Comprehensive Landscape of RRM2 with Immune Infiltration in Pan-Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Cell Culture, Quantitative Real-Time PCR (qRT-PCR) and Western Blot Assays
2.3. Cell Transfection, Cell Proliferation, Colony Formation Assays and IC50 Determination
2.4. RRM2 Data Acquisition and Processing
2.5. Analysis of RRM2 Expression Profiles with Different Public Platforms
2.6. Survival Analysis and Cox Regression Analyses
2.7. RRM2 Genomic Alterations Analysis
2.8. RRM2-Interacting Genes and Protein-Protein Interaction (PPI) Network
2.9. Functional Enrichment Analysis
2.10. Immune Infiltration and Immune-related Genes Analysis
2.11. Statistical Analysis
3. Results
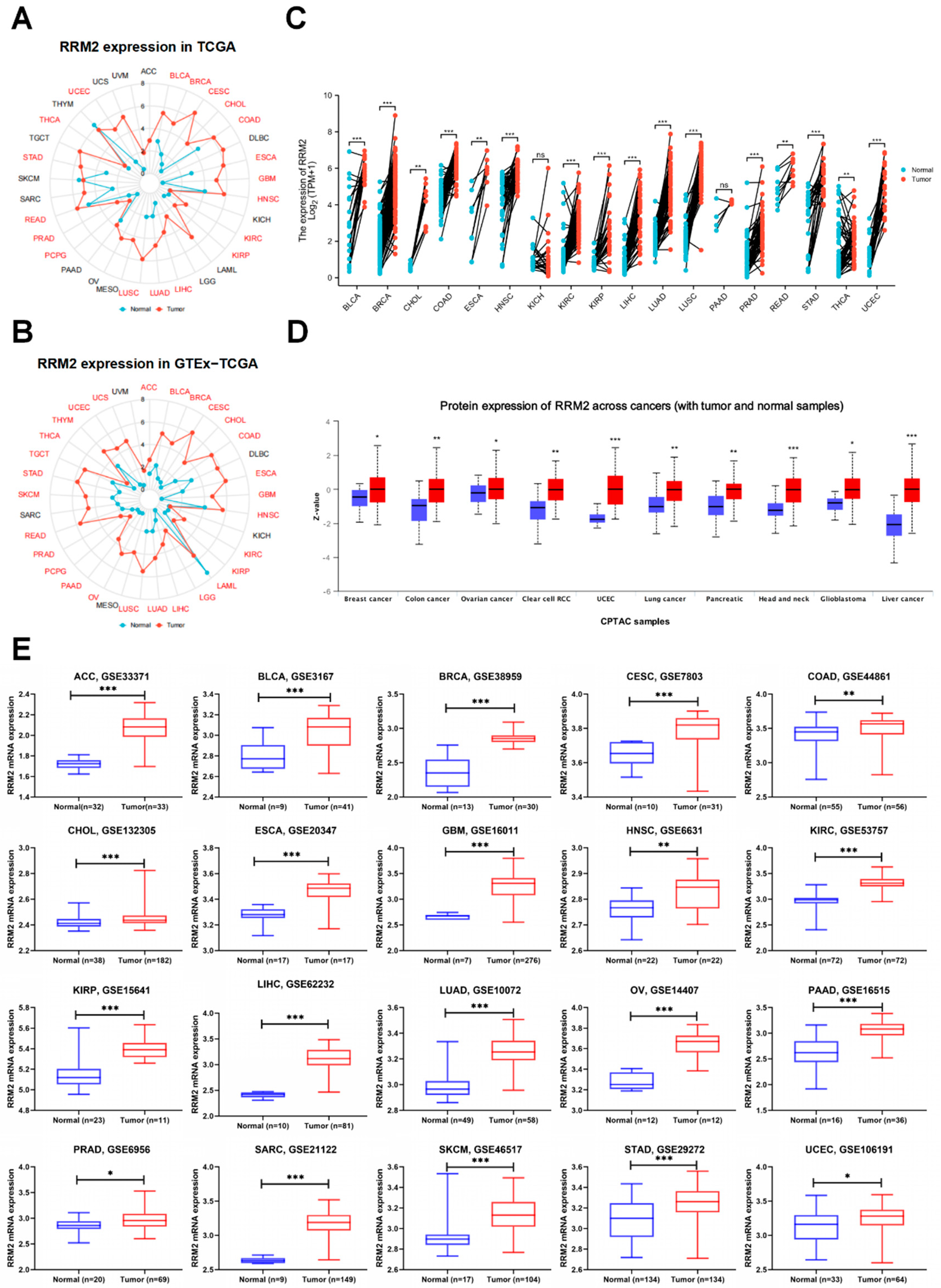
3.1. Overexpression of RRM2 in Pan-Cancer
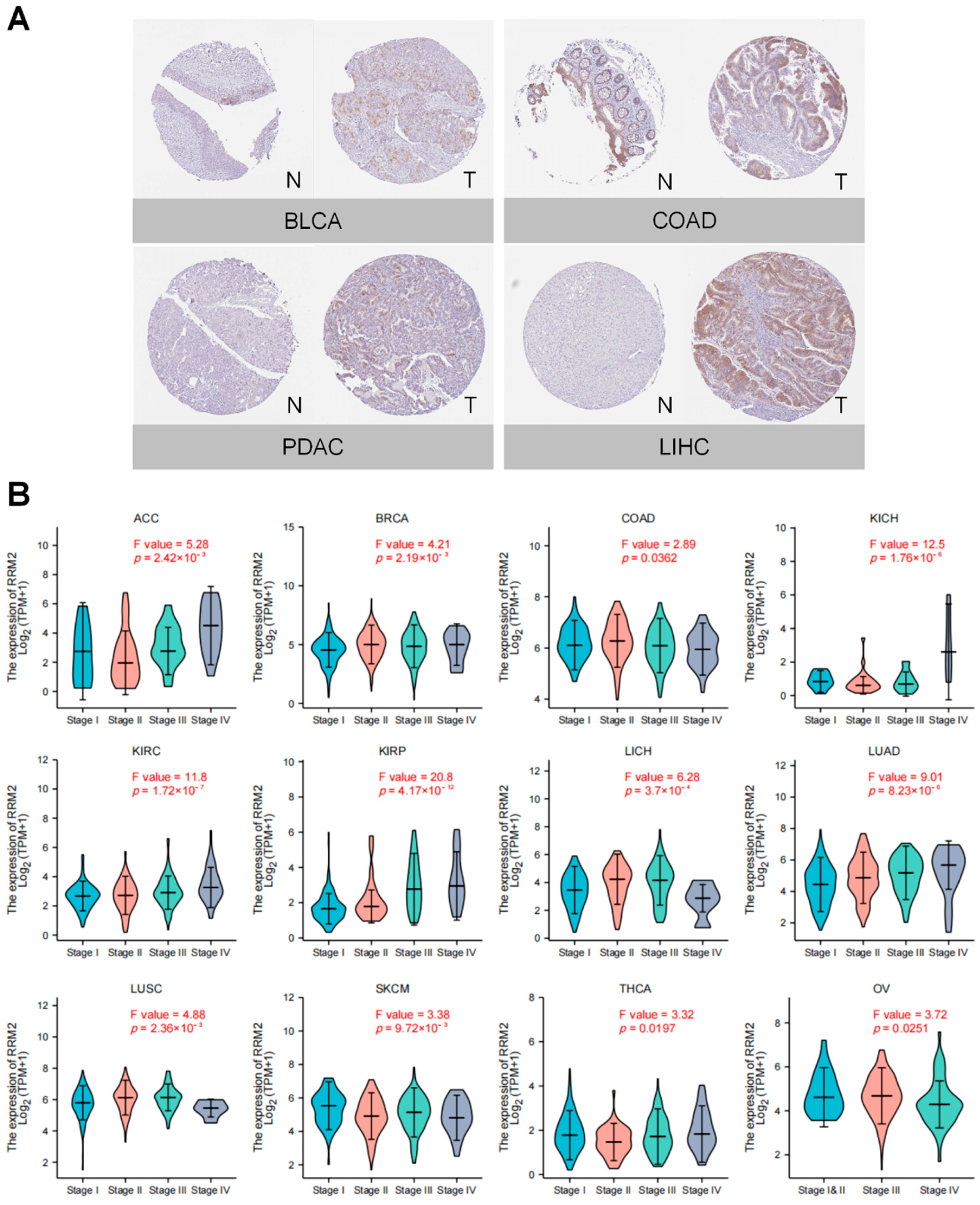
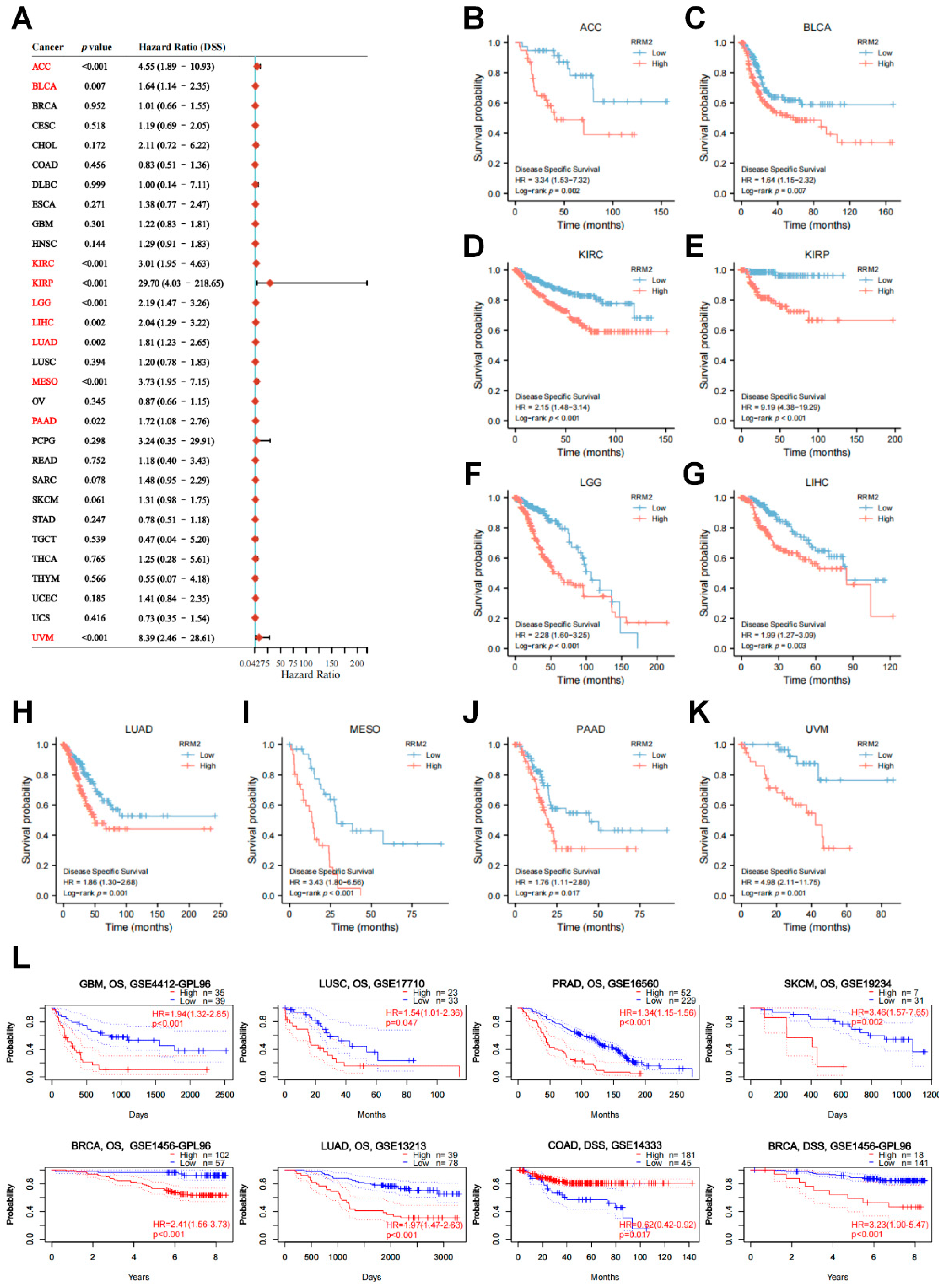
3.2. RRM2 Correlates with Tumor Stages and Prognosis in Pan-Cancer
3.3. Genetic Alteration and Mutation Landscape of RRM2 in Cancers
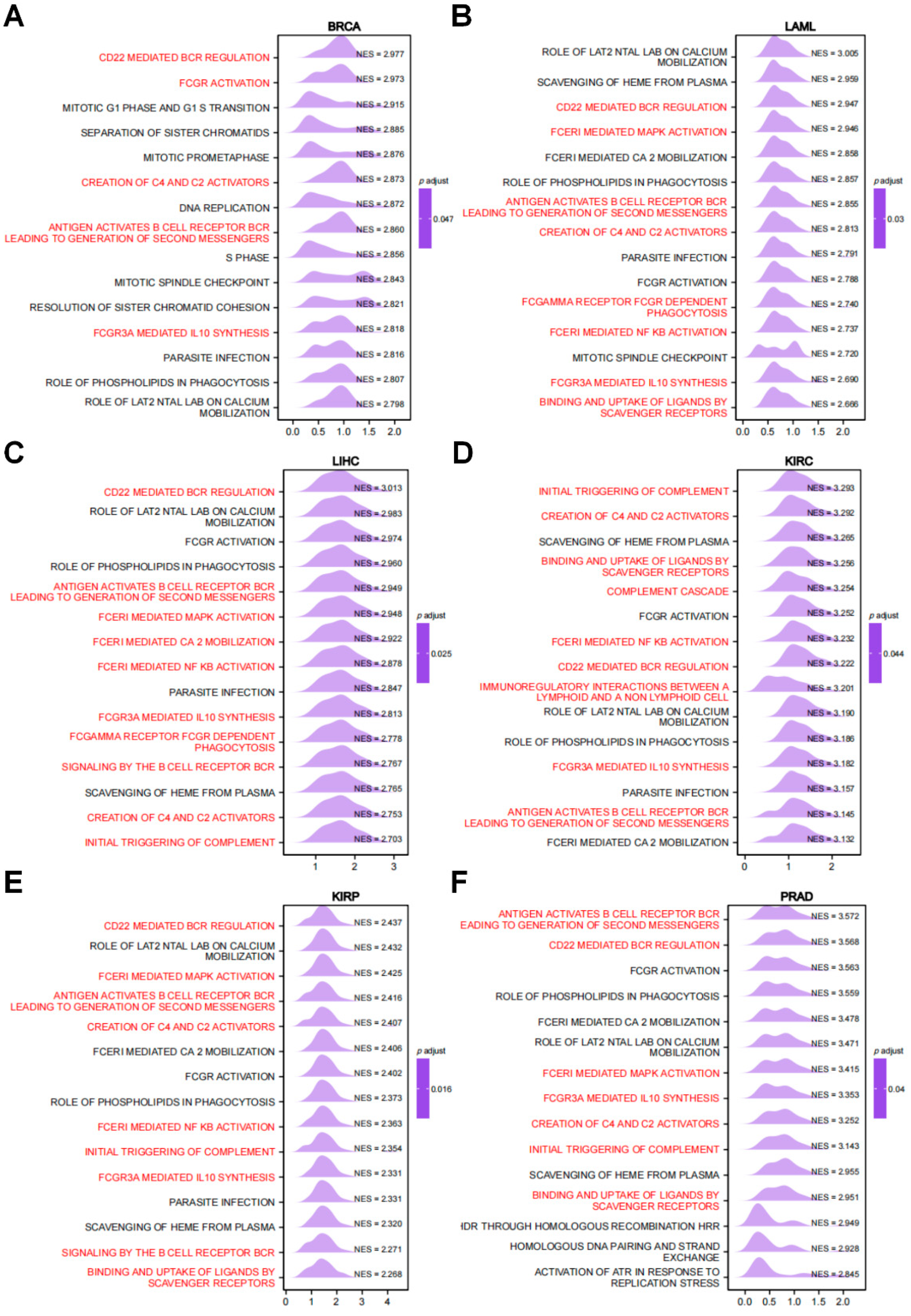
3.4. Network of RRM2-Interacting Genes and Enrichment Analysis of RRM2-Related Partners
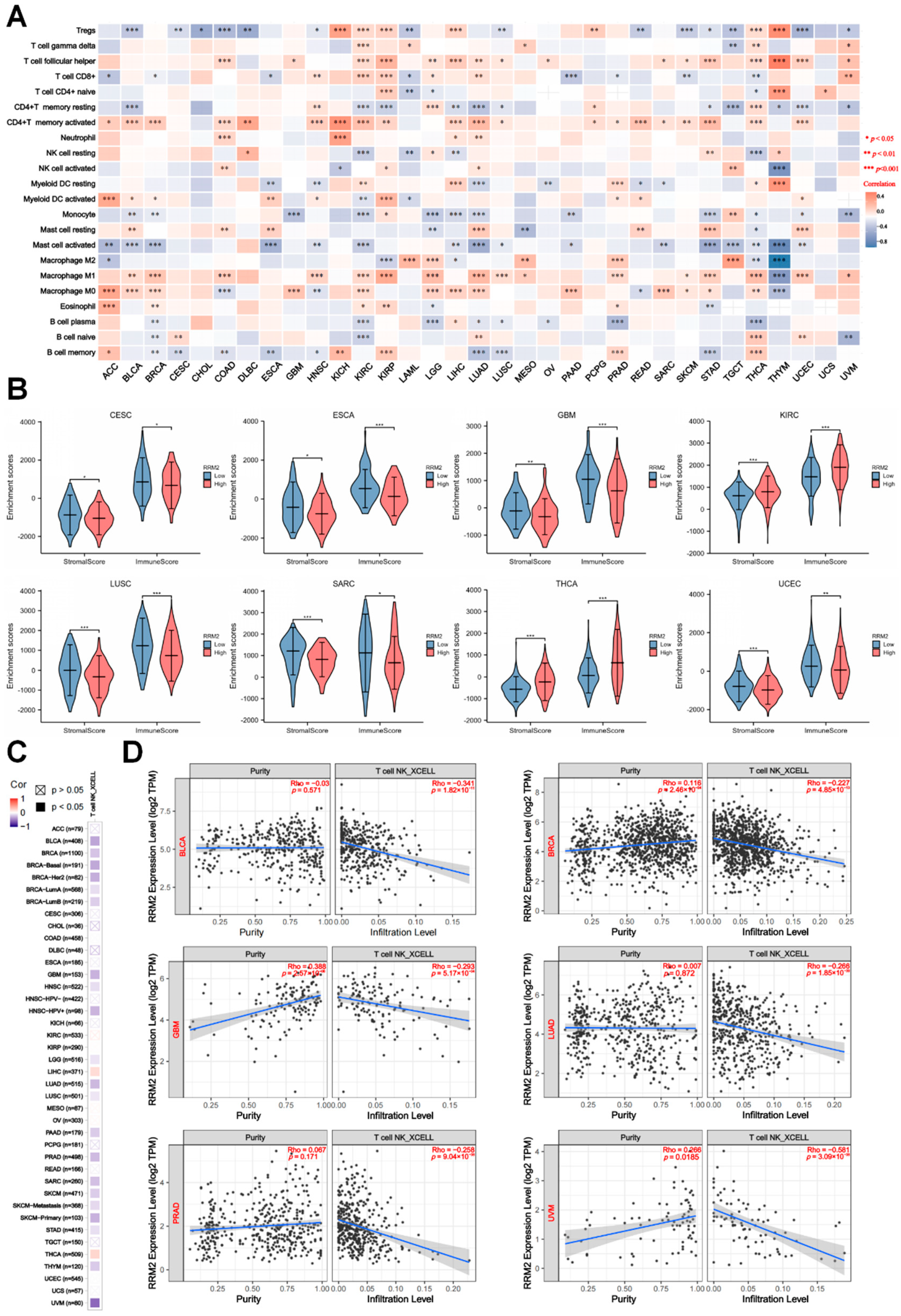
3.5. RRM2 Expression Correlates with Immune cells and Tumor Immune Infiltration
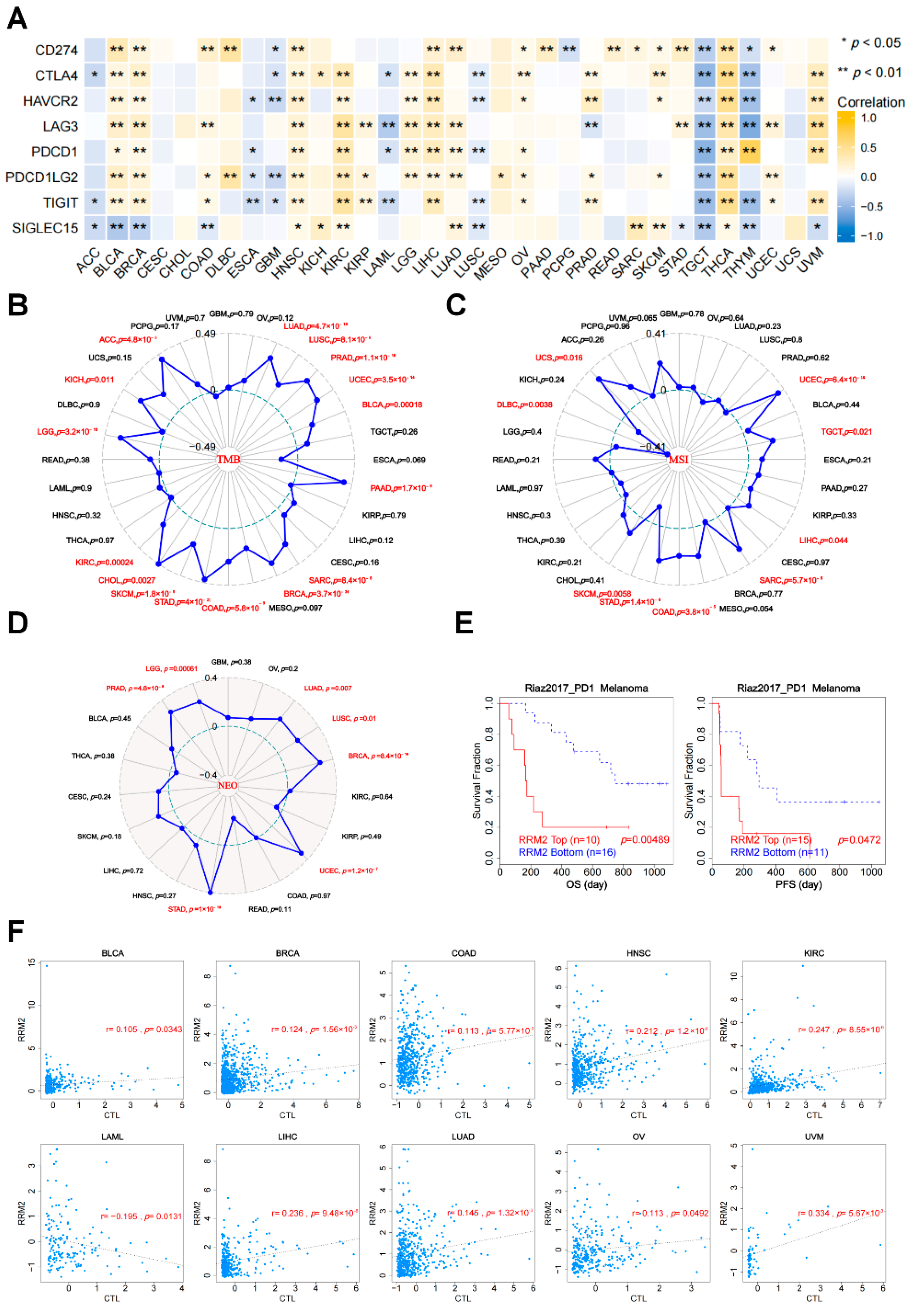
3.6. The Relationships between RRM2 Expression and ICIs, TMB, MSI, NEO and CTL
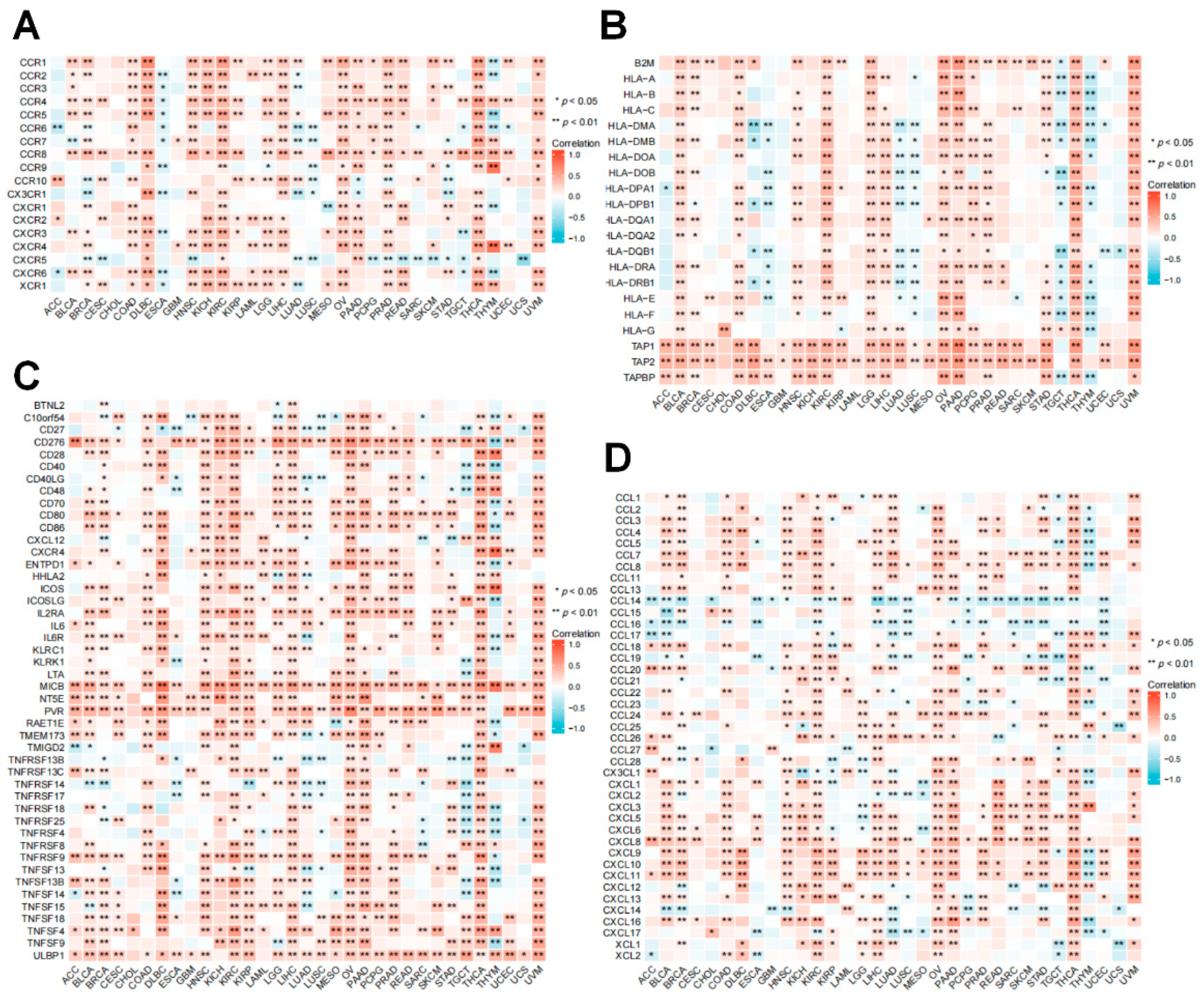
3.7. Relationship between RRM2 Expression with Immune-Related Genes
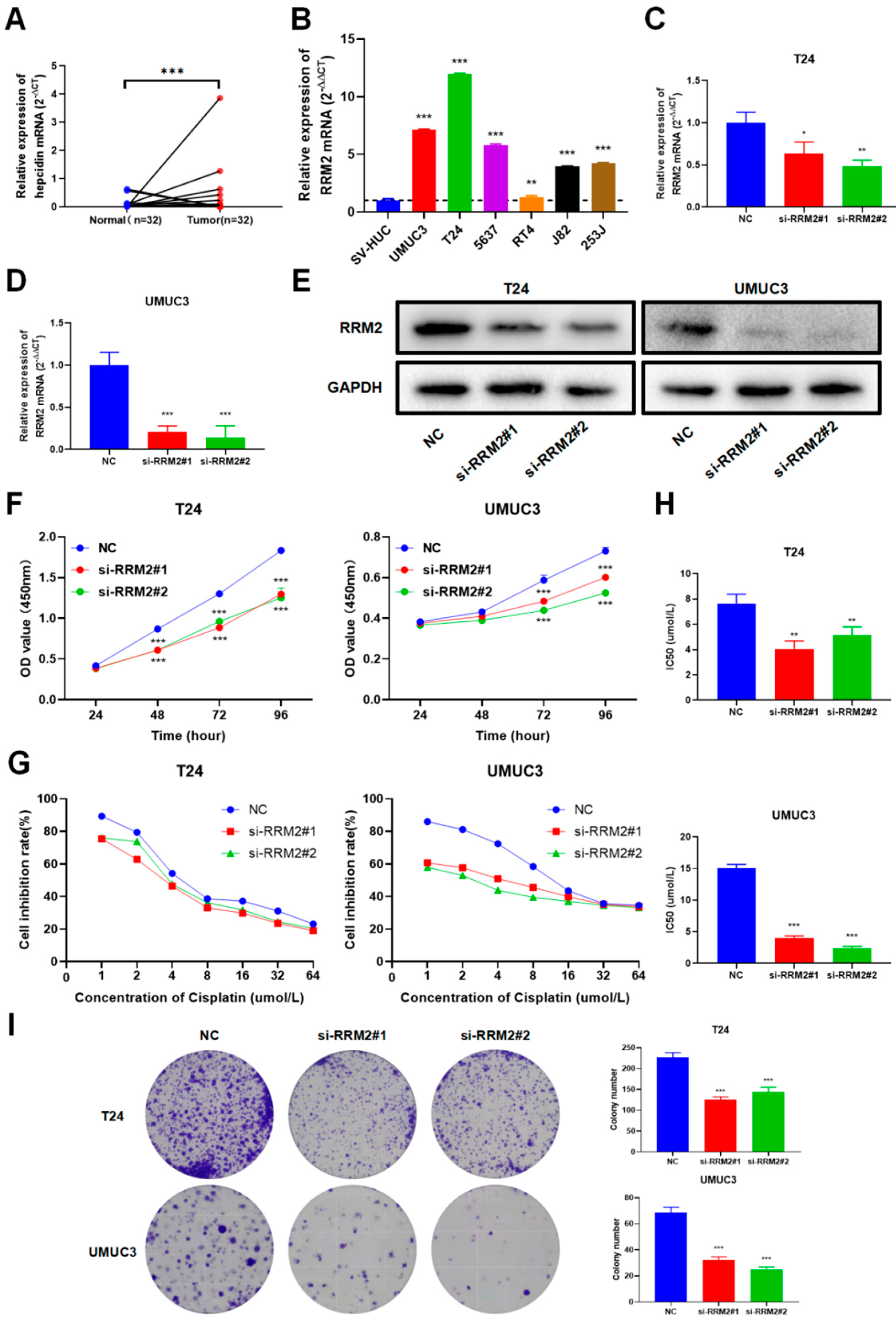
3.8. Validation of RRM2 Expression and Function in BLCA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Humeau, J.; Buqué, A.; Zitvogel, L.; Kroemer, G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2020, 17, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Fan, A.; Wang, B.; Wang, X.; Nie, Y.; Fan, D.; Zhao, X.; Lu, Y. Immunotherapy in colorectal cancer: Current achievements and future perspective. Int. J. Biol. Sci. 2021, 17, 3837–3849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Zheng, H.; Ning, Y.; Zhan, Y.; Liu, S.; Wen, Q.; Fan, S. New insights into the important roles of tumor cell-intrinsic PD-1. Int. J. Biol. Sci. 2021, 17, 2537–2547. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Larkin, J.; Tabernero, J.; Bonini, C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet 2021, 397, 1010–1022. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Jiang, L.; Jin, X.; Ying, S.; Wu, Z.; Wang, L.; Yu, W.; Tong, J.; Zhang, L.; Lou, Y.; et al. Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed. Pharmacother. 2021, 133, 110996. [Google Scholar] [CrossRef]
- Tanaka, H.; Arakawa, H.; Yamaguchi, T.; Shiraishi, K.; Fukuda, S.; Matsui, K.; Takei, Y.; Nakamura, Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 2000, 404, 42–49. [Google Scholar] [CrossRef]
- Chen, G.; Luo, Y.; Warncke, K.; Sun, Y.; Yu, D.S.; Fu, H.; Behera, M.; Ramalingam, S.S.; Doetsch, P.W.; Duong, D.M.; et al. Acetylation regulates ribonucleotide reductase activity and cancer cell growth. Nat. Commun. 2019, 10, 3213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, M.; Groß, M.; Holler, J.M.; Coggins, S.A.; Patil, N.; Leupold, J.H.; Munschauer, M.; Schenone, M.; Hartigan, C.R.; Allgayer, H.; et al. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1-RRM2 axis in cancer. Nat. Commun. 2020, 11, 3214. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, Y.; Zhang, N.; Gao, Y.; Han, L.; Li, S.; Li, J.; Liu, X.; Gong, Y.; Xie, C. RRM2 silencing suppresses malignant phenotype and enhances radiosensitivity via activating cGAS/STING signaling pathway in lung adenocarcinoma. Cell Biosci. 2021, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Hu, S.; Wu, J.; Chen, L.; Lu, J.; Wang, X.; Liu, X.; Zhou, B.; Yen, Y. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: Implication of RRM2 in angiogenesis. Mol. Cancer 2009, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Yang, B.; Zhang, H.; Song, J.; Zhang, Y.; Xing, J.; Yang, Z.; Wei, C.; Xu, T.; Yu, Z.; et al. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int. J. Biol. Sci. 2019, 15, 533–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, A.G.; Feng, H.; Wang, P.X.; Han, D.P.; Chen, X.H.; Zheng, M.H. Emerging roles of the ribonucleotide reductase M2 in colorectal cancer and ultraviolet-induced DNA damage repair. World J. Gastroenterol. 2012, 18, 4704–4713. [Google Scholar] [CrossRef]
- Rahman, M.A.; Amin, A.R.; Wang, D.; Koenig, L.; Nannapaneni, S.; Chen, Z.; Wang, Z.; Sica, G.; Deng, X.; Chen, Z.G.; et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: A potential target for cancer therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 3416–3428. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Wang, T.; Li, N.; Zhang, W.; Hu, Y. The High Expression of RRM2 Can Predict the Malignant Transformation of Endometriosis. Adv. Ther. 2021, 38, 5178–5190. [Google Scholar] [CrossRef]
- Key, J.; Sen, N.E.; Arsović, A.; Krämer, S.; Hülse, R.; Khan, N.N.; Meierhofer, D.; Gispert, S.; Koepf, G.; Auburger, G. Systematic Surveys of Iron Homeostasis Mechanisms Reveal Ferritin Superfamily and Nucleotide Surveillance Regulation to be Modified by PINK1 Absence. Cells 2020, 9, 2229. [Google Scholar] [CrossRef]
- Shen, H.; Wu, H.; Sun, F.; Qi, J.; Zhu, Q. A novel four-gene of iron metabolism-related and methylated for prognosis prediction of hepatocellular carcinoma. Bioengineered 2021, 12, 240–251. [Google Scholar] [CrossRef]
- Mbah, N.E.; Lyssiotis, C.A. Metabolic regulation of ferroptosis in the tumor microenvironment. J. Biol. Chem. 2022, 298, 101617. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Yao, Y.M.; Tian, Y.P. Ferroptosis: A Trigger of Proinflammatory State Progression to Immunogenicity in Necroinflammatory Disease. Front. Immunol. 2021, 12, 701163. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lin, J.; Guo, S.; Xue, X.; Wang, Y.; Qiu, S.; Cui, J.; Ma, L.; Zhang, X.; Wang, J. RRM2 protects against ferroptosis and is a tumor biomarker for liver cancer. Cancer Cell Int. 2020, 20, 587. [Google Scholar] [CrossRef] [PubMed]
- Cieslik, M.; Chinnaiyan, A.M. Global genomics project unravels cancer’s complexity at unprecedented scale. Nature 2020, 578, 39–40. [Google Scholar] [CrossRef] [Green Version]
- Shindo, Y.; Hazama, S.; Tsunedomi, R.; Suzuki, N.; Nagano, H. Novel Biomarkers for Personalized Cancer Immunotherapy. Cancers 2019, 11, 1223. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Hu, Q.; Chen, H.; Liang, K.; Yuan, Y.; Xiang, Y.; Ruan, H.; Zhang, Z.; Song, A.; Zhang, H.; et al. Characterization of Hypoxia-associated Molecular Features to Aid Hypoxia-Targeted Therapy. Nat. Metab. 2019, 1, 431–444. [Google Scholar] [CrossRef]
- Ohmura, S.; Marchetto, A.; Orth, M.F.; Li, J.; Jabar, S.; Ranft, A.; Vinca, E.; Ceranski, K.; Carreño-Gonzalez, M.J.; Romero-Pérez, L.; et al. Translational evidence for RRM2 as a prognostic biomarker and therapeutic target in Ewing sarcoma. Mol. Cancer 2021, 20, 97. [Google Scholar] [CrossRef]
- Fan, H.; Villegas, C.; Wright, J.A. Ribonucleotide reductase R2 component is a novel malignancy determinant that cooperates with activated oncogenes to determine transformation and malignant potential. Proc. Natl. Acad. Sci. USA 1996, 93, 14036–14040. [Google Scholar] [CrossRef] [Green Version]
- Heidel, J.D.; Liu, J.Y.; Yen, Y.; Zhou, B.; Heale, B.S.; Rossi, J.J.; Bartlett, D.W.; Davis, M.E. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 2207–2215. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Pang, J.; Liu, Y.; Zhang, J.; Zhang, C.; Shen, G.; Song, L. Suppression of RRM2 inhibits cell proliferation, causes cell cycle arrest and promotes the apoptosis of human neuroblastoma cells and in human neuroblastoma RRM2 is suppressed following chemotherapy. Oncol. Rep. 2018, 40, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, B.; Su, L.; Hu, S.; Hu, W.; Yip, M.L.; Wu, J.; Gaur, S.; Smith, D.L.; Yuan, Y.C.; Synold, T.W.; et al. A small-molecule blocking ribonucleotide reductase holoenzyme formation inhibits cancer cell growth and overcomes drug resistance. Cancer Res. 2013, 73, 6484–6493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markowitsch, S.D.; Schupp, P.; Lauckner, J.; Vakhrusheva, O.; Slade, K.S.; Mager, R.; Efferth, T.; Haferkamp, A.; Juengel, E. Artesunate Inhibits Growth of Sunitinib-Resistant Renal Cell Carcinoma Cells through Cell Cycle Arrest and Induction of Ferroptosis. Cancers 2020, 12, 3150. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef] [PubMed]
- Petitprez, F.; Meylan, M.; de Reyniès, A.; Sautès-Fridman, C.; Fridman, W.H. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front. Immunol. 2020, 11, 784. [Google Scholar] [CrossRef]
- Bayik, D.; Lathia, J.D. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 2021, 21, 526–536. [Google Scholar] [CrossRef]
- Chen, Y.; Meng, Z.; Zhang, L.; Liu, F. CD2 Is a Novel Immune-Related Prognostic Biomarker of Invasive Breast Carcinoma That Modulates the Tumor Microenvironment. Front. Immunol. 2021, 12, 664845. [Google Scholar] [CrossRef]
- Zhou, B.; Gao, S. Pan-Cancer Analysis of FURIN as a Potential Prognostic and Immunological Biomarker. Front. Mol. Biosci. 2021, 8, 648402. [Google Scholar] [CrossRef]
- Tang, B.; Xu, W.; Wang, Y.; Zhu, J.; Wang, H.; Tu, J.; Weng, Q.; Kong, C.; Yang, Y.; Qiu, R.; et al. Identification of critical ferroptosis regulators in lung adenocarcinoma that RRM2 facilitates tumor immune infiltration by inhibiting ferroptotic death. Clin. Immunol. 2021, 232, 108872. [Google Scholar] [CrossRef]
- Szarkowska, J.; Cwiek, P.; Szymanski, M.; Rusetska, N.; Jancewicz, I.; Stachowiak, M.; Swiatek, M.; Luba, M.; Konopinski, R.; Kubala, S.; et al. RRM2 gene expression depends on BAF180 subunit of SWISNF chromatin remodeling complex and correlates with abundance of tumor infiltrating lymphocytes in ccRCC. Am. J. Cancer Res. 2021, 11, 5965–5978. [Google Scholar]
- Paul, S.; Chhatar, S.; Mishra, A.; Lal, G. Natural killer T cell activation increases iNOS(+)CD206(-) M1 macrophage and controls the growth of solid tumor. J. Immunother. Cancer 2019, 7, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krijgsman, D.; de Vries, N.L.; Skovbo, A.; Andersen, M.N.; Swets, M.; Bastiaannet, E.; Vahrmeijer, A.L.; van de Velde, C.J.H.; Heemskerk, M.H.M.; Hokland, M.; et al. Characterization of circulating T-, NK-, and NKT cell subsets in patients with colorectal cancer: The peripheral blood immune cell profile. Cancer Immunol. Immunother. CII 2019, 68, 1011–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Shen, L.; Wang, Y.; Liu, Q.; Goodwin, T.J.; Li, J.; Dorosheva, O.; Liu, T.; Liu, R.; Huang, L. Synergistic and low adverse effect cancer immunotherapy by immunogenic chemotherapy and locally expressed PD-L1 trap. Nat. Commun. 2018, 9, 2237. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Chen, M.; Zhang, R.; Zhang, W.; Wang, C.; Zhang, Y.; Wei, X.; Guan, Y.; Liu, J.; Feng, K.; et al. Manganese is critical for antitumor immune responses via cGAS-STING and improves the efficacy of clinical immunotherapy. Cell Res. 2020, 30, 966–979. [Google Scholar] [CrossRef]
- Xiong, W.; Zhang, B.; Yu, H.; Zhu, L.; Yi, L.; Jin, X. RRM2 Regulates Sensitivity to Sunitinib and PD-1 Blockade in Renal Cancer by Stabilizing ANXA1 and Activating the AKT Pathway. Adv. Sci. 2021, 8, e2100881. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Cohen, R.; Colle, R.; Pudlarz, T.; Heran, M.; Duval, A.; Svrcek, M.; André, T. Immune Checkpoint Inhibition in Metastatic Colorectal Cancer Harboring Microsatellite Instability or Mismatch Repair Deficiency. Cancers 2021, 13, 1149. [Google Scholar] [CrossRef]
- Verdon, D.J.; Jenkins, M.R. Identification and Targeting of Mutant Peptide Neoantigens in Cancer Immunotherapy. Cancers 2021, 13, 4245. [Google Scholar] [CrossRef]
- Borst, J.; Ahrends, T.; Bąbała, N.; Melief, C.J.M.; Kastenmüller, W. CD4 T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- Morikawa, T.; Maeda, D.; Kume, H.; Homma, Y.; Fukayama, M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology 2010, 57, 885–892. [Google Scholar] [CrossRef]
- Scharadin, T.M.; Zhang, H.; Zimmermann, M.; Wang, S.; Malfatti, M.A.; Cimino, G.D.; Turteltaub, K.; de Vere White, R.; Pan, C.X.; Henderson, P.T. Diagnostic Microdosing Approach to Study Gemcitabine Resistance. Chem. Res. Toxicol. 2016, 29, 1843–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Lu, J.; Niu, Z.; Ding, K.; Bi, D.; Liu, S.; Li, J.; Wu, F.; Zhang, H.; Zhao, Z.; et al. A Potent Chemotherapeutic Strategy with Eg5 Inhibitor against Gemcitabine Resistant Bladder Cancer. PLoS ONE 2015, 10, e0144484. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Characteristics | Odds Ratio (OR) | p Value |
|---|---|---|
| Age (>70 vs. ≤70) | 0.854 (0.579–1.260) | 0.428 |
| Gender (Male vs. Female) | 0.928 (0.598–1.438) | 0.738 |
| T stage (T3&T4 vs. T1&T2) | 1.211 (0.789–1.864) | 0.382 |
| N stage (N1&N2&N3 vs. N0) | 1.088 (0.711–1.669) | 0.697 |
| M stage (M1 vs. M0) | 1.493 (0.436–5.332) | 0.519 |
| Pathologic stage (Stage III&Stage IV vs. Stage I&Stage II) | 1.305 (0.864–1.976) | 0.207 |
| Histologic grade (High Grade vs. Low Grade) | 6.447 (2.140–27.837) | 0.003 ** |
| Characteristics | OS | DSS | ||
|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age (>70 vs. ≤70) | 1.250 (0.780–2.002) | 0.354 | -- | |
| Gender (Male/Female) | -- | -- | ||
| T stage (T3&T4/T1&T2) | 3.066 (1.045–8.995) | 0.041 * | 3.018 (0.872–10.445) | 0.081 |
| N stage (N1&N2&N3/N0) | 2.139 (1.255–3.646) | 0.005 ** | 2.777 (1.417–5.444) | 0.003 ** |
| M stage (M1/M0) | 1.317 (0.504–3.442) | 0.574 | 1.453 (0.489–4.316) | 0.501 |
| Pathologic stage (Stage III&IV/Stage I&II) | 0.511 (0.157–1.663) | 0.265 | 0.520 (0.125–2.155) | 0.367 |
| Histologic grade (High Grade/Low Grade) | -- | -- | ||
| RRM2 (High Expression/Low Expression) | 1.693 (1.052–2.723) | 0.030 * | 2.482 (1.331–4.628) | 0.004 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Song, Q.; Yang, Y.; Wang, L.; Wu, Z. Comprehensive Landscape of RRM2 with Immune Infiltration in Pan-Cancer. Cancers 2022, 14, 2938. https://doi.org/10.3390/cancers14122938
Zhou Z, Song Q, Yang Y, Wang L, Wu Z. Comprehensive Landscape of RRM2 with Immune Infiltration in Pan-Cancer. Cancers. 2022; 14(12):2938. https://doi.org/10.3390/cancers14122938
Chicago/Turabian StyleZhou, Zijian, Qiang Song, Yuanyuan Yang, Lujia Wang, and Zhong Wu. 2022. "Comprehensive Landscape of RRM2 with Immune Infiltration in Pan-Cancer" Cancers 14, no. 12: 2938. https://doi.org/10.3390/cancers14122938






