Liquid Biopsy Landscape in Patients with Primary Upper Tract Urothelial Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
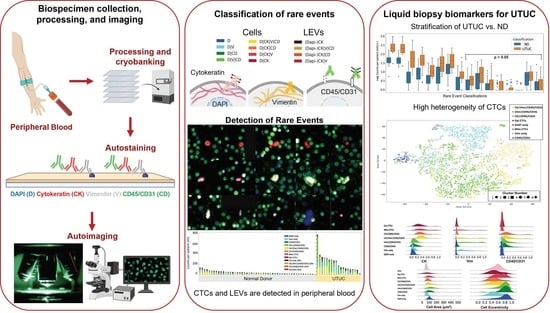
2. Materials and Methods
2.1. Study Design
2.2. Blood Sample Processing
2.3. Blood Sample Staining and Imaging
2.4. Rare-Event Detection and Classification
2.5. Statistical Analysis
3. Results
3.1. Liquid Biopsy Analysis Prior to Surgery
3.2. Rare-Cell Characterization
3.3. LEV Detection
3.4. Correlation with Clinical Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bersanelli, M.; Buti, S.; Giannatempo, P.; Raggi, D.; Necchi, A.; Leonetti, A.; Banna, G.L.; Petrelli, F. Outcome of patients with advanced upper tract urothelial carcinoma treated with immune checkpoint inhibitors: A systematic review and meta-analysis. Crit. Rev. Oncol. 2021, 159, 103241. [Google Scholar] [CrossRef]
- Rouprêt, M.; Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Cowan, N.C.; Dominguez-Escrig, J.L.; Gontero, P.; Mostafid, A.H.; et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2020 Update. Eur. Urol. 2021, 79, 62–79. [Google Scholar] [CrossRef]
- Honda, Y.; Nakamura, Y.; Teishima, J.; Goto, K.; Higaki, T.; Narita, K.; Akagi, M.; Terada, H.; Kaichi, Y.; Fujii, S.; et al. Clinical staging of upper urinary tract urothelial carcinoma for T staging: Review and pictorial essay. Int. J. Urol. 2019, 26, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Petros, F.G. Epidemiology, clinical presentation, and evaluation of upper-tract urothelial carcinoma. Transl. Androl. Urol. 2020, 9, 1794–1798. [Google Scholar] [CrossRef]
- Xylinas, E.; Rink, M.; Margulis, V.; Karakiewicz, P.; Novara, G.; Shariat, S.F. Multifocal carcinoma in situ of the upper tract is associated with high risk of bladder cancer recurrence. Eur. Urol. 2012, 61, 1069–1070. [Google Scholar] [CrossRef]
- Cosentino, M.; Palou, J.; Gaya, J.M.; Breda, A.; Rodriguez-Faba, O.; Villavicencio-Mavrich, H. Upper urinary tract urothelial cell carcinoma: Location as a predictive factor for concomitant bladder carcinoma. World J. Urol. 2013, 31, 141–145. [Google Scholar] [CrossRef]
- Leow, J.; Liu, Z.; Tan, T.W.; Lee, Y.M.; Yeo, E.K.; Chong, Y.-L. Optimal Management of Upper Tract Urothelial Carcinoma: Current Perspectives. Onco Targets Ther. 2020, 13, 1–15. [Google Scholar] [CrossRef]
- Subiela, J.D.; Territo, A.; Mercadé, A.; Balañà, J.; Aumatell, J.; Calderon, J.; Gallioli, A.; González-Padilla, D.A.; Gaya, J.M.; Palou, J.; et al. Diagnostic accuracy of ureteroscopic biopsy in predicting stage and grade at final pathology in upper tract urothelial carcinoma: Systematic review and meta-analysis. Eur. J. Surg. Oncol. 2020, 46, 1989–1997. [Google Scholar] [CrossRef]
- Mori, K.; Katayama, S.; Laukhtina, E.; Schuettfort, V.M.; Pradere, B.; Quhal, F.; Motlagh, R.S.; Mostafaei, H.; Grossmann, N.C.; Rajwa, P.; et al. Discordance between Clinical and Pathological Staging and Grading in Upper Tract Urothelial Carcinoma. Clin. Genitourin. Cancer 2022, 20, 95.e1–95.e6. [Google Scholar] [CrossRef] [PubMed]
- Marchioni, M.; Primiceri, G.; Cindolo, L.; Hampton, L.J.; Grob, M.B.; Guruli, G.; Schips, L.; Shariat, S.F.; Autorino, R. Impact of diagnostic ureteroscopy on intravesical recurrence in patients undergoing radical nephroureterectomy for upper tract urothelial cancer: A systematic review and meta-analysis. BJU Int. 2017, 120, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Katims, A.B.; Say, R.; Derweesh, I.; Uzzo, R.; Minervini, A.; Wu, Z.; Abdollah, F.; Sundaram, C.; Ferro, M.; Rha, K.; et al. Risk Factors for Intravesical Recurrence after Minimally Invasive Nephroureterectomy for Upper Tract Urothelial Cancer (ROBUUST Collaboration). J. Urol. 2021, 206, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Cheng, K.S.; Chen, A.; Xu, Z.; Chen, X.; Tian, J.; Xu, C.; Sun, Y.; Neoh, K.H.; Dai, Y.; et al. Microfluidic Assaying of Circulating Tumor Cells and Its Application in Risk Stratification of Urothelial Bladder Cancer. Front. Oncol. 2021, 11, 701298. [Google Scholar] [CrossRef] [PubMed]
- Soave, A.; Riethdorf, S.; Dahlem, R.; Minner, S.; Weisbach, L.; Engel, O.; Fisch, M.; Pantel, K.; Rink, M. Detection and oncological effect of circulating tumour cells in patients with variant urothelial carcinoma histology treated with radical cystectomy. BJU Int. 2017, 119, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fan, W.; Deng, Q.; Tang, S.; Wang, P.; Xu, P.; Wang, J.; Yu, M. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: A meta-analysis of 30 published studies. Oncotarget 2017, 8, 59527–59538. [Google Scholar] [CrossRef]
- Claps, F.; Mir, M.C.; Zargar, H. Molecular markers of systemic therapy response in urothelial carcinoma. Asian J. Urol. 2021, 8, 376–390. [Google Scholar] [CrossRef]
- Marrinucci, D.; Bethel, K.; Kolatkar, A.; Luttgen, M.S.; Malchiodi, M.; Baehring, F.; Voigt, K.; Lazar, D.; Nieva, J.J.; Bazhenova, L.; et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys. Biol. 2012, 9, 016003. [Google Scholar] [CrossRef]
- Gerdtsson, A.S.; Thiele, J.-A.; Shishido, S.N.; Zheng, S.; Schaffer, R.; Bethel, K.; Curley, S.; Lenz, H.-J.; Hanna, D.L.; Nieva, J.; et al. Single cell correlation analysis of liquid and solid biopsies in metastatic colorectal cancer. Oncotarget 2019, 10, 7016–7030. [Google Scholar] [CrossRef][Green Version]
- Malihi, P.D.; Graf, R.P.; Rodriguez, A.; Ramesh, N.; Lee, J.; Sutton, R.; Jiles, R.; Velasco, C.R.; Sei, E.; Kolatkar, A.; et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clin. Cancer Res. 2020, 26, 4143–4153. [Google Scholar] [CrossRef]
- Thiele, J.A.; Pitule, P.; Hicks, J.; Kuhn, P. Single-Cell Analysis of Circulating Tumor Cells. Methods Mol. Biol. 2019, 1908, 243–264. [Google Scholar]
- Malihi, P.D.; Morikado, M.; Welter, L.; Liu, S.T.; Miller, E.T.; Cadaneanu, R.M.; Knudsen, B.S.; Lewis, M.S.; Carlsson, A.; Velasco, C.R.; et al. Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg. Sci. Phys. Oncol. 2018, 4, 015003. [Google Scholar] [CrossRef]
- Carlsson, A.; Kuhn, P.; Luttgen, M.S.; Keomanee-Dizon, K.; Troncoso, P.; Corn, P.G.; Kolatkar, A.; Hicks, J.B.; Logothetis, C.J.; Zurita, A.J. Paired High-Content Analysis of Prostate Cancer Cells in Bone Marrow and Blood Characterizes Increased Androgen Receptor Expression in Tumor Cell Clusters. Clin. Cancer Res. 2017, 23, 1722–1732. [Google Scholar] [CrossRef]
- Ruiz, C.; Li, J.; Luttgen, M.S.; Kolatkar, A.; Kendall, J.T.; Flores, E.; Topp, Z.; Samlowski, W.E.; McClay, E.; Bethel, K.; et al. Limited genomic heterogeneity of circulating melanoma cells in advanced stage patients. Phys. Biol. 2015, 12, 016008. [Google Scholar] [CrossRef]
- Gerdtsson, A.; Setayesh, S.; Malihi, P.; Ruiz, C.; Carlsson, A.; Nevarez, R.; Matsumoto, N.; Gerdtsson, E.; Zurita, A.; Logothetis, C.; et al. Large Extracellular Vesicle Characterization and Association with Circulating Tumor Cells in Metastatic Castrate Resistant Prostate Cancer. Cancers 2021, 13, 1056. [Google Scholar] [CrossRef]
- Chai, S.; Matsumoto, N.; Storgard, R.; Peng, C.-C.; Aparicio, A.; Ormseth, B.; Rappard, K.; Cunningham, K.; Kolatkar, A.; Nevarez, R.; et al. Platelet-Coated Circulating Tumor Cells Are a Predictive Biomarker in Patients with Metastatic Castrate-Resistant Prostate Cancer. Mol. Cancer Res. 2021, 19, 2036–2045. [Google Scholar] [CrossRef]
- Shishido, S.N.; Sayeed, S.; Courcoubetis, G.; Djaladat, H.; Miranda, G.; Pienta, K.J.; Nieva, J.; Hansel, D.E.; Desai, M.; Gill, I.S.; et al. Characterization of Cellular and Acellular Analytes from Pre-Cystectomy Liquid Biopsies in Patients Newly Diagnosed with Primary Bladder Cancer. Cancers 2022, 14, 758. [Google Scholar] [CrossRef]
- Spearman, C. The Proof and Measurement of Association between Two Things. Am. J. Psychol. 1987, 100, 441–471. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual Comparisons of Grouped Data by Ranking Methods. J. Econ. Èntomol. 1946, 39, 269–270. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Ward, M.P.; Kane, L.E.; Norris, L.A.; Mohamed, B.M.; Kelly, T.; Bates, M.; Clarke, A.; Brady, N.; Martin, C.M.; Brooks, R.D. Platelets, immune cells and the coagulation cascade; friend or foe of the circulating tumour cell? Mol. Cancer 2021, 20, 59. [Google Scholar] [CrossRef] [PubMed]
- Foerster, B.; Moschini, M.; Abufaraj, M.; Soria, F.; Gust, K.M.; Rouprêt, M.; Karakiewicz, P.I.; Briganti, A.; Rink, M.; Kluth, L.; et al. Predictive and Prognostic Value of Preoperative Thrombocytosis in Upper Tract Urothelial Carcinoma. Clin. Genitourin. Cancer 2017, 15, e1039–e1045. [Google Scholar] [CrossRef]
- Hu, L.; Su, L.; Cheng, H.; Mo, C.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Fan, T.; Lin, B.; et al. Single-Cell RNA Sequencing Reveals the Cellular Origin and Evolution of Breast Cancer in BRCA1 Mutation Carriers. Cancer Res. 2021, 81, 2600–2611. [Google Scholar] [CrossRef]
- Dago, A.E.; Stepansky, A.; Carlsson, A.; Luttgen, M.; Kendall, J.; Baslan, T.; Kolatkar, A.; Wigler, M.; Bethel, K.; Gross, M.; et al. Rapid Phenotypic and Genomic Change in Response to Therapeutic Pressure in Prostate Cancer Inferred by High Content Analysis of Single Circulating Tumor Cells. PLoS ONE 2014, 9, e101777. [Google Scholar] [CrossRef]
- Shishido, S.N.; Welter, L.; Rodriguez-Lee, M.; Kolatkar, A.; Xu, L.; Ruiz, C.; Gerdtsson, A.S.; Restrepo-Vassalli, S.; Carlsson, A.; Larsen, J.; et al. Preanalytical Variables for the Genomic Assessment of the Cellular and Acellular Fractions of the Liquid Biopsy in a Cohort of Breast Cancer Patients. J. Mol. Diagn. 2020, 22, 319–337. [Google Scholar] [CrossRef]
- Welter, L.; Xu, L.; McKinley, D.; Dago, A.E.; Prabakar, R.K.; Restrepo-Vassalli, S.; Xu, K.; Rodriguez-Lee, M.; Kolatkar, A.; Nevarez, R.; et al. Treatment response and tumor evolution: Lessons from an extended series of multianalyte liquid biopsies in a metastatic breast cancer patient. Cold Spring Harb. Mol. Case Stud. 2020, 6, a005819. [Google Scholar] [CrossRef]
- Shishido, S.N.; Masson, R.; Xu, L.; Welter, L.; Prabakar, R.K.; Souza, A.D.; Spicer, D.; Kang, I.; Jayachandran, P.; Hicks, J.; et al. Disease characterization in liquid biopsy from HER2-mutated, non-amplified metastatic breast cancer patients treated with neratinib. NPJ Breast Cancer 2022, 8, 22. [Google Scholar] [CrossRef]
- Birkenkamp-Demtröder, K.; Christensen, E.; Nordentoft, I.K.; Knudsen, M.; Taber, A.; Høyer, S.; Lamy, P.; Agerbaek, M.; Jensen, J.B.; Dyrskjøt, L. Monitoring Treatment Response and Metastatic Relapse in Advanced Bladder Cancer by Liquid Biopsy Analysis. Eur. Urol. 2018, 73, 535–540. [Google Scholar] [CrossRef]
- Christensen, E.; Birkenkamp-Demtröder, K.; Nordentoft, I.; Høyer, S.; van der Keur, K.; van Kessel, K.; Zwarthoff, E.; Agerbaek, M.; Ørntoft, T.F.; Jensen, J.B.; et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur. Urol. 2017, 71, 961–969. [Google Scholar] [CrossRef]
- Patel, K.; Van Der Vos, K.E.; Smith, C.G.; Mouliere, F.; Tsui, D.; Morris, J.; Chandrananda, D.; Marass, F.; Van Den Broek, D.; Neal, D.; et al. Association of Plasma and Urinary Mutant DNA with Clinical Outcomes in Muscle Invasive Bladder Cancer. Sci. Rep. 2017, 7, 5554. [Google Scholar] [CrossRef] [PubMed]



| Variable | Category | Value |
|---|---|---|
| Age | Median (range), year | 66.5 (43–88) |
| BMI | Median (range), kg/m2 | 26.05 (17.6–37.8) |
| Gender | Male | 17 (85%) |
| Female | 3 (15%) | |
| Smoker | Never | 8 (40%) |
| Former | 8 (40%) | |
| Current | 4 (20%) | |
| CCI | 0 | 10 (50%) |
| ≥1 | 10 (50%) | |
| Previous Bladder Cancer | Yes | 15 (75%) |
| No | 5 (25%) | |
| Type of Surgery | Nephroureterectomy +/− Bladder Cuff Excision | 17 (85%) |
| Distal Ureterectomy | 2 (10%) | |
| Laser Ablation | 1 (5%) | |
| Histology | Pure Urothelial | 17 (85%) |
| Urothelial with Variant Histology | 2 (10%) | |
| N/A * | 1 (5%) | |
| Pathology Grade | Low | 3 (15%) |
| High | 16 (80%) | |
| N/A * | 1 (5%) | |
| RNU pTstage | pT0 | 0 (0%) |
| pTa | 10 (50%) | |
| pTis | 0 (0%) | |
| pT1 | 1 (5%) | |
| pT2 | 0 (0%) | |
| pT3 | 7 (35%) | |
| pT4 | 1 (5%) | |
| N/A * | 1 (5%) | |
| Lymph Node Status | Node+ | 16 (80%) |
| Node− | 3 (15%) | |
| N/A * | 1 (5%) | |
| Neoadjuvant Chemo | Yes | 7 (35%) |
| No | 13 (65%) | |
| Adjuvant Chemo | Yes | 1 (5%) |
| No | 19 (95%) | |
| Recurrence (Bladder) | Yes | 3 (15%) |
| No | 17 (85%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shishido, S.N.; Ghoreifi, A.; Sayeed, S.; Courcoubetis, G.; Huang, A.; Ye, B.; Mrutyunjaya, S.; Gill, I.S.; Kuhn, P.; Mason, J.; et al. Liquid Biopsy Landscape in Patients with Primary Upper Tract Urothelial Carcinoma. Cancers 2022, 14, 3007. https://doi.org/10.3390/cancers14123007
Shishido SN, Ghoreifi A, Sayeed S, Courcoubetis G, Huang A, Ye B, Mrutyunjaya S, Gill IS, Kuhn P, Mason J, et al. Liquid Biopsy Landscape in Patients with Primary Upper Tract Urothelial Carcinoma. Cancers. 2022; 14(12):3007. https://doi.org/10.3390/cancers14123007
Chicago/Turabian StyleShishido, Stephanie N., Alireza Ghoreifi, Salmaan Sayeed, George Courcoubetis, Amy Huang, Brandon Ye, Sankalp Mrutyunjaya, Inderbir S. Gill, Peter Kuhn, Jeremy Mason, and et al. 2022. "Liquid Biopsy Landscape in Patients with Primary Upper Tract Urothelial Carcinoma" Cancers 14, no. 12: 3007. https://doi.org/10.3390/cancers14123007
APA StyleShishido, S. N., Ghoreifi, A., Sayeed, S., Courcoubetis, G., Huang, A., Ye, B., Mrutyunjaya, S., Gill, I. S., Kuhn, P., Mason, J., & Djaladat, H. (2022). Liquid Biopsy Landscape in Patients with Primary Upper Tract Urothelial Carcinoma. Cancers, 14(12), 3007. https://doi.org/10.3390/cancers14123007