2D-DIGE-MS Proteomics Approaches for Identification of Gelsolin and Peroxiredoxin 4 with Lymph Node Metastasis in Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples and Assessment of CRC Tissue Microarrays (TMAs)
2.2. Chemical Reagents and Antibodies
2.3. Protein Extraction, Protein Labeling, and Gel Image Analysis
2.4. Protein Identification by Mass Spectrometry (MS)
2.5. Human Colon Cancer (CRC) Cell Cultures and Maintenance
2.6. Generation of Stably and Transiently Expressing Gelsolin and Peroxiredoxin-4 Colorectal Cancer Cell Clone
2.7. Cell Cycle Distribution Analysis
2.8. Preparation of Total Cell Extracts and Immunoblot Analyses
2.9. Matrigel Invasion and Scratch Assays
2.10. Establishment of a Subcutaneous Tumor Xenograft
2.11. Chromatin Immunoprecipitation (ChIP) Analysis
2.12. Statistical Analysis
3. Results
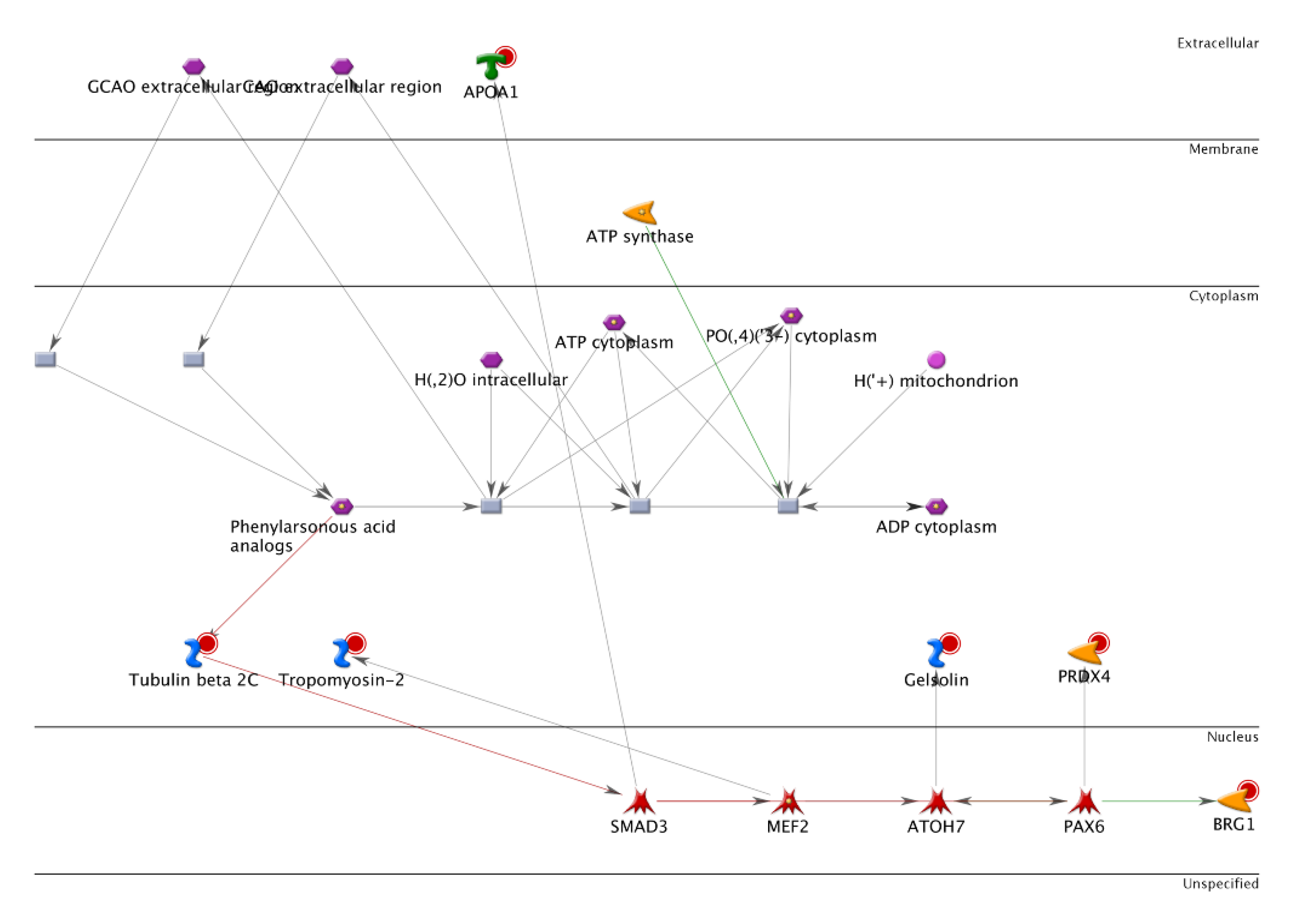
3.1. Identification of Differentially Expressed Proteins Associated with Lymph Node Metastasis Stage I and Stage IV of Human Colorectal Cancer by Two-Dimensional Difference Gel Electrophoresis Proteome Map
3.2. Proteomic Profiling of the Lymph Node Metastasized Colorectal Cancer Tissues by Mass Spectrometry
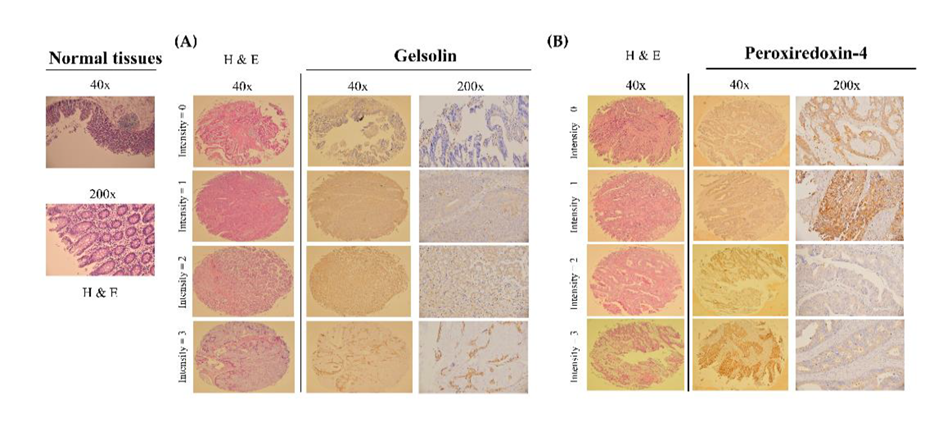
3.3. Validation of the Gelsolin and Peroxiredoxin-4 Expression Patterns on Tissue Microarrays
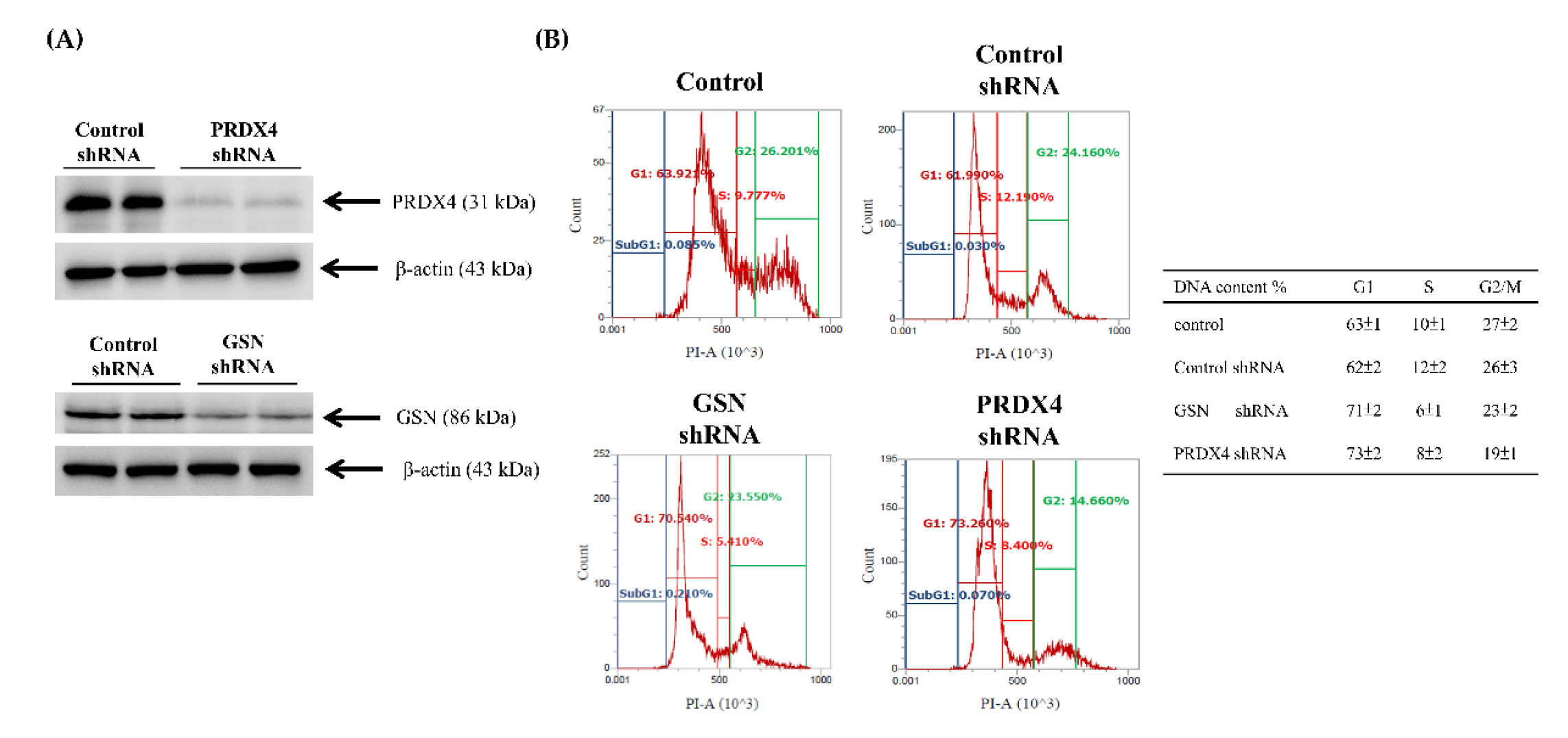
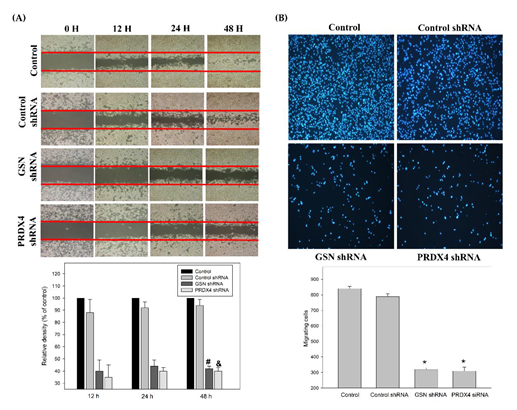
3.4. Effects of Gelsolin and Peroxiredoxin-4 on Cell Biologic Processes
3.5. Effects of Gelsolin and Peroxiredoxin-4 on Epithelial–Mesenchymal Transition in Colorectal Cancer
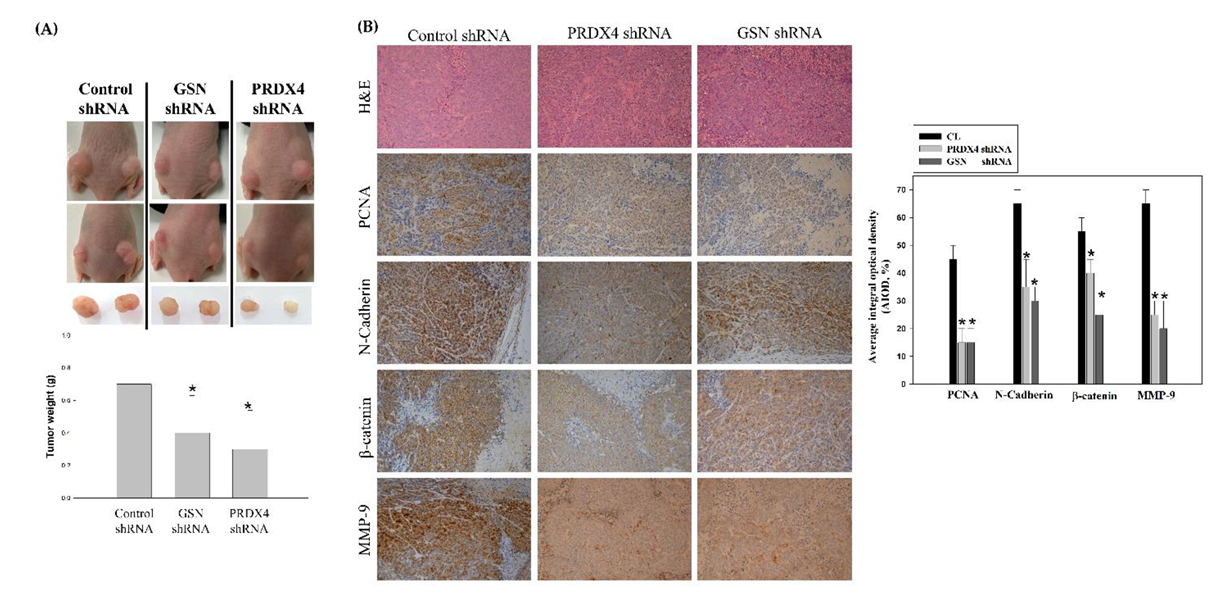
3.6. Suppression of Colorectal Cancer Tumorigenesis by Loss of Gelsolin and Peroxiredoxin-4 In Vivo
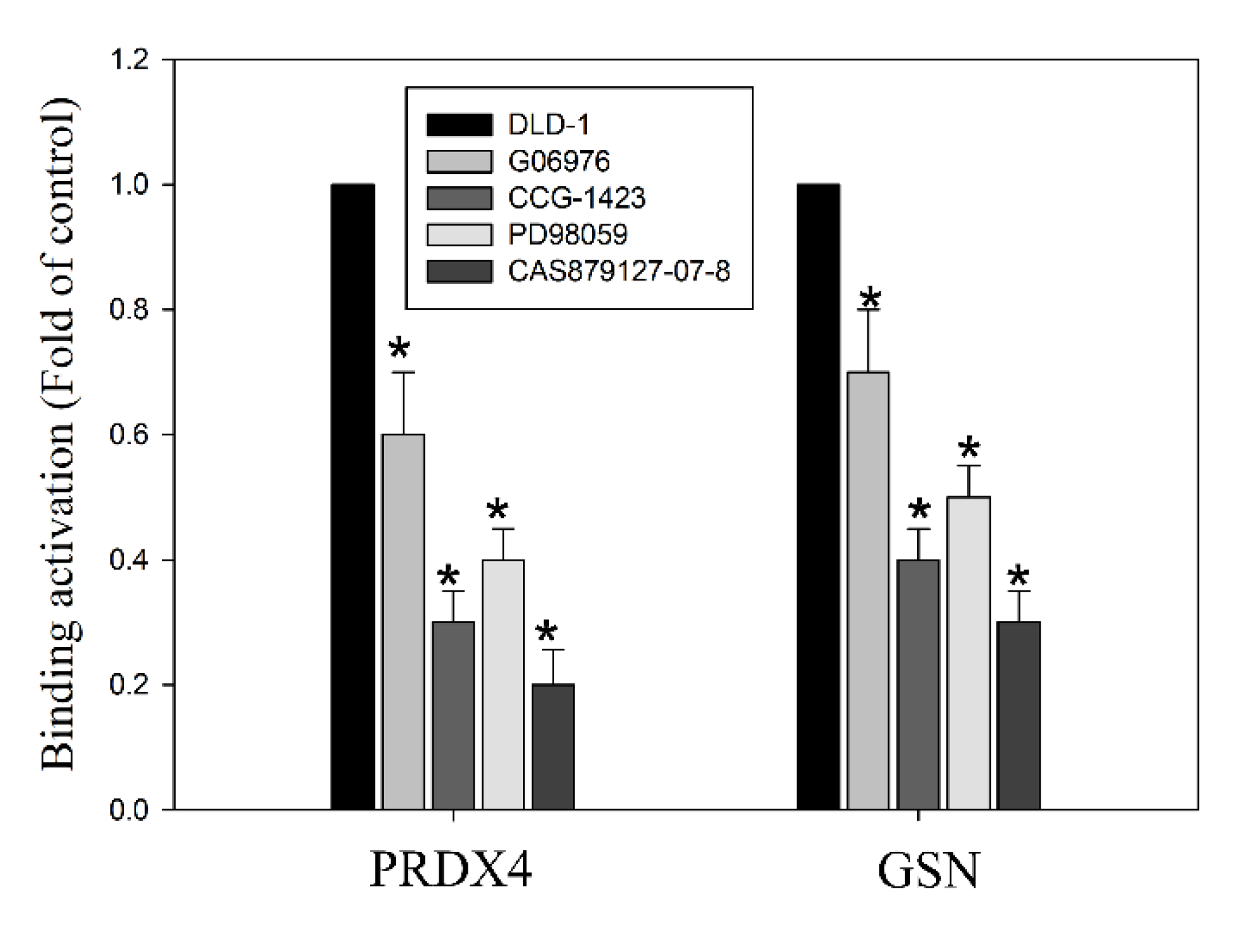
3.7. Histone Modification of Gelsolin and Peroxiredoxin-4 Promoters by EGFR/RhoA/PKC/ERK Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compton, C.C. Optimal pathologic staging: Defining stage II disease. Clin. Cancer Res. 2007, 13, 6862s–6870s. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustavsson, B.; Carlsson, G.; Machover, D.; Petrelli, N.; Roth, A.; Schmoll, H.J.; Tveit, K.M.; Gibson, F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin. Colorectal. Cancer 2015, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, Y.; Hayashi, N.; Sakamoto, Y.; Ohuchi, M.; Tokunagam, R.; Kurashige, J.; Hiyoshi, Y.; Baba, Y.; Iwagami, S.; Yoshida, N.; et al. Predictors of long-term survival in patients with stage IV colorectal cancer with multi-organ metastases: A single-center retrospective analysis. Int. J. Clin. Oncol. 2015, 20, 1140–1146. [Google Scholar] [CrossRef]
- Lamprecht, S.; Kaller, M.; Schmidt, E.M.; Blaj, C.; Schiergens, T.S.; Engel, J.; Jung, A.; Hermeking, H.; Grunewald, T.G.P.; Kirchner, T.; et al. PBX3 Is Part of an EMT Regulatory Network and Indicates Poor Outcome in Colorectal Cancer. Clin. Cancer Res. 2018, 24, 1974–1986. [Google Scholar] [CrossRef] [Green Version]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [Green Version]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef]
- Murphy, L.O.; Blenis, J. MAPK signal specificity: The right place at the right time. Trends Biochem. Sci. 2006, 31, 268–275. [Google Scholar] [CrossRef]
- Kang, J.H.; Toita, R.; Kim, C.W.; Katayama, Y. Protein kinase C (PKC) isozyme-specific substrates and their design. Biotechnol. Adv. 2012, 30, 1662–1672. [Google Scholar] [CrossRef]
- Wang, S.; Li, C.; Wang, W.; Xing, C. PBX3 promotes gastric cancer invasion and metastasis by inducing epithelial-mesenchymal transition. Oncol. Lett. 2016, 12, 3485–3491. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wu, Z.; Wong, G.; Pectasides, E.; Nagaraja, A.; Stachler, M.; Zhang, H.; Chen, T.; Zhang, H.; Liu, J.B.; et al. CDK4/6 or MAPK blockade enhances efficacy of EGFR inhibition in oesophageal squamous cell carcinoma. Nat. Commun. 2017, 8, 13897. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.H.; Zhou, X.L.; Shi, Y.L.; Yang, J. Roles of p38 MAPK and JNK in TGF-beta1-induced human alveolar epithelial to mesenchymal transition. Arch. Med. Res. 2013, 44, 93–98. [Google Scholar] [CrossRef]
- Geisler, C.; Gaisa, N.T.; Pfister, D.; Fuessel, S.; Kristiansen, G.; Braunschweig, T.; Gostek, S.; Beine, B.; Diehl, H.C.; Jackson, A.M.; et al. Identification and validation of potential new biomarkers for prostate cancer diagnosis and prognosis using 2D-DIGE and MS. Biomed. Res. Int. 2015, 2015, 454256. [Google Scholar] [CrossRef]
- Lin, Y.; Buckhaults, P.J.; Lee, J.R.; Xiong, H.; Farrell, C.; Podolsky, R.H.; Schade, R.R.; Dynan, W.S. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia 2009, 11, 864–873. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.S.; Huang, C.Y.; Hsieh, M.C.; Kuo, Y.H.; Tung, S.Y.; Shen, C.H.; Hsieh, Y.Y.; Teng, C.C.; Lee, K.C.; Lee, K.F.; et al. Expression of PRDX6 Correlates with Migration and Invasiveness of Colorectal Cancer Cells. Cell Physiol. Biochem. 2018, 51, 2616–2630. [Google Scholar] [CrossRef]
- Pimentel-Nunes, P.; Afonso, L.; Lopes, P.; Roncon-Albuquerque, R., Jr.; Goncalves, N.; Henrique, R.; Moreira-Dias, L.; Leite-Moreira, A.F.; Dinis-Ribeiro, M. Increased expression of toll-like receptors (TLR) 2, 4 and 5 in gastric dysplasia. Pathol. Oncol. Res. 2011, 17, 677–683. [Google Scholar] [CrossRef]
- Lihong, H.; Linlin, G.; Yiping, G.; Yang, S.; Xiaoyu, Q.; Zhuzhu, G.; Xiaohan, Y.; Xin, Z.; Liyan, X.; Shujuan, S. Proteomics approaches for identification of tumor relevant protein targets in pulmonary squamous cell carcinoma by 2D-DIGE-MS. PLoS ONE 2014, 9, e95121. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.C.; Kuo, H.C.; Hsieh, M.C.; Huang, C.Y.; Teng, C.C.; Tung, S.Y.; Shen, C.H.; Lee, K.F.; Yang, Y.L.; Lee, K.C. Identification of Two Novel CIL-102 Upregulations of ERP29 and FUMH to Inhibit the Migration and Invasiveness of Colorectal Cancer Cells by Using the Proteomic Approach. Biomolecules 2021, 11, 1280. [Google Scholar] [CrossRef]
- Lee, K.C.; Lee, K.F.; Tung, S.Y.; Huang, W.S.; Lee, L.Y.; Chen, W.P.; Chen, C.C.; Teng, C.C.; Shen, C.H.; Hsieh, M.C.; et al. Induction Apoptosis of Erinacine A in Human Colorectal Cancer Cells Involving the Expression of TNFR, Fas, and Fas Ligand via the JNK/p300/p50 Signaling Pathway with Histone Acetylation. Front. Pharmacol. 2019, 10, 1174. [Google Scholar] [CrossRef]
- Lee, K.C.; Kuo, H.C.; Shen, C.H.; Lu, C.C.; Huang, W.S.; Hsieh, M.C.; Huang, C.Y.; Kuo, Y.H.; Hsieh, Y.Y.; Teng, C.C.; et al. A proteomics approach to identifying novel protein targets involved in erinacine A-mediated inhibition of colorectal cancer cells’ aggressiveness. J. Cell Mol. Med. 2017, 21, 588–599. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.S.; Kuo, Y.H.; Kuo, H.C.; Hsieh, M.C.; Huang, C.Y.; Lee, K.C.; Lee, K.F.; Shen, C.H.; Tung, S.Y.; Teng, C.C. CIL-102-Induced Cell Cycle Arrest and Apoptosis in Colorectal Cancer Cells via Upregulation of p21 and GADD45. PLoS ONE 2017, 12, e0168989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.C.; Huang, W.S.; Lee, K.F.; Lee, K.C.; Hsieh, M.C.; Huang, C.Y.; Lee, L.Y.; Lee, B.O.; Teng, C.C.; Shen, C.H.; et al. Inhibitory effect of Erinacines A on the growth of DLD-1 colorectal cancer cells is induced by generation of reactive oxygen species and activation of p70S6K and p21. J. Funct. Foods 2016, 21, 474–484. [Google Scholar] [CrossRef]
- Teng, C.C.; Kuo, H.C.; Sze, C.I. Quantitative proteomic analysis of the inhibitory effects of CIL-102 on viability and invasiveness in human glioma cells. Toxicol. Appl. Pharmacol. 2013, 272, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Tung, S.Y.; Lee, K.C.; Lee, K.F.; Yang, Y.L.; Huang, W.S.; Lee, L.Y.; Chen, W.P.; Chen, C.C.; Teng, C.C.; Shen, C.H.; et al. Apoptotic mechanisms of gastric cancer cells induced by isolated erinacine S through epigenetic histone H3 methylation of FasL and TRAIL. Food Funct. 2021, 12, 3455–3468. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.C.; Tung, S.Y.; Lee, K.C.; Lee, K.F.; Huang, W.S.; Shen, C.H.; Hsieh, M.C.; Huang, C.Y.; Sheen, J.M.; Kuo, H.C. Novel regulator role of CIL-102 in the epigenetic modification of TNFR1/TRAIL to induce cell apoptosis in human gastric cancer. Food Chem. Toxicol. 2021, 147, 111856. [Google Scholar] [CrossRef]
- Pan, P.J.; Wang, J.C.; Tsai, C.C.; Kuo, H.C. Identification of early response to hypertonic dextrose prolotherapy markers in knee osteoarthritis patients by an inflammation-related cytokine array. J. Chin. Med. Assoc. 2022, 85, 525–531. [Google Scholar] [CrossRef]
- Deng, R.; Hao, J.; Han, W.; Ni, Y.; Huang, X.; Hu, Q. Gelsolin regulates proliferation, apoptosis, migration and invasion in human oral carcinoma cells. Oncol. Lett. 2015, 9, 2129–2134. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zheng, J.; Yan, Y.; Liu, J.; Zhou, Y. Gelsolin Can Be a Prognostic Biomarker and Correlated with Immune Infiltrates in Gastric Cancer. Int. J. Gen. Med. 2022, 15, 927–936. [Google Scholar] [CrossRef]
- Elko, E.A.; Manuel, A.M.; White, S.; Zito, E.; van der Vliet, A.; Anathy, V.; Janssen-Heininger, Y.M.W. Oxidation of peroxiredoxin-4 induces oligomerization and promotes interaction with proteins governing protein folding and endoplasmic reticulum stress. J. Biol. Chem. 2021, 296, 100665. [Google Scholar] [CrossRef]
- Jia, W.; Chen, P.; Cheng, Y. PRDX4 and Its Roles in Various Cancers. Technol. Cancer Res. Treat. 2019, 18, 1533033819864313. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.; Lin, C.; Zhong, L.L.; Zhao, L.; Zhang, G.; Lu, A.; Wu, J.; Bian, Z. Targeting histone methylation for colorectal cancer. Therap. Adv. Gastroenterol. 2017, 10, 114–131. [Google Scholar] [CrossRef] [Green Version]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Chen, Z.; Sasaki, K.; Ishihara, S. ASO Author Reflections: Obturator Lymph Node Metastasis of Low Rectal Cancer. Ann. Surg. Oncol. 2022, 29, 4220–4221. [Google Scholar] [CrossRef]
- Cabello, C.M.; Bair, W.B., 3rd; Wondrak, G.T. Experimental therapeutics: Targeting the redox Achilles heel of cancer. Curr. Opin. Investig. Drugs 2007, 8, 1022–1037. [Google Scholar]
- Basak, D.; Uddin, M.N.; Hancock, J. The Role of Oxidative Stress and Its Counteractive Utility in Colorectal Cancer (CRC). Cancers 2020, 12, 3336. [Google Scholar] [CrossRef]
- Kim, J.C.; Ha, Y.J.; Tak, K.H.; Roh, S.A.; Kwon, Y.H.; Kim, C.W.; Yoon, Y.S.; Lee, J.L.; Park, Y.; Kim, S.K.; et al. Opposite functions of GSN and OAS2 on colorectal cancer metastasis, mediating perineural and lymphovascular invasion, respectively. PLoS ONE 2018, 13, e0202856. [Google Scholar] [CrossRef] [Green Version]
- Almazi, J.G.; Pockney, P.; Gedye, C.; Smith, N.D.; Hondermarck, H.; Verrills, N.M.; Dun, M.D. Cell-Free DNA Blood Collection Tubes Are Appropriate for Clinical Proteomics: A Demonstration in Colorectal Cancer. Proteom. Clin. Appl. 2018, 12, e1700121. [Google Scholar] [CrossRef] [Green Version]
- Palande, K.K.; Beekman, R.; van der Meeren, L.E.; Beverloo, H.B.; Valk, P.J.; Touw, I.P. The antioxidant protein peroxiredoxin 4 is epigenetically down regulated in acute promyelocytic leukemia. PLoS ONE 2011, 6, e16340. [Google Scholar] [CrossRef] [Green Version]
- Yi, N.; Xiao, M.B.; Ni, W.K.; Jiang, F.; Lu, C.H.; Ni, R.Z. High expression of peroxiredoxin 4 affects the survival time of colorectal cancer patients, but is not an independent unfavorable prognostic factor. Mol. Clin. Oncol. 2014, 2, 767–772. [Google Scholar] [CrossRef] [Green Version]
- Sreekumar, R.; Emaduddin, M.; Al-Saihati, H.; Moutasim, K.; Chan, J.; Spampinato, M.; Bhome, R.; Yuen, H.M.; Mescoli, C.; Vitale, A.; et al. Protein kinase C inhibitors override ZEB1-induced chemoresistance in HCC. Cell Death Dis. 2019, 10, 703. [Google Scholar] [CrossRef]
- Ungefroren, H.; Witte, D.; Lehnert, H. The role of small GTPases of the Rho/Rac family in TGF-beta-induced EMT and cell motility in cancer. Dev. Dyn. 2018, 247, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225.e12. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ren, B.; Yang, J.; Wang, H.; Yang, G.; Xu, R.; You, L.; Zhao, Y. The role of histone methylation in the development of digestive cancers: A potential direction for cancer management. Signal Transduct. Target. Ther. 2020, 5, 143. [Google Scholar] [CrossRef]




| Stage I (Non-Metastasis; n = 8) | Stage IV (Metastasis; n = 8) | |
|---|---|---|
| Age (years) | 64 | 64 |
| Sex | ||
| Male | 3 | 5 |
| Female | 5 | 3 |
| Histologic grade | ||
| Well | 5 | 0 |
| Moderate | 3 | 6 |
| Poor | 0 | 2 |
| Primary site | ||
| Proximal colon | 1 | 4 |
| Distal colon | 3 | 2 |
| Rectum | 4 | 2 |
| Primary tumor | ||
| T1 | 3 | 0 |
| T2 | 5 | 1 |
| T3 | - | 4 |
| T4 | - | 3 |
| Lymph node metastasis | ||
| nil | 8 | 0 |
| N1(1–3) | 0 | 2 |
| N2 (>4) | 0 | 6 |
| Spot | Protein Name | Mr/PI | Accession No | MASCOT Score | Matched Peptides |
|---|---|---|---|---|---|
| 1 | Tropomyosin beta chain | 32/4.5 | TPM2_HUMAN | 304 | 15 |
| 2 | Translationally-controlled | 19/4.6 | TCTP_HUMAN | 125 | 6 |
| 3 | Proteasome subunit alpha type-5 | 26/4.5 | PSA5_HUMAN | 71 | 71 |
| 4 | Glucosidase 2 subunit beta | 60/4.1 | GLU2B_HUMAN | 75 | 3 |
| 5 | Transcription activator BRG1 | 85/8.4 | SMCA4_HUMAN | 42 | 4 |
| 6 | E3 ubiquitin-protein ligase HERC2 | 53/5.8 | HERC2_HUMAN | 41 | 4 |
| 7 | Tubulin beta-4B chain | 50/4.6 | TBB4B_HUMAN | 472 | 18 |
| 8 | N-alpha-acetyltransferase 35, NatC auxiliary subunit | 84/6.6 | NAA35_HUMAN | 51 | 4 |
| 9 | 14-3-3 protein zeta/delta | 27/4.5 | 1433Z_HUMAN | 268 | 14 |
| 10 | Apolipoprotein A-I | 30/5.4 | APOA1_HUMAN | 350 | 18 |
| 11 | Rho GDP-dissociation inhibitor 1 | 23/4.8 | GDIR1_HUMAN | 199 | 9 |
| 12 | ATP synthase subunit d | 18/5.0 | ATP5H_HUMAN | 72 | 4 |
| 13 | Elongation factor Tu | 49/7.9 | EFTU_HUMAN | 323 | 15 |
| 14 | Selenium-binding protein 1 | 52/5.9 | SBP1_HUMAN | 149 | 5 |
| 15 | Gelsolin | 86/5.8 | GSN_HUMAN | 330 | 17 |
| 16 | Peroxiredoxin-4 | 30/5.8 | PRDX4_HUMAN | 65 | 4 |
| 17 | Proteasome activator complex subunit 2 | 27/5.4 | PSME2_HUMAN | 186 | 8 |
| 18 | Heterogeneous nuclear ribonucleoprotein F | 45/5.2 | HNRPF_HUMAN | 95 | 4 |
| No. | Protein | Staining (Score) | Node-Negative CRC (n) | Node-Positive CRC (n) |
|---|---|---|---|---|
| 1 | Peroxiredoxin-4 | None (0) | 0 (0.0%) | 3 (13.6%) |
| Low (1–3) | 12 (85.7%) | 4 (18.2%) | ||
| High (4–6) | 2 (14.3%) | 15 (68.2%) | ||
| Total (n) | 14 (100%) | 22 (100%) | ||
| p | 0.04 | |||
| 2 | Gelsolin | None (0) | 0 (0.0%) | 1 (4.5%) |
| Low (1–3) | 13 (92.9%) | 12 (54.5%) | ||
| High (4–6) | 1 (7.1%) | 9 (40.9%) | ||
| Total (n) | 14 (100%) | 22 (100%) | ||
| p | 0.02 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-Y.; Lee, K.-C.; Tung, S.-Y.; Huang, W.-S.; Teng, C.-C.; Lee, K.-F.; Hsieh, M.-C.; Kuo, H.-C. 2D-DIGE-MS Proteomics Approaches for Identification of Gelsolin and Peroxiredoxin 4 with Lymph Node Metastasis in Colorectal Cancer. Cancers 2022, 14, 3189. https://doi.org/10.3390/cancers14133189
Huang C-Y, Lee K-C, Tung S-Y, Huang W-S, Teng C-C, Lee K-F, Hsieh M-C, Kuo H-C. 2D-DIGE-MS Proteomics Approaches for Identification of Gelsolin and Peroxiredoxin 4 with Lymph Node Metastasis in Colorectal Cancer. Cancers. 2022; 14(13):3189. https://doi.org/10.3390/cancers14133189
Chicago/Turabian StyleHuang, Cheng-Yi, Ko-Chao Lee, Shui-Yi Tung, Wen-Shin Huang, Chih-Chuan Teng, Kam-Fai Lee, Meng-Chiao Hsieh, and Hsing-Chun Kuo. 2022. "2D-DIGE-MS Proteomics Approaches for Identification of Gelsolin and Peroxiredoxin 4 with Lymph Node Metastasis in Colorectal Cancer" Cancers 14, no. 13: 3189. https://doi.org/10.3390/cancers14133189





