevirathne, S.; Richman, S.D.; Quirke, P.; Campo, L.; Domingo, E.; et al. A Robust Multiplex Immunofluorescence and Digital Pathology Workflow for the Characterisation of the Tumour Immune Microenvironment. Mol. Oncol. 2020, 14, 2384–2402. [Google Scholar] [CrossRef] [PubMed]
- Humphries, M.P.; Maxwell, P.; Salto-Tellez, M. QuPath: The Global Impact of an Open Source Digital Pathology System. Comput. Struct. Biotechnol. J. 2021, 19, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Berben, L.; Wildiers, H.; Marcelis, L.; Antoranz, A.; Bosisio, F.; Hatse, S.; Floris, G. Computerised Scoring Protocol for Identification and Quantification of Different Immune Cell Populations in Breast Tumour Regions by the Use of QuPath Software. Histopathology 2020, 77, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Loughrey, M.B.; Bankhead, P.; Coleman, H.G.; Hagan, R.S.; Craig, S.; McCorry, A.M.B.; Gray, R.T.; McQuaid, S.; Dunne, P.D.; Hamilton, P.W.; et al. Validation of the Systematic Scoring of Immunohistochemically Stained Tumour Tissue Microarrays Using QuPath Digital Image Analysis. Histopathology 2018, 73, 327–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jhun, I.; Shepherd, D.; Hung, Y.P.; Madrigal, E.; Le, L.P.; Mino-Kenudson, M. Digital Image Analysis for Estimating Stromal CD8+ Tumor-Infiltrating Lymphocytes in Lung Adenocarcinoma. J. Pathol. Inform. 2021, 12, 28. [Google Scholar] [CrossRef]
- Apaolaza, P.S.; Petropoulou, P.I.; Rodriguez-Calvo, T. Whole-Slide Image Analysis of Human Pancreas Samples to Elucidate the Immunopathogenesis of Type 1 Diabetes Using the QuPath Software. Front. Mol. Biosci. 2021, 8, 503. [Google Scholar] [CrossRef]
- Courtney, J.M.; Morris, G.P.; Cleary, E.M.; Howells, D.W.; Sutherland, B.A. An Automated Approach to Improve the Quantification of Pericytes and Microglia in Whole Mouse Brain Sections. eNeuro 2021, 8, ENEURO.0177–21.2021. [Google Scholar] [CrossRef]
- Linette, G.P.; Carreno, B.M. Tumor-Infiltrating Lymphocytes in the Checkpoint Inhibitor Era. Curr. Hematol. Malig. Rep. 2019, 14, 286–291. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, S.K.; Rana, B.; Rana, A. Tumor-Infiltrating CD8(+) T Cell Antitumor Efficacy and Exhaustion: Molecular Insights. Drug Discov. Today 2021, 26, 951–967. [Google Scholar] [CrossRef]
- Sanmamed, M.F.; Nie, X.; Desai, S.S.; Villaroel-Espindola, F.; Badri, T.; Zhao, D.; Kim, A.W.; Ji, L.; Zhang, T.; Quinlan, E.; et al. A Burned-out Cd8+ t-Cell Subset Expands in the Tumor Microenvironment and Curbs Cancer Immunotherapy. Cancer Discov. 2021, 11, 1700–1715. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, M.E.; Rodriguez, I.; Garasa, S.; Barbes, B.; Solorzano, J.L.; Perez-Gracia, J.L.; Labiano, S.; Sanmamed, M.F.; Azpilikueta, A.; Bolaños, E.; et al. Abscopal Effects of Radiotherapy Are Enhanced by Combined Immunostimulatory MAbs and Are Dependent on CD8 T Cells and Crosspriming. Cancer Res. 2016, 76, 5994–6005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, C.; Li, Z.; Guo, S.; Chen, P.; Chen, X.; Zhou, Q.; Chen, J.; Yu, X.; Wu, X.; Ma, W.; et al. Tumor PD-L1 Expression Is Correlated with Increased TILs and Poor Prognosis in Penile Squamous Cell Carcinoma. Oncoimmunology 2017, 6, e1269047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohneis, P.; Boral, S.; Kaufmann, A.M.; Lehmann, A.; Schewe, C.; Dietel, M.; Anagnostopoulos, I.; Jöhrens, K. Human Papilloma Virus Status of Penile Squamous Cell Carcinoma Is Associated with Differences in Tumour-Infiltrating T Lymphocytes. Virchows Arch. 2015, 466, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Lérias, S.; Esteves, S.; Silva, F.; Cunha, M.; Cochicho, D.; Martins, L.; Félix, A. CD274 (PD-L1), CDKN2A (P16), TP53, and EGFR Immunohistochemical Profile in Primary, Recurrent and Metastatic Vulvar Cancer. Mod. Pathol. 2020, 33, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Trafalis, D.T.; Alifieris, C.E.; Kalantzis, A.; Verigos, K.E.; Vergadis, C.; Sauvage, S. Evidence for Efficacy of Treatment with the Anti-PD-1 Mab Nivolumab in Radiation and Multichemorefractory Advanced Penile Squamous Cell Carcinoma. J. Immunother. 2018, 41, 300–305. [Google Scholar] [CrossRef]
- Mantovani, G.; Fragomeni, S.M.; Inzani, F.; Fagotti, A.; Della Corte, L.; Gentileschi, S.; Tagliaferri, L.; Zannoni, G.F.; Scambia, G.; Garganese, G. Molecular Pathways in Vulvar Squamous Cell Carcinoma: Implications for Target Therapeutic Strategies. J. Cancer Res. Clin. Oncol. 2018, 146, 1647–1658. [Google Scholar] [CrossRef]
- Iseas, S.; Golubicki, M.; Robbio, J.; Ruiz, G.; Guerra, F.; Mariani, J.; Salanova, R.; Cabanne, A.; Eleta, M.; Gonzalez, J.V.; et al. A Clinical and Molecular Portrait of Non-Metastatic Anal Squamous Cell Carcinoma. Transl. Oncol. 2021, 14, 101084. [Google Scholar] [CrossRef]
- Wessely, A.; Heppt, M.V.; Kammerbauer, C.; Steeb, T.; Kirchner, T.; Flaig, M.J.; French, L.E.; Berking, C.; Schmoeckel, E.; Reinholz, M. Evaluation of PD-L1 Expression and HPV Genotyping in Anal Squamous Cell Carcinoma. Cancers 2020, 12, 2516. [Google Scholar] [CrossRef]
- Denis, C.; Sakalihasan, S.; Frères, P.; Withofs, N.; Sautois, B. Cemiplimab for Cisplatin Resistant Metastatic Penile Cancer. Case Rep. Oncol. 2021, 14, 972–976. [Google Scholar] [CrossRef]
- Hahn, A.W.; Chahoud, J.; Campbell, M.T.; Karp, D.D.; Wang, J.; Stephen, B.; Tu, S.M.; Pettaway, C.A.; Naing, A. Pembrolizumab for Advanced Penile Cancer: A Case Series from a Phase II Basket Trial. Investig. New Drugs 2021, 39, 1405–1410. [Google Scholar] [CrossRef]
- How, J.A.; Jazaeri, A.A.; Soliman, P.T.; Fleming, N.D.; Gong, J.; Piha-Paul, S.A.; Janku, F.; Stephen, B.; Naing, A. Pembrolizumab in Vaginal and Vulvar Squamous Cell Carcinoma: A Case Series from a Phase II Basket Trial. Sci. Rep. 2021, 11, 3667. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Piha-Paul, S.A.; Munster, P.; Pishvaian, M.J.; van Brummelen, E.M.J.; Cohen, R.B.; Gomez-Roca, C.; Ejadi, S.; Stein, M.; Chan, E.; et al. Safety and Antitumor Activity of the Anti-PD-1 Antibody Pembrolizumab in Patients with Recurrent Carcinoma of the Anal Canal. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2017, 28, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Guren, M.G.; Khan, K.; Brown, G.; Renehan, A.G.; Steigen, S.E.; Deutsch, E.; Martinelli, E.; Arnold, D. Anal Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2021, 32, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Ngoi, N.Y.L.; Lim, D.; Poon, M.L.M.; Thian, Y.L.; Lim, Y.W.; Lim, S.E.; Tong, P.; Lum, J.H.Y.; Ng, J.; et al. PD-L1 Expressing Recurrent Clear Cell Carcinoma of the Vulva with Durable Partial Response to Pembrolizumab: A Case Report. Onco. Targets. Ther. 2021, 14, 3921–3928. [Google Scholar] [CrossRef]
- Su, X.; Zhang, J.; Fu, C.; Xiao, M.; Wang, C. Recurrent Metastatic Penile Cancer Patient with Positive Pd-L1 Expression Obtained Significant Benefit from Immunotherapy: A Case Report and Literature Review. Onco. Targets. Ther. 2020, 13, 3319–3324. [Google Scholar] [CrossRef] [Green Version]
- Tostes, F.T.; Fernandes, I.; Segatelli, V.; Callegaro, D.; Carmagnani Pestana, R. Response to Pembrolizumab in Advanced Anal Squamous Cell Carcinoma With High TMB and PD-L1 and PD-L2 Amplification. Clin. Colorectal Cancer 2021, 20, 350–353. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, R.; Zhang, B.; Zhou, H.; Lin, Z.; Yao, T. Pembrolizumab in FIGO IVB Verrucous Carcinoma of the Vulva: A Case Report. Front. Oncol. 2021, 11, 1689. [Google Scholar] [CrossRef]
- Yeku, O.; Russo, A.L.; Lee, H.; Spriggs, D. A Phase 2 Study of Combined Chemo-Immunotherapy with Cisplatin-Pembrolizumab and Radiation for Unresectable Vulvar Squamous Cell Carcinoma. J. Transl. Med. 2020, 18, 350. [Google Scholar] [CrossRef]
- Reiser, J.; Banerjee, A. Effector, Memory, and Dysfunctional CD8+ T Cell Fates in the Antitumor Immune Response. J. Immunol. Res. 2016, 2016, 8941260. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef]
- Fu, T.; Dai, L.J.; Wu, S.Y.; Xiao, Y.; Ma, D.; Jiang, Y.Z.; Shao, Z.M. Spatial Architecture of the Immune Microenvironment Orchestrates Tumor Immunity and Therapeutic Response. J. Hematol. Oncol. 2021, 14, 98. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).