High Expression of PRNP Predicts Poor Prognosis in Korean Patients with Gastric Cancer
Abstract
Simple Summary
Abstract
1. Introduction
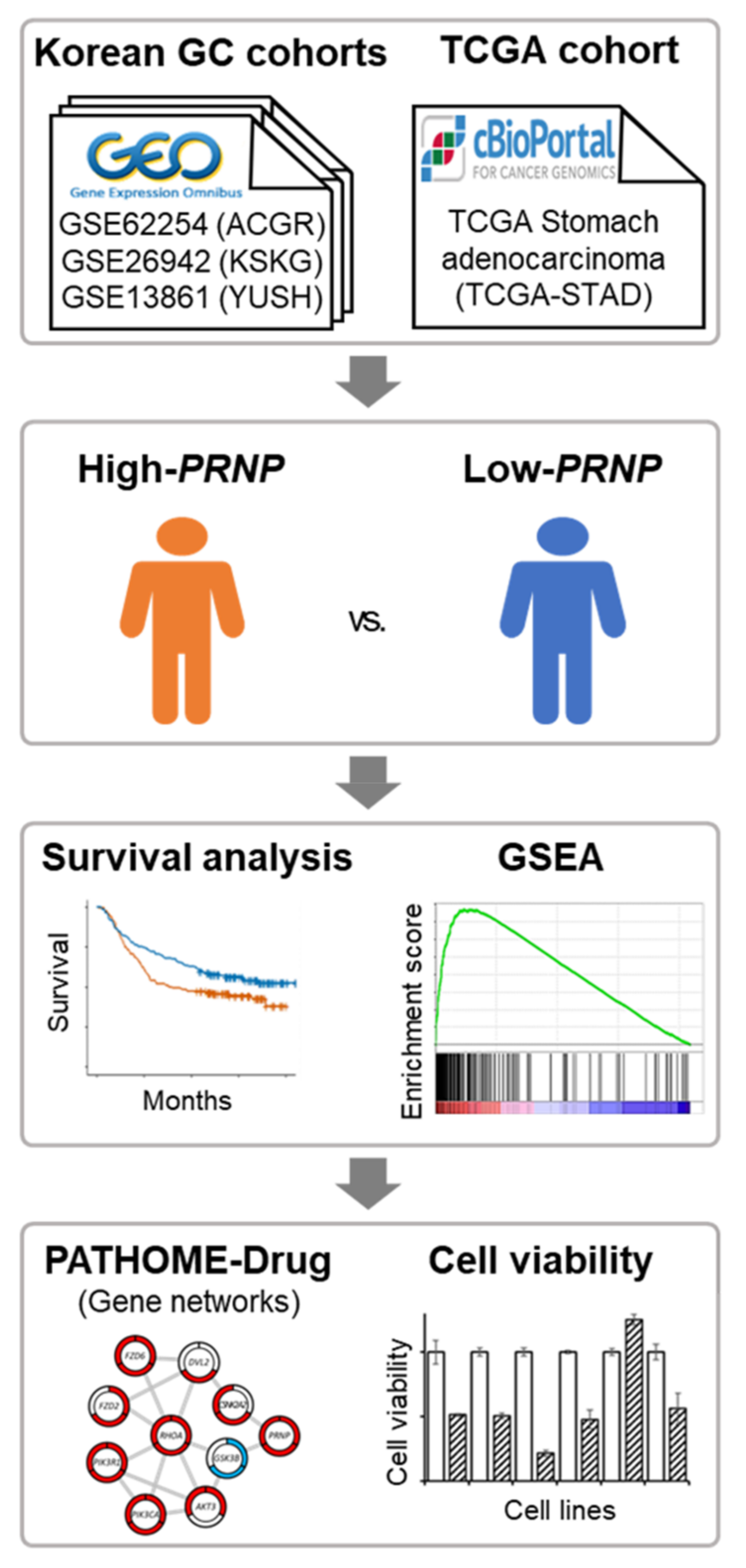
2. Materials and Methods
2.1. Collection of mRNA Expression Data and Clinical Information
2.2. Kaplan–Meier Survival Analysis and Cox Proportional Hazards Model
2.3. Gene Set Enrichment Analysis (GSEA)
2.4. Differential Expression Gene Network
2.5. RNA Extraction and Real-Time qPCR
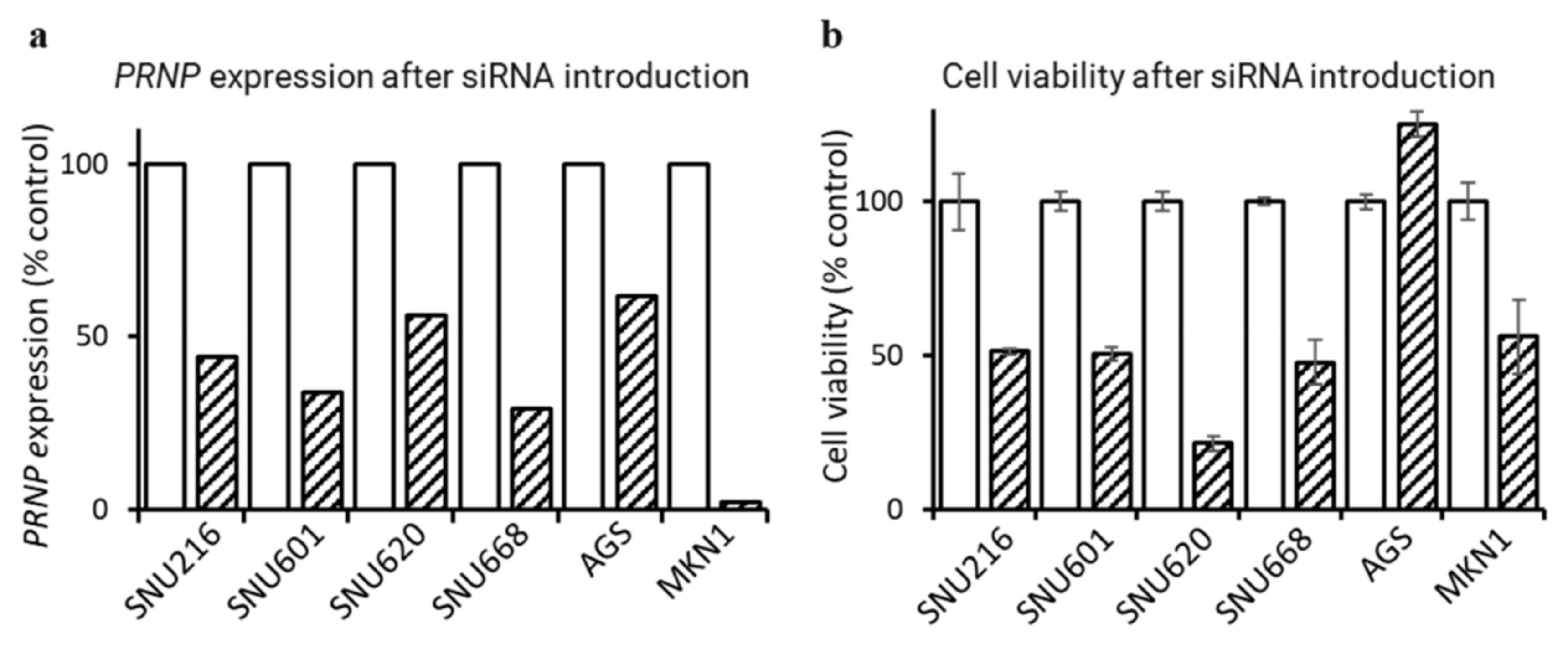
2.6. siPRNP Transfection and MTS Cell Viability Assay
3. Results
3.1. Overview
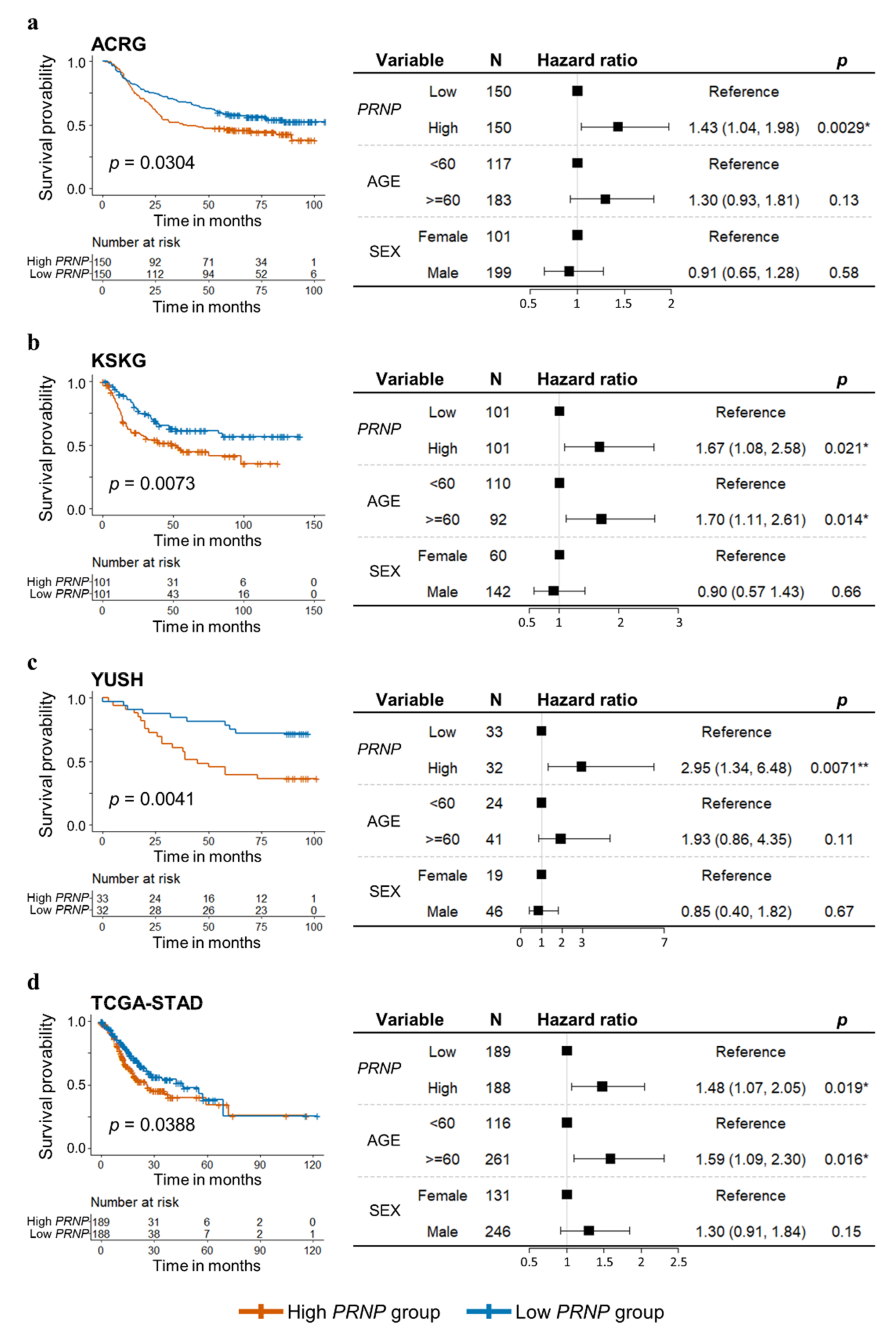
3.2. High Levels of PRNP Expression Are an Independent Prognostic Factor for GC
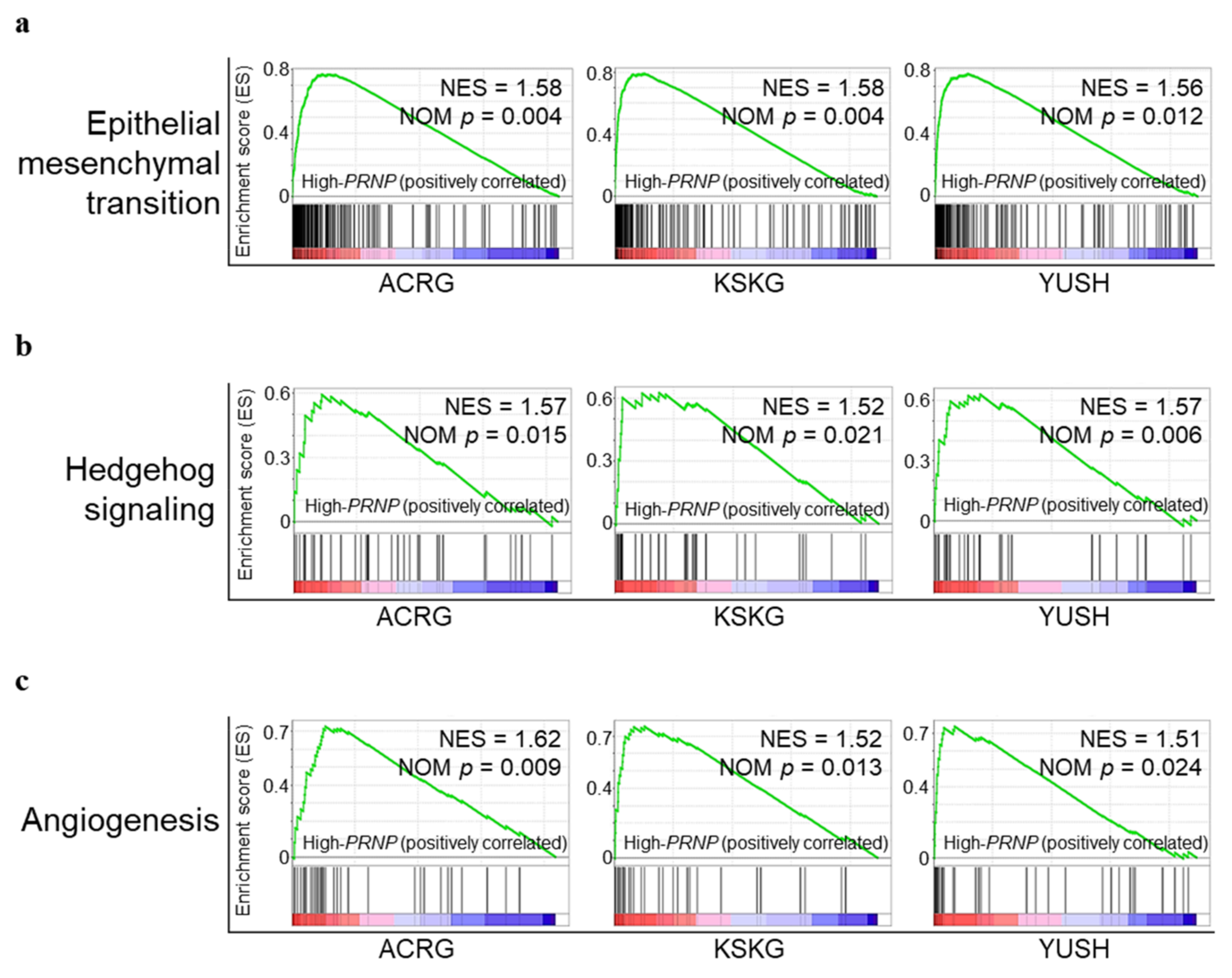
3.3. Upregulation of PRNP in GC Is Associated with Epithelial Mesenchymal Transition, Hedgehog Signaling, and Angiogenesis
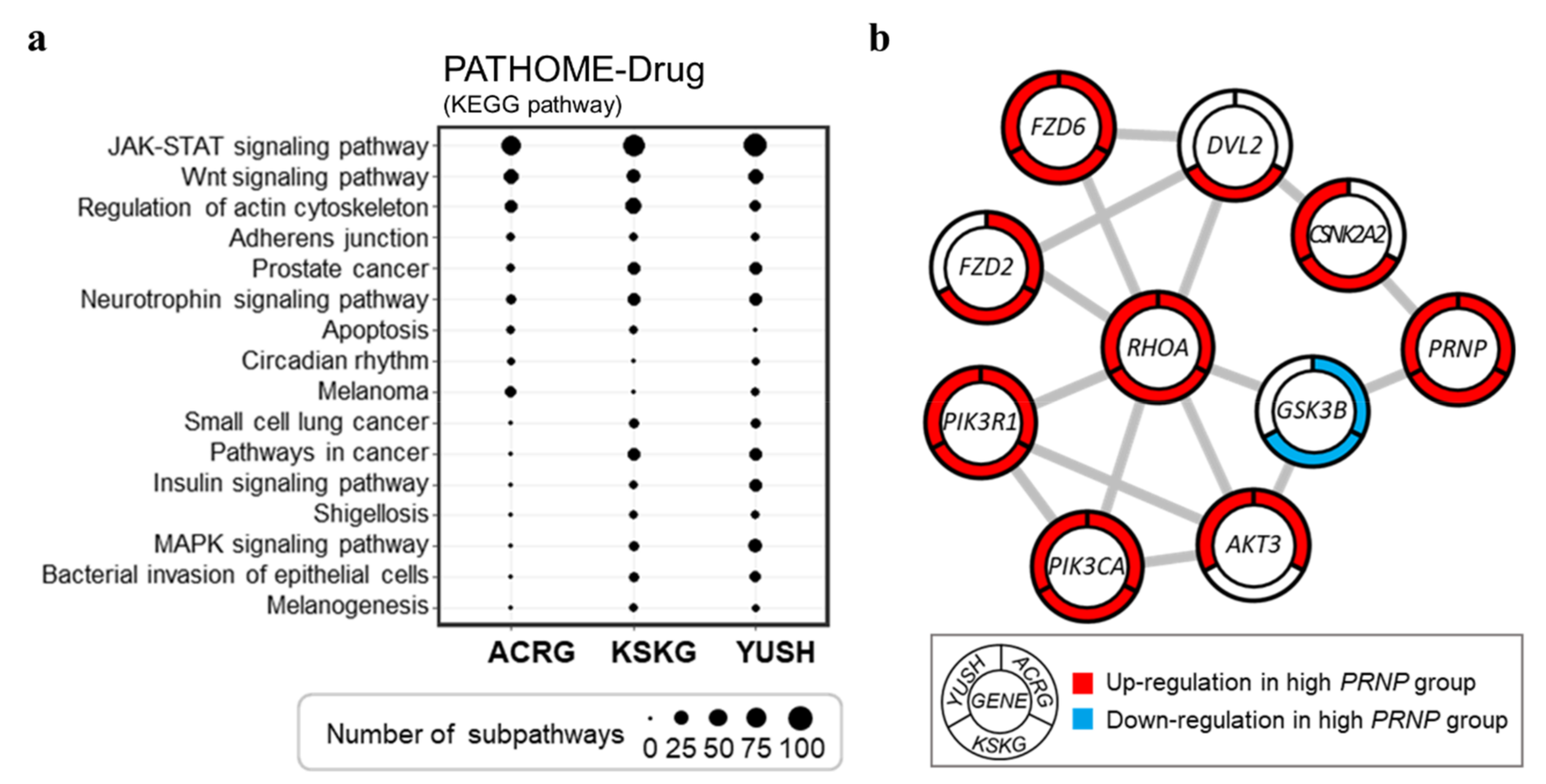
3.4. Networks of Altered Sub-Pathway Genes Reveal Potential Interactions between PRNP and RHOA
3.5. Downregulation of PRNP by siRNA Suppresses GC Cell Proliferation
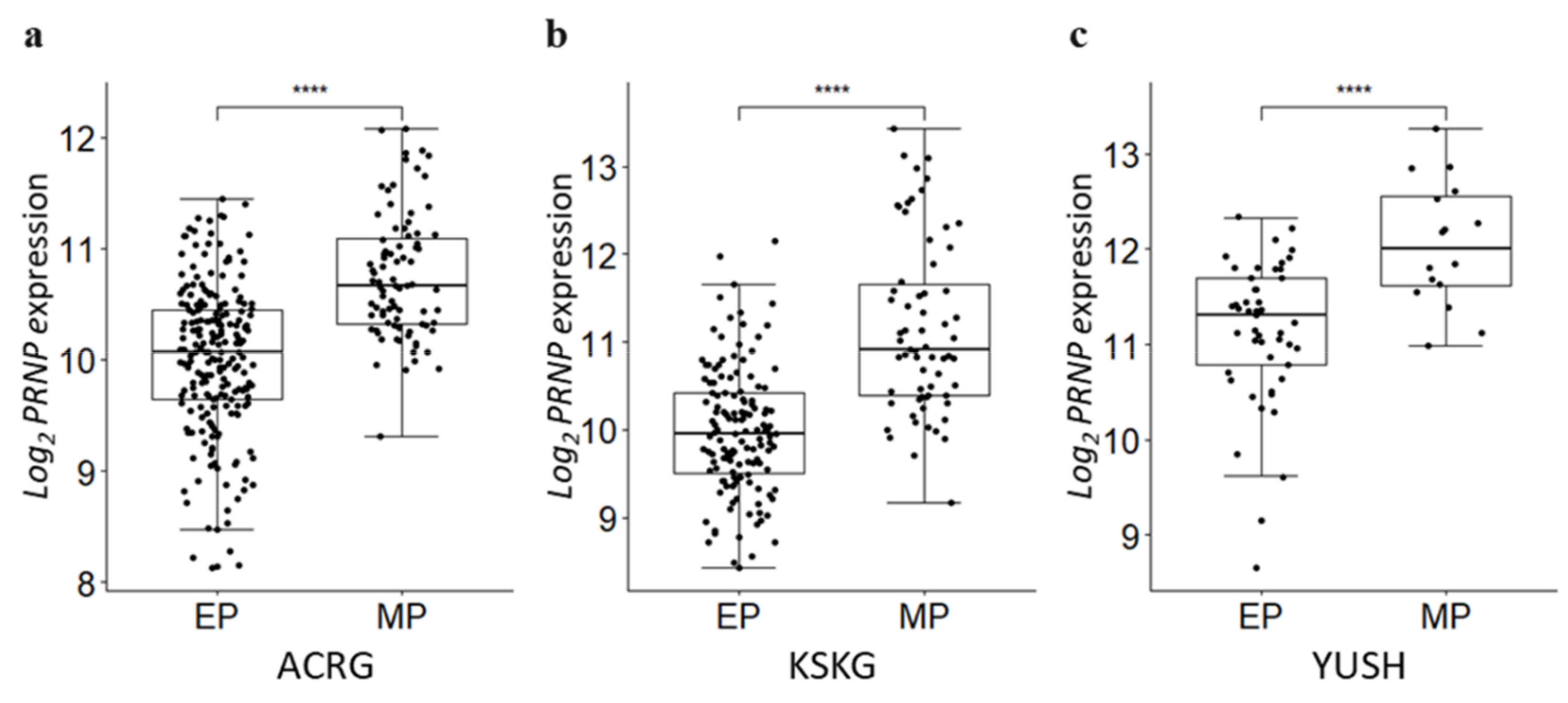
3.6. PRNP Is Upregulated in the Mesenchymal Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, S.; Won, Y.J.; Lee, J.J.; Jung, K.W.; Kong, H.J.; Im, J.S.; Seo, H.G. The Community of Population-Based Regional Cancer Registries Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2018. Cancer Res. Treat. 2021, 53, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Pellino, A.; Riello, E.; Nappo, F.; Brignola, S.; Murgioni, S.; Ahcene-Djaballah, S.; Lonardi, S.; Zagonel, V.; Rugge, M.; Loupakis, F.; et al. Targeted therapies in metastatic gastric cancer: Current knowledge and future perspectives. World J. Gastroenterol. 2019, 25, 5773–5788. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897. [Google Scholar] [CrossRef]
- Riley, R.D.; Sauerbrei, W.; Altman, D.G. Prognostic markers in cancer: The evolution of evidence from single studies to meta-analysis, and beyond. Br. J. Cancer 2009, 100, 1219–1229. [Google Scholar] [CrossRef]
- Kretzschmar, H.A.; Stowring, L.E.; Westaway, D.; Stubblebine, W.H.; Prusiner, S.B.; DeArmond, S.J. Molecular Cloning of a Human Prion Protein cDNA. DNA 1986, 5, 315–324. [Google Scholar] [CrossRef]
- Prusiner, S.B. Shattuck lecture—Neurodegenerative Diseases and Prions. N. Engl. J. Med. 2001, 344, 1516–1526. [Google Scholar] [CrossRef]
- Halliez, S.; Passet, B.; Martin-Lannerã©E, S.; Hernandez-Rapp, J.; Laude, H.; Mouillet-Richard, S.; Vilotte, J.-L.; Beringue, V.; Martin-Lannerée, S.; Béringue, V. To develop with or without the prion protein. Front. Cell Dev. Biol. 2014, 2, 58. [Google Scholar] [CrossRef][Green Version]
- Ding, M.; Chen, Y.; Lang, Y.; Cui, L. The Role of Cellular Prion Protein in Cancer Biology: A Potential Therapeutic Target. Front. Oncol. 2021, 11, 742949. [Google Scholar] [CrossRef]
- Go, G.; Lee, S.H. The Cellular Prion Protein: A Promising Therapeutic Target for Cancer. Int. J. Mol. Sci. 2020, 21, 9208. [Google Scholar] [CrossRef] [PubMed]
- Roucou, X.; Giannopoulos, P.N.; Zhang, Y.; Jodoin, J.; Goodyer, C.G.; Leblanc, A. Cellular prion protein inhibits proapoptotic Bax conformational change in human neurons and in breast carcinoma MCF-7 cells. Cell Death Differ. 2005, 12, 783–795. [Google Scholar] [CrossRef]
- Meslin, F.; Conforti, R.; Mazouni, C.; Morel, N.; Tomasic, G.; Drusch, F.; Yacoub, M.; Sabourin, J.C.; Grassi, J.; Delaloge, S.; et al. Efficacy of adjuvant chemotherapy according to Prion protein expression in patients with estrogen receptor-negative breast cancer. Ann. Oncol. 2007, 18, 1793–1798. [Google Scholar] [CrossRef]
- De Lacerda, T.C.S.; Costa-Silva, B.; Giudice, F.S.; Dias, M.V.S.; de Oliveira, G.P.; Teixeira, B.L.; dos Santos, T.G.; Martins, V.R. Prion protein binding to HOP modulates the migration and invasion of colorectal cancer cells. Clin. Exp. Metastasis 2016, 33, 441–451. [Google Scholar] [CrossRef]
- Sauer, H.; Dagdanova, A.; Hescheler, J.; Wartenberg, M. Redox-regulation of intrinsic prion expression in multicellular prostate tumor spheroids. Free Radic. Biol. Med. 1999, 27, 1276–1283. [Google Scholar] [CrossRef]
- Li, C.; Yu, S.; Nakamura, F.; Yin, S.; Xu, J.; Petrolla, A.A.; Singh, N.; Tartakoff, A.; Abbott, D.; Xin, W.; et al. Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J. Clin. Investig. 2009, 119, 2725–2736. [Google Scholar] [CrossRef]
- Li, Q.-Q.; Cao, X.-X.; Xu, J.-D.; Chen, Q.; Wang, W.-J.; Tang, F.; Chen, Z.-Q.; Liu, X.-P.; Xu, Z.-D. The role of P-glycoprotein/cellular prion protein interaction in multidrug-resistant breast cancer cells treated with paclitaxel. Cell Mol. Life Sci. 2009, 66, 504–515. [Google Scholar] [CrossRef]
- Tang, Z.; Ma, J.; Zhang, W.; Gong, C.; He, J.; Wang, Y.; Yu, G.; Yuan, C.; Wang, X.; Sun, Y.; et al. The Role of Prion Protein Expression in Predicting Gastric Cancer Prognosis. J. Cancer 2016, 7, 984–990. [Google Scholar] [CrossRef][Green Version]
- Liang, J.; Pan, Y.; Zhang, D.; Guo, C.; Shi, Y.; Wang, J.; Chen, Y.; Wang, X.; Liu, J.; Guo, X.; et al. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007, 21, 2247–2256. [Google Scholar] [CrossRef]
- Pan, Y.; Zhao, L.; Liang, J.; Liu, J.; Shi, Y.; Liu, N.; Zhang, G.; Jin, H.; Gao, J.; Xie, H.; et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006, 20, 1886–1888. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [CrossRef]
- Oh, S.C.; Sohn, B.H.; Cheong, J.-H.; Kim, S.B.; Lee, J.E.; Park, K.C.; Lee, S.H.; Park, J.-L.; Park, Y.-Y.; Lee, H.-S.; et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat. Commun. 2018, 9, 1777. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Cho, J.Y.; Lim, J.Y.; Cheong, J.H.; Park, Y.-Y.; Yoon, S.-L.; Kim, S.M.; Kim, S.-B.; Kim, H.; Hong, S.W.; Park, Y.N.; et al. Gene Expression Signature–Based Prognostic Risk Score in Gastric Cancer. Clin. Cancer Res. 2011, 17, 1850–1857. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Therneau, T. A package for survival analysis in S. R Package Version 2015, 2, 7. [Google Scholar]
- White, M.C.; Holman, D.M.; Goodman, R.A.; Richardson, L.C. Cancer Risk Among Older Adults: Time for Cancer Prevention to Go Silver. Gerontologist 2019, 59, S1–S6. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Nam, S.; Lee, S.; Park, S.; Lee, J.; Park, A.; Kim, Y.H.; Park, T. PATHOME-Drug: A subpathway-based polypharmacology drug-repositioning method. Bioinformatics 2021, 38, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Demchak, B.; Ideker, T. Cytoscape tools for the web age: D3.js and Cytoscape.js exporters. F1000Res 2014, 3, 143. [Google Scholar] [CrossRef] [PubMed]
- Amack, J.D. Cellular dynamics of EMT: Lessons from live in vivo imaging of embryonic development. Cell Commun. Signal. 2021, 19, 79. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, Y.; Xie, J. The Hedgehog pathway: Role in cell differentiation, polarity and proliferation. Arch. Toxicol. 2015, 89, 179–191. [Google Scholar] [CrossRef]
- Steelman, L.S.; Pohnert, S.C.; Shelton, J.G.; Franklin, R.A.; Bertrand, F.E.; McCubrey, J.A. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 2004, 18, 189–218. [Google Scholar] [CrossRef]
- Hadjihannas, M.V.; Bernkopf, D.B.; Brückner, M.; Behrens, J. Cell cycle control of Wnt/β-catenin signalling by conductin/axin2 through CDC20. EMBO Rep. 2012, 13, 347–354. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 2007, 1773, 642–652. [Google Scholar] [CrossRef]
- Chang, H.R.; Nam, S.; Lee, J.; Kim, J.-H.; Jung, H.R.; Park, H.S.; Park, S.; Ahn, Y.Z.; Huh, I.; Balch, C.; et al. Systematic approach identifies RHOA as a potential biomarker therapeutic target for Asian gastric cancer. Oncotarget 2016, 7, 81435–81451. [Google Scholar] [CrossRef]
- Katsuno, Y.; Lamouille, S.; Derynck, R. TGF-β signaling and epithelial–mesenchymal transition in cancer progression. Curr. Opin. Oncol. 2013, 25, 76–84. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef]
- Besnier, L.S.; Cardot, P.; Da Rocha, B.; Simon, A.; Loew, D.; Klein, C.; Riveau, B.; Lacasa, M.; Clair, C.; Rousset, M.; et al. The cellular prion protein PrPcis a partner of the Wnt pathway in intestinal epithelial cells. Mol. Biol. Cell 2015, 26, 3313–3328. [Google Scholar] [CrossRef]
- Nam, S.; Kim, J.H.; Lee, D.H. RHOA in Gastric Cancer: Functional Roles and Therapeutic Potential. Front. Genet. 2019, 10, 438. [Google Scholar] [CrossRef]
- Son, H.-J.; Moon, A. Epithelial-mesenchymal Transition and Cell Invasion. Toxicol. Res. 2010, 26, 245–252. [Google Scholar] [CrossRef]
- Shinto, O.; Yashiro, M.; Kawajiri, H.; Shimizu, K.; Shimizu, T.; Miwa, A.; Hirakawa, K. Inhibitory effect of a TGFβ receptor type-I inhibitor, Ki26894, on invasiveness of scirrhous gastric cancer cells. Br. J. Cancer 2010, 102, 844–851. [Google Scholar] [CrossRef]
- Lopes, M.H.; Santos, T.G.; Rodrigues, B.R.; Queiroz-Hazarbassanov, N.; Cunha, I.W.; Wasilewska-Sampaio, A.P.; Costa-Silva, B.; Marchi, F.A.; Bleggi-Torres, L.F.; I Sanematsu, P.; et al. Disruption of prion protein–HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival. Oncogene 2015, 34, 3305–3314. [Google Scholar] [CrossRef]
- Zhou, L.; Shang, Y.; Liu, C.; Li, J.; Hu, H.; Liang, C.; Han, Y.; Zhang, W.; Liang, J.; Wu, K. Overexpression of PrPc, combined with MGr1-Ag/37LRP, is predictive of poor prognosis in gastric cancer. Int. J. Cancer 2014, 135, 2329–2337. [Google Scholar] [CrossRef]
- Sy, M.-S.; Altekruse, S.F.; Li, C.; Lynch, C.F.; Goodman, M.T.; Hernandez, B.Y.; Zhou, L.; Saber, M.S.; Hewitt, S.M.; Xin, W. Association of prion protein expression with pancreatic adenocarcinoma survival in the SEER residual tissue repository. Cancer Biomark. 2011, 10, 251–258. [Google Scholar] [CrossRef]
- Du, L.; Rao, G.; Wang, H.; Li, B.; Tian, W.; Cui, J.; He, L.; Laffin, B.; Tian, X.; Hao, C.; et al. CD44-Positive Cancer Stem Cells Expressing Cellular Prion Protein Contribute to Metastatic Capacity in Colorectal Cancer. Cancer Res. 2013, 73, 2682–2694. [Google Scholar] [CrossRef]
- Santos, E.M.; Fraga, C.A.D.C.; Xavier, A.R.E.D.O.; Xavier, M.A.D.S.; Souza, M.G.; de Jesus, S.F.; de Paula, A.M.B.; Farias, L.C.; Santos, S.H.S.; Santos, T.G.; et al. Prion protein is associated with a worse prognosis of head and neck squamous cell carcinoma. J. Oral Pathol. Med. 2021, 50, 985–994. [Google Scholar] [CrossRef]
- Lin, S.-C.; Lin, C.-H.; Shih, N.-C.; Liu, H.-L.; Wang, W.-C.; Lin, K.-Y.; Liu, Z.-Y.; Tseng, Y.-J.; Chang, H.-K.; Lin, Y.-C.; et al. Cellular prion protein transcriptionally regulated by NFIL3 enhances lung cancer cell lamellipodium formation and migration through JNK signaling. Oncogene 2020, 39, 385–398. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, P.; Lei, S.; Deng, F.; Xiao, G.G.; Liu, Y.; Chen, X.; Li, L.; Wu, S.; Chen, Y.; et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008, 40, 426–436. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, M.; Moon, S.; Eom, H.J.; Lim, S.M.; Kim, Y.H.; Nam, S. High Expression of PRNP Predicts Poor Prognosis in Korean Patients with Gastric Cancer. Cancers 2022, 14, 3173. https://doi.org/10.3390/cancers14133173
Choi M, Moon S, Eom HJ, Lim SM, Kim YH, Nam S. High Expression of PRNP Predicts Poor Prognosis in Korean Patients with Gastric Cancer. Cancers. 2022; 14(13):3173. https://doi.org/10.3390/cancers14133173
Chicago/Turabian StyleChoi, Minseok, SeongRyeol Moon, Hyo Jin Eom, Seung Mook Lim, Yon Hui Kim, and Seungyoon Nam. 2022. "High Expression of PRNP Predicts Poor Prognosis in Korean Patients with Gastric Cancer" Cancers 14, no. 13: 3173. https://doi.org/10.3390/cancers14133173
APA StyleChoi, M., Moon, S., Eom, H. J., Lim, S. M., Kim, Y. H., & Nam, S. (2022). High Expression of PRNP Predicts Poor Prognosis in Korean Patients with Gastric Cancer. Cancers, 14(13), 3173. https://doi.org/10.3390/cancers14133173





