Urinary microRNAs and Their Significance in Prostate Cancer Diagnosis: A 5-Year Update
Abstract
:Simple Summary
Abstract
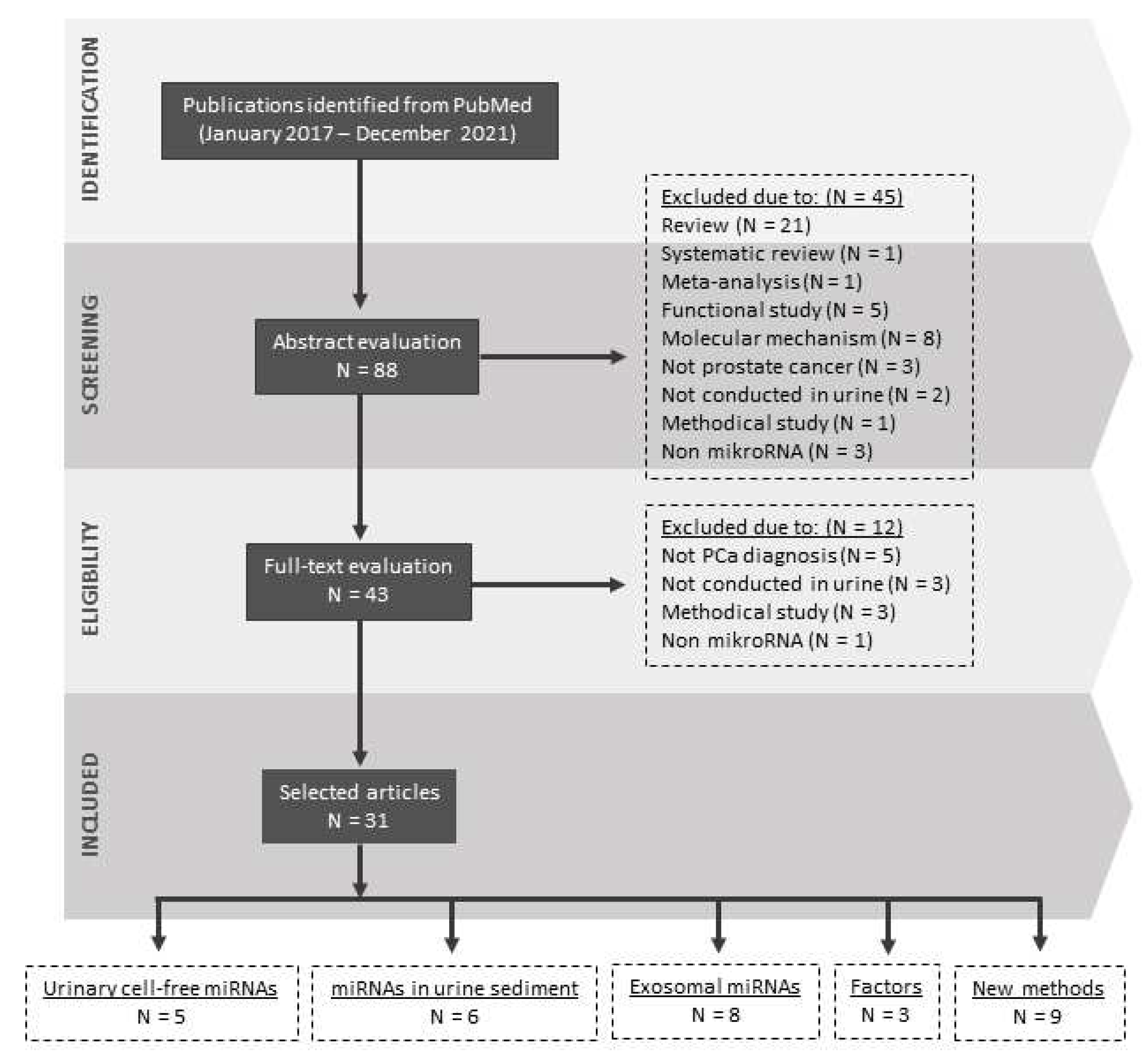
1. Introduction
2. microRNAs in Prostate Cancer
3. Urinary miRNAs
4. Urinary miRNAs and Prostate Cancer Diagnostics
4.1. Urinary Cell-Free miRNAs
4.2. miRNAs in Urine Sediment
4.3. Exosomal miRNAs
5. New Frontiers in Urinary miRNA-Based PCa Detection
5.1. Factors Influencing Urinary miRNA Analysis
5.2. Alternative Methods of Detection
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- James, N.D.; de Bono, J.S.; Spears, M.R.; Clarke, N.W.; Mason, M.D.; Dearnaley, D.P.; Ritchie, A.W.S.; Amos, C.L.; Gilson, C.; Jones, R.J.; et al. Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N. Engl. J. Med. 2017, 377, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Kyriakopoulos, C.E.; Chen, Y.H.; Carducci, M.A.; Liu, G.; Jarrard, D.F.; Hahn, N.M.; Shevrin, D.H.; Dreicer, R.; Hussain, M.; Eisenberger, M.; et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer: Long-Term Survival Analysis of the Randomized Phase III E3805 CHAARTED Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 1080–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, C.; Castro, E.; Fizazi, K.; Heidenreich, A.; Ost, P.; Procopio, G.; Tombal, B.; Gillessen, S. Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Raman, S.S.; Mirak, S.A.; Kwan, L.; Bajgiran, A.M.; Hsu, W.; Maehara, C.K.; Ahuja, P.; Faiena, I.; Pooli, A.; et al. Detection of Individual Prostate Cancer Foci via Multiparametric Magnetic Resonance Imaging. Eur. Urol. 2019, 75, 712–720. [Google Scholar] [CrossRef] [Green Version]
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef]
- Moyer, V.A. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2012, 157, 120–134. [Google Scholar] [CrossRef] [Green Version]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer 2017, 16, 80. [Google Scholar] [CrossRef]
- Liu, B.; Shyr, Y.; Cai, J.; Liu, Q. Interplay between miRNAs and host genes and their role in cancer. Brief. Funct. Genom. 2018, 18, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Gambari, R.; Brognara, E.; Spandidos, D.A.; Fabbri, E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Yeh, Y.; Guo, Q.; Connelly, Z.; Cheng, S.; Yang, S.; Prieto-Dominguez, N.; Yu, X. Wnt/Beta-Catenin Signaling and Prostate Cancer Therapy Resistance. Adv. Exp. Med. Biol. 2019, 1210, 351–378. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, P.E.; Tindall, D.J. Androgen receptor signaling in prostate cancer development and progression. J. Carcinog. 2011, 10, 20. [Google Scholar] [CrossRef]
- Suh, J.; Payvandi, F.; Edelstein, L.C.; Amenta, P.S.; Zong, W.X.; Gélinas, C.; Rabson, A.B. Mechanisms of constitutive NF-kappaB activation in human prostate cancer cells. Prostate 2002, 52, 183–200. [Google Scholar] [CrossRef]
- Kroon, P.; Berry, P.A.; Stower, M.J.; Rodrigues, G.; Mann, V.M.; Simms, M.; Bhasin, D.; Chettiar, S.; Li, C.; Li, P.K.; et al. JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res. 2013, 73, 5288–5298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alwanian, W.M.; Tyner, A.L. Protein tyrosine kinase 6 signaling in prostate cancer. Am. J. Clin. Exp. Urol. 2020, 8, 1–8. [Google Scholar] [PubMed]
- Fernandes, R.C.; Hickey, T.E.; Tilley, W.D.; Selth, L.A. Interplay between the androgen receptor signaling axis and microRNAs in prostate cancer. Endocr. Relat. Cancer 2019, 26, R237–R257. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Guo, J.X.; Shao, Z.Q. miR-21 targets and inhibits tumor suppressor gene PTEN to promote prostate cancer cell proliferation and invasion: An experimental study. Asian Pac. J. Trop. Med. 2017, 10, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Reis, S.T.; Pontes-Junior, J.; Antunes, A.A.; Dall’Oglio, M.F.; Dip, N.; Passerotti, C.C.; Rossini, G.A.; Morais, D.R.; Nesrallah, A.J.; Piantino, C.; et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012, 12, 14. [Google Scholar] [CrossRef]
- Ren, D.; Yang, Q.; Dai, Y.; Guo, W.; Du, H.; Song, L.; Peng, X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol. Cancer 2017, 16, 117. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, M.; Wang, Y.; Luo, J. Curcumin sensitizes prostate cancer cells to radiation partly via epigenetic activation of miR-143 and miR-143 mediated autophagy inhibition. J. Drug Target. 2017, 25, 645–652. [Google Scholar] [CrossRef]
- Ren, D.; Wang, M.; Guo, W.; Huang, S.; Wang, Z.; Zhao, X.; Du, H.; Song, L.; Peng, X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014, 358, 763–778. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Li, Y.; Wang, Z.; Banerjee, S.; Ahmad, A.; Kim, H.R.; Sarkar, F.H. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells 2009, 27, 1712–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Lu, Y.; Cui, D.; Li, E.; Zhu, Y.; Zhao, Y.; Zhao, F.; Xia, S. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol. Rep. 2014, 31, 910–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fendler, A.; Stephan, C.; Yousef, G.M.; Kristiansen, G.; Jung, K. The translational potential of microRNAs as biofluid markers of urological tumours. Nat. Rev. Urol. 2016, 13, 734–752. [Google Scholar] [CrossRef]
- Cheng, L.; Sun, X.; Scicluna, B.J.; Coleman, B.M.; Hill, A.F. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014, 86, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Mlcochova, H.; Hezova, R.; Stanik, M.; Slaby, O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol. Oncol. 2014, 32, 41.e1–41.e9. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef] [Green Version]
- Haj-Ahmad, T.A.; Abdalla, M.A.; Haj-Ahmad, Y. Potential Urinary miRNA Biomarker Candidates for the Accurate Detection of Prostate Cancer among Benign Prostatic Hyperplasia Patients. J. Cancer 2014, 5, 182–191. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Ramalinga, M.; Chijioke, J.; Marian, C.; Oermann, E.K.; Uhm, S.; Kim, J.S.; Chen, L.N.; et al. MicroRNA profiling in prostate cancer--the diagnostic potential of urinary miR-205 and miR-214. PLoS ONE 2013, 8, e76994. [Google Scholar] [CrossRef] [Green Version]
- Paiva, R.M.; Zauli, D.A.G.; Neto, B.S.; Brum, I.S. Urinary microRNAs expression in prostate cancer diagnosis: A systematic review. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2020, 22, 2061–2073. [Google Scholar] [CrossRef]
- Juracek, J.; Slaby, O. Urinary MicroRNAs as Emerging Class of Noninvasive Biomarkers. Methods Mol. Biol. 2020, 2115, 221–247. [Google Scholar] [CrossRef] [PubMed]
- Byun, Y.J.; Piao, X.M.; Jeong, P.; Kang, H.W.; Seo, S.P.; Moon, S.K.; Lee, J.Y.; Choi, Y.H.; Lee, H.Y.; Kim, W.T.; et al. Urinary microRNA-1913 to microRNA-3659 expression ratio as a non-invasive diagnostic biomarker for prostate cancer. Investig. Clin. Urol. 2021, 62, 340–348. [Google Scholar] [CrossRef]
- Fredsøe, J.; Rasmussen, A.K.I.; Thomsen, A.R.; Mouritzen, P.; Høyer, S.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. Eur. Urol. Focus 2018, 4, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredsøe, J.; Rasmussen, A.K.I.; Laursen, E.B.; Cai, Y.; Howard, K.A.; Pedersen, B.G.; Borre, M.; Mouritzen, P.; Ørntoft, T.; Sørensen, K.D. Independent Validation of a Diagnostic Noninvasive 3-MicroRNA Ratio Model (uCaP) for Prostate Cancer in Cell-Free Urine. Clin. Chem. 2019, 65, 540–548. [Google Scholar] [CrossRef] [Green Version]
- Lekchnov, E.A.; Amelina, E.V.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Murashov, I.S.; Pashkovskaya, O.A.; Gorizkii, A.M.; Zheravin, A.A.; et al. Searching for the Novel Specific Predictors of Prostate Cancer in Urine: The Analysis of 84 miRNA Expression. Int. J. Mol. Sci. 2018, 19, 4088. [Google Scholar] [CrossRef] [Green Version]
- Konoshenko, M.Y.; Lekchnov, E.A.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Pak, S.V.; Laktionov, P.P. The Panel of 12 Cell-Free MicroRNAs as Potential Biomarkers in Prostate Neoplasms. Diagnostics 2020, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanoğlu, S.; Göncü, B.S.; Yücesan, E.; Atasoy, S.; Kayali, Y.; Özten KandaŞ, N. Investigating differential miRNA expression profiling using serum and urine specimens for detecting potential biomarker for early prostate cancer diagnosis. Turk. J. Med. Sci. 2021, 51, 1764–1774. [Google Scholar] [CrossRef]
- Guelfi, G.; Cochetti, G.; Stefanetti, V.; Zampini, D.; Diverio, S.; Boni, A.; Mearini, E. Next Generation Sequencing of urine exfoliated cells: An approach of prostate cancer microRNAs research. Sci. Rep. 2018, 8, 7111. [Google Scholar] [CrossRef] [PubMed]
- Ghorbanmehr, N.; Gharbi, S.; Korsching, E.; Tavallaei, M.; Einollahi, B.; Mowla, S.J. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate 2019, 79, 88–95. [Google Scholar] [CrossRef]
- Nayak, B.; Khan, N.; Garg, H.; Rustagi, Y.; Singh, P.; Seth, A.; Dinda, A.K.; Kaushal, S. Role of miRNA-182 and miRNA-187 as potential biomarkers in prostate cancer and its correlation with the staging of prostate cancer. Int. Braz J. Urol Off. J. Braz. Soc. Urol. 2020, 46, 614–623. [Google Scholar] [CrossRef]
- Borkowetz, A.; Lohse-Fischer, A.; Scholze, J.; Lotzkat, U.; Thomas, C.; Wirth, M.P.; Fuessel, S.; Erdmann, K. Evaluation of MicroRNAs as Non-Invasive Diagnostic Markers in Urinary Cells from Patients with Suspected Prostate Cancer. Diagnostics 2020, 10, 578. [Google Scholar] [CrossRef] [PubMed]
- Foj, L.; Ferrer, F.; Serra, M.; Arévalo, A.; Gavagnach, M.; Giménez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Gracia, T.; Wang, X.; Su, Y.; Norgett, E.E.; Williams, T.L.; Moreno, P.; Micklem, G.; Karet Frankl, F.E. Urinary Exosomes Contain MicroRNAs Capable of Paracrine Modulation of Tubular Transporters in Kidney. Sci. Rep. 2017, 7, 40601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Möller, A.; Lobb, R.J. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 2020, 20, 697–709. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef]
- Ku, A.; Fredsøe, J.; Sørensen, K.D.; Borre, M.; Evander, M.; Laurell, T.; Lilja, H.; Ceder, Y. High-Throughput and Automated Acoustic Trapping of Extracellular Vesicles to Identify microRNAs With Diagnostic Potential for Prostate Cancer. Front. Oncol. 2021, 11, 631021. [Google Scholar] [CrossRef]
- Danarto, R.; Astuti, I.; Umbas, R.; Haryana, S.M. Urine miR-21-5p and miR-200c-3p as potential non-invasive biomarkers in patients with prostate cancer. Turk. J. Urol. 2020, 46, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Bonnu, C.H.; Ramadhani, A.N.; Saputro, R.B.; Sesotyosari, S.L.; Danarto, R.; Astuti, I.; Haryana, S.M. The Potential of hsa-mir-106b-5p as Liquid Biomarker in Prostate Cancer Patients in Indonesia. Asian Pac. J. Cancer Prev. APJCP 2021, 22, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.; Kaul, D.; Mavuduru, R.S.; Kakkar, N.; Bhatia, A. Urinary-exosomal miR-2909: A novel pathognomonic trait of prostate cancer severity. J. Biotechnol. 2017, 259, 135–139. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Fujita, K.; Tomiyama, E.; Hatano, K.; Hayashi, Y.; Wang, C.; Ishizuya, Y.; Yamamoto, Y.; Hayashi, T.; Kato, T.; et al. MiR-30b-3p and miR-126-3p of urinary extracellular vesicles could be new biomarkers for prostate cancer. Transl. Androl. Urol. 2021, 10, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, L.X.; Diao, Y.J.; Wang, J.; Ye, Y.; Hao, X.K. Identification of Urinary Exosomal miRNAs for the Non-Invasive Diagnosis of Prostate Cancer. Cancer Manag. Res. 2021, 13, 25–35. [Google Scholar] [CrossRef]
- Wang, W.W.; Sorokin, I.; Aleksic, I.; Fisher, H.; Kaufman, R.P., Jr.; Winer, A.; McNeill, B.; Gupta, R.; Tilki, D.; Fleshner, N.; et al. Expression of Small Noncoding RNAs in Urinary Exosomes Classifies Prostate Cancer into Indolent and Aggressive Disease. J. Urol. 2020, 204, 466–475. [Google Scholar] [CrossRef]
- Lee, J.; Kwon, M.H.; Kim, J.A.; Rhee, W.J. Detection of exosome miRNAs using molecular beacons for diagnosing prostate cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, S52–S63. [Google Scholar] [CrossRef] [Green Version]
- Davey, M.; Benzina, S.; Savoie, M.; Breault, G.; Ghosh, A.; Ouellette, R.J. Affinity Captured Urinary Extracellular Vesicles Provide mRNA and miRNA Biomarkers for Improved Accuracy of Prostate Cancer Detection: A Pilot Study. Int. J. Mol. Sci. 2020, 21, 8330. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Bryzgunova, O.E.; Lekchnov, E.A.; Amelina, E.V.; Yarmoschuk, S.V.; Pak, S.V.; Laktionov, P.P. The Influence of Radical Prostatectomy on the Expression of Cell-Free MiRNA. Diagnostics 2020, 10, 600. [Google Scholar] [CrossRef]
- Scott, G.K.; Mattie, M.D.; Berger, C.E.; Benz, S.C.; Benz, C.C. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006, 66, 1277–1281. [Google Scholar] [CrossRef] [Green Version]
- Höglund, K.; Bogstedt, A.; Fabre, S.; Aziz, A.; Annas, P.; Basun, H.; Minthon, L.; Lannfelt, L.; Blennow, K.; Andreasen, N. Longitudinal stability evaluation of biomarkers and their correlation in cerebrospinal fluid and plasma from patients with Alzheimer’s disease. J. Alzheimer’s Dis. JAD 2012, 32, 939–947. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.; Belmonte, K.C.; Kasten, T.; Bateman, R.; Kim, J. Intra- and Inter-individual Variability of microRNA Levels in Human Cerebrospinal Fluid: Critical Implications for Biomarker Discovery. Sci. Rep. 2017, 7, 12720. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Olkhov-Mitsel, E.; Xie, H.; Yao, C.Q.; Zhao, F.; Jahangiri, S.; Cuizon, C.; Scarcello, S.; Jeyapala, R.; Watson, J.D.; et al. Temporal Stability and Prognostic Biomarker Potential of the Prostate Cancer Urine miRNA Transcriptome. J. Natl. Cancer Inst. 2020, 112, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Gofrit, O.N.; Katz, R.; Shapiro, A.; Yutkin, V.; Pizov, G.; Zorn, K.C.; Duvdevani, M.; Landau, E.H.; Pode, D. Gross hematuria in patients with prostate cancer: Etiology and management. ISRN Surg. 2013, 2013, 685327. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Edelman, J.J.; Kao, S.C.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondal, T.; Jensby Nielsen, S.; Baker, A.; Andreasen, D.; Mouritzen, P.; Wrang Teilum, M.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, W.; Du, Y.; Guo, Q.; Mao, Y.; Zhou, X.; Hua, D. Serum miR-16 as a potential biomarker for human cancer diagnosis: Results from a large-scale population. J. Cancer Res. Clin. Oncol. 2019, 145, 787–796. [Google Scholar] [CrossRef]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef]
- Bazzell, B.G.; Rainey, W.E.; Auchus, R.J.; Zocco, D.; Bruttini, M.; Hummel, S.L.; Byrd, J.B. Human Urinary mRNA as a Biomarker of Cardiovascular Disease. Circulation. Genom. Precis. Med. 2018, 11, e002213. [Google Scholar] [CrossRef] [Green Version]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. Off. J. Jpn. Urol. Assoc. 2018, 25, 770–779. [Google Scholar] [CrossRef] [Green Version]
- Freitas, D.; Balmaña, M.; Poças, J.; Campos, D.; Osório, H.; Konstantinidi, A.; Vakhrushev, S.Y.; Magalhães, A.; Reis, C.A. Different isolation approaches lead to diverse glycosylated extracellular vesicle populations. J. Extracell. Vesicles 2019, 8, 1621131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musante, L.; Saraswat, M.; Ravidà, A.; Byrne, B.; Holthofer, H. Recovery of urinary nanovesicles from ultracentrifugation supernatants. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc. -Eur. Ren. Assoc. 2013, 28, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Wachalska, M.; Koppers-Lalic, D.; van Eijndhoven, M.; Pegtel, M.; Geldof, A.A.; Lipinska, A.D.; van Moorselaar, R.J.; Bijnsdorp, I.V. Protein Complexes in Urine Interfere with Extracellular Vesicle Biomarker Studies. J. Circ. Biomark. 2016, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Zaporozhchenko, I.A.; Lekchnov, E.A.; Amelina, E.V.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Pashkovskaya, O.A.; Zheravin, A.A.; Pak, S.V.; Rykova, E.Y.; et al. Data analysis algorithm for the development of extracellular miRNA-based diagnostic systems for prostate cancer. PLoS ONE 2019, 14, e0215003. [Google Scholar] [CrossRef] [Green Version]
- Markert, L.; Holdmann, J.; Klinger, C.; Kaufmann, M.; Schork, K.; Turewicz, M.; Eisenacher, M.; Savelsbergh, A. Small RNAs as biomarkers to differentiate benign and malign prostate diseases: An alternative for transrectal punch biopsy of the prostate? PLoS ONE 2021, 16, e0247930. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Allelein, S.; Pandey, R.; Medina-Perez, P.; Osman, E.; Kuhlmeier, D.; Soleymani, L. Two-Step Competitive Hybridization Assay: A Method for Analyzing Cancer-Related microRNA Embedded in Extracellular Vesicles. Anal. Chem. 2021, 93, 15913–15921. [Google Scholar] [CrossRef]
- Kim, J.; Shim, J.S.; Han, B.H.; Kim, H.J.; Park, J.; Cho, I.J.; Kang, S.G.; Kang, J.Y.; Bong, K.W.; Choi, N. Hydrogel-based hybridization chain reaction (HCR) for detection of urinary exosomal miRNAs as a diagnostic tool of prostate cancer. Biosens. Bioelectron. 2021, 192, 113504. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.; Cho, Y.S.; Kim, Y.; Tae, J.H.; No, T.I.; Shim, J.S.; Jeong, Y.; Kang, S.H.; Lee, K.H. Electrical Cartridge Sensor Enables Reliable and Direct Identification of MicroRNAs in Urine of Patients. ACS Sens. 2021, 6, 833–841. [Google Scholar] [CrossRef]
- Yasui, T.; Yanagida, T.; Ito, S.; Konakade, Y.; Takeshita, D.; Naganawa, T.; Nagashima, K.; Shimada, T.; Kaji, N.; Nakamura, Y.; et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci. Adv. 2017, 3, e1701133. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Koo, K.M.; Wang, Y.; Trau, M. Native MicroRNA Targets Trigger Self-Assembly of Nanozyme-Patterned Hollowed Nanocuboids with Optimal Interparticle Gaps for Plasmonic-Activated Cancer Detection. Small 2019, 15, e1904689. [Google Scholar] [CrossRef] [PubMed]
| Author, Year | Urine Fraction | Screening (Method/Samples) | Validation (Method/Samples) | Proposed Biomarkers/Comments | Reference |
|---|---|---|---|---|---|
| Byun, 2021 | urine supernatant | Agilent Human miRNA Microarray/14 PCa, 5 BPH | qPCR/ cohort 1: 9 PCa, 8 BPH; cohort 2: 44 PCa, 39 BPH | ↑ miR-1913 to miR-3659 ratio | [32] |
| Fredsøe, 2018 | urine supernatant | RT-qPCR array/188 PCa, 20 BPH | RT-qPCR array/197 PCa, 20 BPH | ↑ miR-222-3p, miR-24-3p, miR-30c-5p/diagnostic model | [33] |
| Fredsøe, 2019 | urine supernatant | RT-qPCR array/404 PCa, 42 BPH; merged cohorts from previous study | RT-qPCR array/cohort 1: 214 PCa, 99 BPH; cohort 2: 139 PCa, 148 BPH | ↑ miR-222-3p, miR-24-3p, miR-30c-5p/diagnostic model | [34] |
| Lekchnov, 2018 | urine supernatant, urine Evs | RT-qPCR array/10 PCa, 10 HC, 10 BPH | - | supernatant: ↑ miR-107-miR-26b-5p, ↑ miR-375-3p-miR-26b-5p; Evs: miR-20a-5p-miR-16-5p, miR-30b-5p-miR-16-5p, miR-31-5p-miR-16-5p, miR-24-3p-miR-200b-3p/miRNA pairs | [35] |
| Konoshenko, 2020 | urine supernatant, urine Evs | based on previous study [35], RT-qPCR array | qPCR/10 PCa, 11 HC, 8 BPH | ↑ miR-125b-miR-30e, ↑ miR-200-miR-30e, ↑ miR-205-miR-30e, ↑ miR-31-miR-30e, ↑ miR-660-miR-30e, ↑ miR-19b-miR-92a/miRNA ratios | [36] |
| Hasanoğlu, 2021 | urine sediment | Affymetrix GeneChip miRNA 4.0 Arrays/8 PCa, 30 HC | qPCR/8 PCa, 30 HC | ↑ miR-320a | [37] |
| Guelfi, 2018 | urine sediment/ exfoliated cells | small RNA sequencing/11 PCa, 11 HC | qPCR/11 PCa, 11 HC | ↓ let-7 family | [38] |
| Ghorbanmehr, 2019 | whole urine | - | qPCR/23 PCa, 22 BPH, 20 HC | ↑ miR-21-5p, ↑ miR-141-3p, ↑ miR-205-5p | [39] |
| Nayak, 2020 | urine sediment | - | qPCR/33 PCa, 30 HC | ↑ miR-182, ↓ miR-187/only in tissue | [40] |
| Borkowetz, 2020 | urine sediment | - | qPCR/50 suspected PCa (26 PCa, 24 tumor-free) | ↓ miR-16, ↓ miR-195 | [41] |
| Foj, 2017 | urinary sediment, urinary Evs | - | qPCR/60 PCa, 10 HC | Sediment: ↑ miR-21, ↑ miR-375, ↑ miR-141, ↓ miR-214; Evs: ↑ miR-21, ↑ miR-375, ↑ let-7c | [42] |
| Author, Year | EVs Isolation Method | Screening (Method/Samples) | Validation (Method/Samples) | Proposed Biomarkers | Reference |
|---|---|---|---|---|---|
| Xu, 2017 | hydrostatic filtration dialysis, ultracentrifugation | qPCR/60 PCa, 37 BPH, 24 HC | - | ↑ miR-145-5p | [49] |
| Ku, 2021 | automated acoustic trapping | NGS/46 PCa GG ≥ 4, 127 PCa GG ≤ 3 + Bx-negative samples | In silico, TCGA prostate dataset/497 subjects | ↓ miR-1, ↑ miR-23b, ↑ miR-27a | [50] |
| Danarto, 2020 | Exiqon miRCURY | qPCR/60 PCa, 20 BPH | - | ↑ miR-21-5p, ↓ miR-200c-3p | [51] |
| Bonnu, 2021 | QIAGEN exosomal Kit | NanoString nCounter Expression Assay/2 PCa, 2 BPH—tissue samples | qPCR/10 PCa, 10 BPH | ↑ has-mir-106b-5p | [52] |
| Wani, 2017 | Exiqon miRCURY | qPCR/90 PCa, 10 BPH, 60 BCa, 50 HC | - | ↑ miR-2909, ↑ miR-615-3p | [53] |
| Matsuzaki, 2021 | differential centrifugation | Affymetrix miRNA microarray 2.0/10 PCa, 4 HC | qPCR/28 PCa, 25 HC | ↑ miR-30b-3p, ↑ miR-126-3p | [54] |
| Li, 2021 | ExoQuick-TC | small RNA sequencing/6 PCa, 3 HC | qPCR/47 PCa, 29 BPH, 25 HC | ↓ miR-375, ↑ miR-451a, ↑ miR-486-3p, ↑ miR-486-5p | [55] |
| Wang, 2020 | Exosome RNA Isolation Kit (Norgen Biotek) | Affymetrix GeneChip miRNA 4.0 Arrays/146 PCa, 89 HC | qPCR OpenArray/868 PCa, 568 HC | Sentinel PCa, Sentinel CS and Sentinel HG | [56] |
| Factor | Effect/Consequence | Significant miRNAs/Comments | Reference |
|---|---|---|---|
| anti-cancer treatment (radical prostatectomy) | miRNA level alteration | miR-19b, miR-30e, miR-31, miR-125b, miR-200b, miR-205, miR-375, miR-378, miR-425, miR-660 | [59] |
| intraindividual variability | changes in level within one subject across repeated measurements | miR-3195, let-7b-5p, miR-144-3p, miR-451a, miR-148a-3p, miR-512-5p, miR-431-5p/intrastable miRNAs | [60] |
| hemolysis | variation in miRNAs enriched in RBC | miR-16, miR-17, miR-92a, miR-106a, miR-210, miR-451 | [65,66] |
| inappropriate reference gene | unreliable data normalization | miR-16 | [68] |
| EV separation method | enrichment of different EV subpopulations and content | - | [72] |
| presence of non-EV components | decrease in EV yield and change in levels of miRNA | miR-21, miR-375 and miR-204 | [74] |
| Author, Year | Method/Approach | Advantage | Disadvantage | Reference |
|---|---|---|---|---|
| Bryzgunova, 2019 | qPCR data evaluation using four-block data analysis algorithm | simplification of miRNA expression, analysis in more urine fractions, compensation of heterogeneity | algorithm based on the analysis of a smaller group of patients, disadvantages connected to qPCR method | [75] |
| Markert, 2021 | machine learning classification algorithm for data analysis | low dependence on the (error-free) measurability of a single marker | algorithm based on the analysis of a small sample size | [76] |
| Lee, 2018 | bi-labeled molecular beacons | direct detection | unknown effect of urine on technology, suitable exosomes isolation | [57] |
| Saha, 2021 | two-step competitive hybridization assay | direct detection, high sensitivity | one marker per analysis, signal normalization | [77] |
| Kim, 2021 | hydrogel-based hybridization chain reaction | analysis without target amplification, low urine volume, ratiometric analysis | instrumentation, needs to be validated on extended cohorts | [78] |
| Kim, 2021 | graphene-based electrical sensor | label-free detection, durability, dynamic range | instrumentation, limited number of measured biomarkers | [79] |
| Yasui, 2017 | electrostatic collection of EVs + standard screening methods | standardized, high efficiency EV collection, small urine volume (1 mL) | only improving EVs extraction, disadvantages connected to subsequent method | [80] |
| Li, 2019 | detection of miRNA-driven self-assembly nanospheres | quantification without pre-processing step, high sensitivity and specificity | synthesis of nanospheres, instrumentation | [81] |
| Davey, 2020 | multi-marker system | detection in EVs, unified peptide-mediated EV capture, combination of different types of markers | disadvantages connected to qPCR method | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juracek, J.; Madrzyk, M.; Stanik, M.; Slaby, O. Urinary microRNAs and Their Significance in Prostate Cancer Diagnosis: A 5-Year Update. Cancers 2022, 14, 3157. https://doi.org/10.3390/cancers14133157
Juracek J, Madrzyk M, Stanik M, Slaby O. Urinary microRNAs and Their Significance in Prostate Cancer Diagnosis: A 5-Year Update. Cancers. 2022; 14(13):3157. https://doi.org/10.3390/cancers14133157
Chicago/Turabian StyleJuracek, Jaroslav, Marie Madrzyk, Michal Stanik, and Ondrej Slaby. 2022. "Urinary microRNAs and Their Significance in Prostate Cancer Diagnosis: A 5-Year Update" Cancers 14, no. 13: 3157. https://doi.org/10.3390/cancers14133157
APA StyleJuracek, J., Madrzyk, M., Stanik, M., & Slaby, O. (2022). Urinary microRNAs and Their Significance in Prostate Cancer Diagnosis: A 5-Year Update. Cancers, 14(13), 3157. https://doi.org/10.3390/cancers14133157







