Combination of Whole-Body Baseline CT Radiomics and Clinical Parameters to Predict Response and Survival in a Stage-IV Melanoma Cohort Undergoing Immunotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Workflow Overview
2.2. Patient Selection
- Stage-IV melanoma;
- First-line treatment with a PD-1 checkpoint inhibitor, a CTLA-4 checkpoint inhibitor, or combination of both;
- Available baseline CT scans prior to treatment initiation;
- Available demographic data, follow-up data, and clinical metadata.
2.3. CT Imaging and Lesion Segmentation
2.4. Radiomic Feature Extraction and Aggregation
2.5. Machine-Learning Model
- Pre-processing: Ordinal encoding of nominal clinical features [24], imputation of missing clinical feature values (0.5 for binary features, median for all other features), standard normalization (zero mean, unit variance) of all features;
- Feature selection using FCBF [22]: Applied only to radiomic features, clinical features were always used;
- Training: Fit of an extremely randomized forest [25];
- Validation: Prediction of outcome on the current validation set and comparison to true outcome using AUC.
2.6. Performance Evaluation
3. Results
3.1. Demographics, Response after Three Months, and Survival after Six and Twelve Months
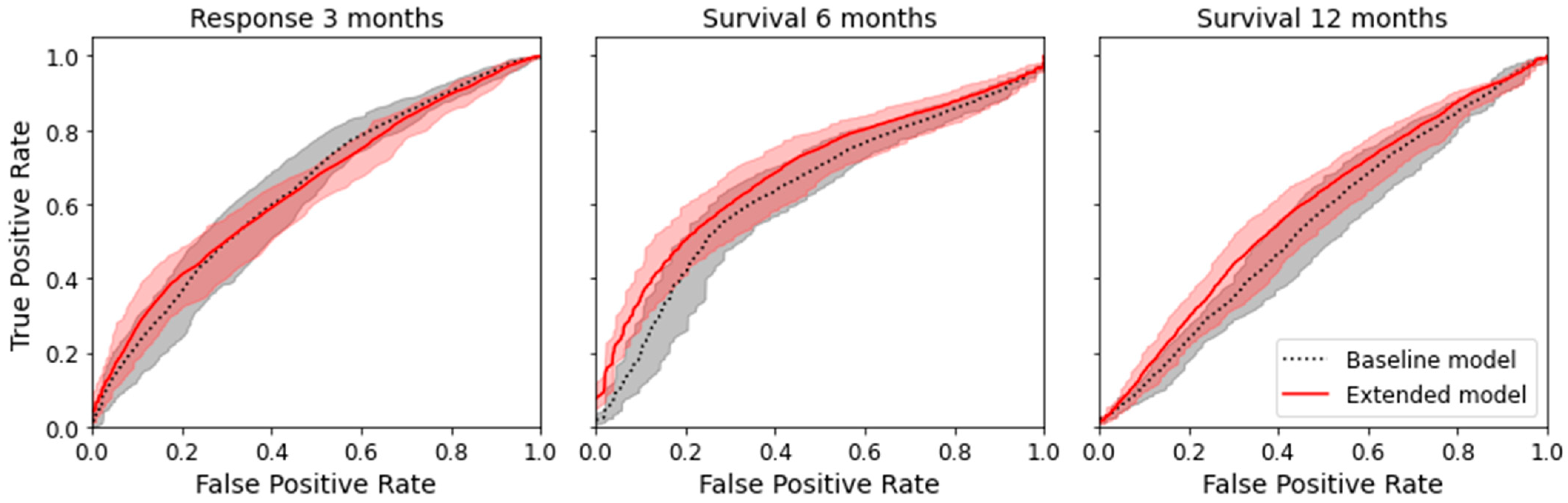
3.2. Prediction of Therapy Response and Survival Using Clinical Data and Radiomic Features
3.2.1. Machine-Learning Model Comparison
3.2.2. Feature Selection
3.3. Low-Risk vs. High-Risk Stratification for Twelve-Month Survival Prediction
4. Discussion
4.1. Prediction of Therapy Response
4.2. Prediction of Six-Month and Twelve-Month Survival
4.3. Risk Stratification for Twelve-Month Survival
4.4. Feature Selection/Radiomic Biomarker
4.5. Strengths
4.6. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALM | acral lentiginous melanoma |
| AUC | area under the curve |
| BRAF | v-Raf murine sarcoma viral oncogene homolog B1 |
| CI | confidence interval |
| CMMR | Central Malignant Melanoma Registry |
| CR | complete response |
| CT | computed tomography |
| CTLA-4 | cytotoxic T-lymphocyte-associated protein 4 |
| CV | cross-validation |
| FCBF | fast correlation-based filter for feature selection |
| IQR | interquartile range |
| LDH | lactate dehydrogenase |
| LMM | lentigo maligna melanoma |
| LoG | Laplacian of Gaussian |
| MEK | mitogen-activated protein kinase kinase |
| ML | machine learning |
| mRMR | minimum redundancy–maximum relevance feature selection |
| n | number |
| NM | nodular melanoma |
| OS | overall survival |
| PACS | picture archiving and communication system |
| PD | progressive disease |
| PD-1 | programmed death 1 |
| PET | positron emission tomography |
| PFS | progression-free survival |
| PR | partial response |
| RAF | rapidly accelerated fibrosarcoma |
| RAS | rat sarcoma |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| ROC | receiver-operating characteristic |
| SD | stable disease |
| SSM | superficial spreading melanoma |
References
- Schadendorf, D.; van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef] [Green Version]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; de Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: A multicentre, double-blind, phase 3 randomised controlled trial. Lancet 2015, 386, 444–451. [Google Scholar] [CrossRef]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015, 372, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018, 19, 603–615. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Wagstaff, J.; Schadendorf, D.; Ferrucci, P.F.; et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2017, 377, 1345–1356. [Google Scholar] [CrossRef]
- Schadendorf, D.; Hodi, F.S.; Robert, C.; Weber, J.S.; Margolin, K.; Hamid, O.; Patt, D.; Chen, T.T.; Berman, D.M.; Wolchok, J.D. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J. Clin. Oncol. 2015, 33, 1889–1894. [Google Scholar] [CrossRef] [Green Version]
- Schadendorf, D.; Larkin, J.; Wolchok, J.; Hodi, F.S.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.J.; Cowey, C.L.; Lao, C.; et al. Health-related quality of life results from the phase III CheckMate 067 study. Eur. J. Cancer 2017, 82, 80–91. [Google Scholar] [CrossRef] [Green Version]
- Diem, S.; Kasenda, B.; Martin-Liberal, J.; Lee, A.; Chauhan, D.; Gore, M.; Larkin, J. Prognostic score for patients with advanced melanoma treated with ipilimumab. Eur. J. Cancer 2015, 51, 2785–2791. [Google Scholar] [CrossRef]
- Diem, S.; Kasenda, B.; Spain, L.; Martin-Liberal, J.; Marconcini, R.; Gore, M.; Larkin, J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br. J. Cancer 2016, 114, 256–261. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Durot, C.; Mule, S.; Soyer, P.; Marchal, A.; Grange, F.; Hoeffel, C. Metastatic melanoma: Pretreatment contrast-enhanced CT texture parameters as predictive biomarkers of survival in patients treated with pembrolizumab. Eur. Radiol. 2019, 29, 3183–3191. [Google Scholar] [CrossRef]
- Trebeschi, S.; Drago, S.G.; Birkbak, N.J.; Kurilova, I.; Calin, A.M.; Delli Pizzi, A.; Lalezari, F.; Lambregts, D.M.J.; Rohaan, M.W.; Parmar, C.; et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [Green Version]
- Basler, L.; Gabrys, H.S.; Hogan, S.A.; Pavic, M.; Bogowicz, M.; Vuong, D.; Tanadini-Lang, S.; Forster, R.; Kudura, K.; Huellner, M.W.; et al. Radiomics, Tumor Volume, and Blood Biomarkers for Early Prediction of Pseudoprogression in Patients with Metastatic Melanoma Treated with Immune Checkpoint Inhibition. Clin. Cancer Res. 2020, 26, 4414–4425. [Google Scholar] [CrossRef] [Green Version]
- Guerrisi, A.; Loi, E.; Ungania, S.; Russillo, M.; Bruzzaniti, V.; Elia, F.; Desiderio, F.; Marconi, R.; Solivetti, F.M.; Strigari, L. Novel cancer therapies for advanced cutaneous melanoma: The added value of radiomics in the decision making process-A systematic review. Cancer Med. 2020, 9, 1603–1612. [Google Scholar] [CrossRef]
- Guerrisi, A.; Russillo, M.; Loi, E.; Ganeshan, B.; Ungania, S.; Desiderio, F.; Bruzzaniti, V.; Falcone, I.; Renna, D.; Ferraresi, V.; et al. Exploring CT Texture Parameters as Predictive and Response Imaging Biomarkers of Survival in Patients With Metastatic Melanoma Treated With PD-1 Inhibitor Nivolumab: A Pilot Study Using a Delta-Radiomics Approach. Front. Oncol. 2021, 11, 704607. [Google Scholar] [CrossRef]
- Wang, Z.L.; Mao, L.L.; Zhou, Z.G.; Si, L.; Zhu, H.T.; Chen, X.; Zhou, M.J.; Sun, Y.S.; Guo, J. Pilot Study of CT-Based Radiomics Model for Early Evaluation of Response to Immunotherapy in Patients With Metastatic Melanoma. Front. Oncol. 2020, 10, 1524. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Enke, J.S.; Moltz, J.H.; Anastasi, M.; Kunz, W.G.; Schmidt, C.; Maurus, S.; Mühlberg, A.; Katzmann, A.; Sühling, M.; Hahn, H.; et al. Radiomics Features of the Spleen as Surrogates for CT-Based Lymphoma Diagnosis and Subtype Differentiation. Cancers 2022, 14, 713. [Google Scholar] [CrossRef]
- Meier, F.; Will, S.; Ellwanger, U.; Schlagenhauff, B.; Schittek, B.; Rassner, G.; Garbe, C. Metastatic pathways and time courses in the orderly progression of cutaneous melanoma. Br. J. Dermatol. 2002, 147, 62–70. [Google Scholar] [CrossRef]
- Yu, L.; Liu, H. Feature Selection for High-Dimensional Data: A Fast Correlation-Based Filter Solution. In Proceedings of the 20th International Conference on Machine Learning (ICML-03), Washington, DC, USA, 21–24 August 2003; Volume 2, pp. 856–863. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Wright, M.N.; Konig, I.R. Splitting on categorical predictors in random forests. PeerJ 2019, 7, e6339. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely Randomized Trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Probst, P.; Boulesteix, A.L.; Bischl, B. Tunability: Importance of Hyperparameters of Machine Learning Algorithms. J. Mach. Learn. Res. 2019, 20, 1934–1965. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Tsamardinos, I.; Greasidou, E.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018, 107, 1895–1922. [Google Scholar] [CrossRef] [Green Version]
- Davidson-Pilon, C. lifelines: Survival analysis in Python. J. Open Source Softw. 2019, 4, 1317. [Google Scholar] [CrossRef] [Green Version]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics 2017, 37, 1483–1503. [Google Scholar] [CrossRef]
- Mackin, D.; Fave, X.; Zhang, L.; Fried, D.; Yang, J.; Taylor, B.; Rodriguez-Rivera, E.; Dodge, C.; Jones, A.K.; Court, L. Measuring Computed Tomography Scanner Variability of Radiomics Features. Investig. Radiol. 2015, 50, 757–765. [Google Scholar] [CrossRef]
- Berenguer, R.; Pastor-Juan, M.D.R.; Canales-Vazquez, J.; Castro-Garcia, M.; Villas, M.V.; Mansilla Legorburo, F.; Sabater, S. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology 2018, 288, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [Green Version]
- Trojaniello, C.; Luke, J.J.; Ascierto, P.A. Therapeutic Advancements Across Clinical Stages in Melanoma, With a Focus on Targeted Immunotherapy. Front. Oncol. 2021, 11, 670726. [Google Scholar] [CrossRef]
- Smith, A.D.; Gray, M.R.; del Campo, S.M.; Shlapak, D.; Ganeshan, B.; Zhang, X.; Carson, W.E., III. Predicting Overall Survival in Patients With Metastatic Melanoma on Antiangiogenic Therapy and RECIST Stable Disease on Initial Posttherapy Images Using CT Texture Analysis. AJR Am. J. Roentgenol. 2015, 205, W283–W293. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Iuga, A.I.; Carolus, H.; Hoink, A.J.; Brosch, T.; Klinder, T.; Maintz, D.; Persigehl, T.; Baessler, B.; Pusken, M. Automated detection and segmentation of thoracic lymph nodes from CT using 3D foveal fully convolutional neural networks. BMC Med. Imaging 2021, 21, 69. [Google Scholar] [CrossRef]
- Vorontsov, E.; Cerny, M.; Regnier, P.; Di Jorio, L.; Pal, C.J.; Lapointe, R.; Vandenbroucke-Menu, F.; Turcotte, S.; Kadoury, S.; Tang, A. Deep Learning for Automated Segmentation of Liver Lesions at CT in Patients with Colorectal Cancer Liver Metastases. Radiol. Artif. Intell. 2019, 1, 180014. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Bluemke, D.A.; Moy, L.; Bredella, M.A.; Ertl-Wagner, B.B.; Fowler, K.J.; Goh, V.J.; Halpern, E.F.; Hess, C.P.; Schiebler, M.L.; Weiss, C.R. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers-From the Radiology Editorial Board. Radiology 2020, 294, 487–489. [Google Scholar] [CrossRef] [Green Version]




| Clinical Data | ||
| Age (years) [median, (IQR)] | 70 (22) | |
| Gender (female) [n, %] | 109 (42%) | |
| Localization of primary tumor [n, %] | Head/neck | 50 (19%) |
| Torso | 63 (24%) | |
| Upper extremity | 30 (11%) | |
| Lower extremity | 71 (27%) | |
| Other | 13 (5%) | |
| n/a | 35 (13%) | |
| Histological subtype [n, %] | SSM | 71 (27%) |
| NM | 62 (24%) | |
| LMM | 13 (5%) | |
| ALM | 29 (11%) | |
| Mucosal | 13 (5%) | |
| Occult | 61 (23%) | |
| n/a | 13 (5%) | |
| BRAF V600E mutation status [n, %] | BRAF wildtype | 180 (69%) |
| BRAF mutation | 74 (28%) | |
| n/a | 8 (3%) | |
| Baseline LDH [n, %] | Normal (<250 U/l) | 164 (63%) |
| Elevated (≥250 U/l) | 85 (32%) | |
| n/a | 13 (5%) | |
| Baseline S100 [n, %] | Normal (<0.1 µg/l) | 117 (45%) |
| Elevated (≥0.1 µg/l) | 125 (48%) | |
| n/a | 20 (8%) | |
| Number of metastatic organs [n, %] | 1–3 | 232 (89%) |
| >3 | 30 (11%) | |
| Cerebral metastases [n, %] | 48 (18%) | |
| Hepatic metastases [n, %] | 85 (32%) | |
| Immunotherapy [n, %] | PD1 | 146 (56%) |
| PD1+CTLA4 | 116 (44%) | |
| Patient Outcome | ||
| Response after 3 months (RECIST 1.1) [n, %] | CR | 10 (4%) |
| PR | 72 (27%) | |
| SD | 42 (16%) | |
| PD | 96 (37%) | |
| n/a | 42 (16%) | |
| Survival after 6 months [n, %] | Yes | 181 (69%) |
| No | 49 (19%) | |
| n/a | 32 (12%) | |
| Survival after 12 months [n, %] | Yes | 115 (44%) |
| No | 73 (28%) | |
| n/a | 74 (28%) | |
| Lesion counts [n lesions, n patients] | All | 6404, 262 |
| Lung | 2738, 157 | |
| Liver | 1120, 79 | |
| Soft tissue/skin | 1111, 110 | |
| Lymph nodes | 876, 154 | |
| Skeletal | 172, 42 | |
| Spleen | 97, 12 | |
| Heart | 8, 3 | |
| Other | 238, 54 | |
| Binary Endpoint | |||
|---|---|---|---|
| Response at 3 Months | Survival at 6 Months | Survival at 12 Months | |
| n cases, (class 0, class 1) | 220 (138, 82) | 230 (49, 181) | 188 (73, 115) |
| Baseline model (clinical features), (AUC (95% CI)) | 0.656 (0.587, 0.719) | 0.620 (0.545, 0.692) | 0.558 (0.481, 0.629) |
| Extended model (clinical and radiomic features), (AUC (95% CI)) | 0.641 (0.581, 0.700) | 0.664 (0.598, 0.729) | 0.600 (0.526, 0.667) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peisen, F.; Hänsch, A.; Hering, A.; Brendlin, A.S.; Afat, S.; Nikolaou, K.; Gatidis, S.; Eigentler, T.; Amaral, T.; Moltz, J.H.; et al. Combination of Whole-Body Baseline CT Radiomics and Clinical Parameters to Predict Response and Survival in a Stage-IV Melanoma Cohort Undergoing Immunotherapy. Cancers 2022, 14, 2992. https://doi.org/10.3390/cancers14122992
Peisen F, Hänsch A, Hering A, Brendlin AS, Afat S, Nikolaou K, Gatidis S, Eigentler T, Amaral T, Moltz JH, et al. Combination of Whole-Body Baseline CT Radiomics and Clinical Parameters to Predict Response and Survival in a Stage-IV Melanoma Cohort Undergoing Immunotherapy. Cancers. 2022; 14(12):2992. https://doi.org/10.3390/cancers14122992
Chicago/Turabian StylePeisen, Felix, Annika Hänsch, Alessa Hering, Andreas S. Brendlin, Saif Afat, Konstantin Nikolaou, Sergios Gatidis, Thomas Eigentler, Teresa Amaral, Jan H. Moltz, and et al. 2022. "Combination of Whole-Body Baseline CT Radiomics and Clinical Parameters to Predict Response and Survival in a Stage-IV Melanoma Cohort Undergoing Immunotherapy" Cancers 14, no. 12: 2992. https://doi.org/10.3390/cancers14122992
APA StylePeisen, F., Hänsch, A., Hering, A., Brendlin, A. S., Afat, S., Nikolaou, K., Gatidis, S., Eigentler, T., Amaral, T., Moltz, J. H., & Othman, A. E. (2022). Combination of Whole-Body Baseline CT Radiomics and Clinical Parameters to Predict Response and Survival in a Stage-IV Melanoma Cohort Undergoing Immunotherapy. Cancers, 14(12), 2992. https://doi.org/10.3390/cancers14122992







