Evaluation Criteria for Chromosome Instability Detection by FISH to Predict Malignant Progression in Premalignant Glottic Laryngeal Lesions
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Data and Tissue Material
2.2. Histopathological Assessment
2.3. Detection of Chromosomes 1 and 7 Alterations by FISH Analysis
2.4. Evaluation of FISH Results
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Defining CI in FFPE Tissue Sections and Application to Premalignant Laryngeal Lesions
3.3. Logistic Regression Analysis for Histopathology, CR-FISH, and PAN-FISH
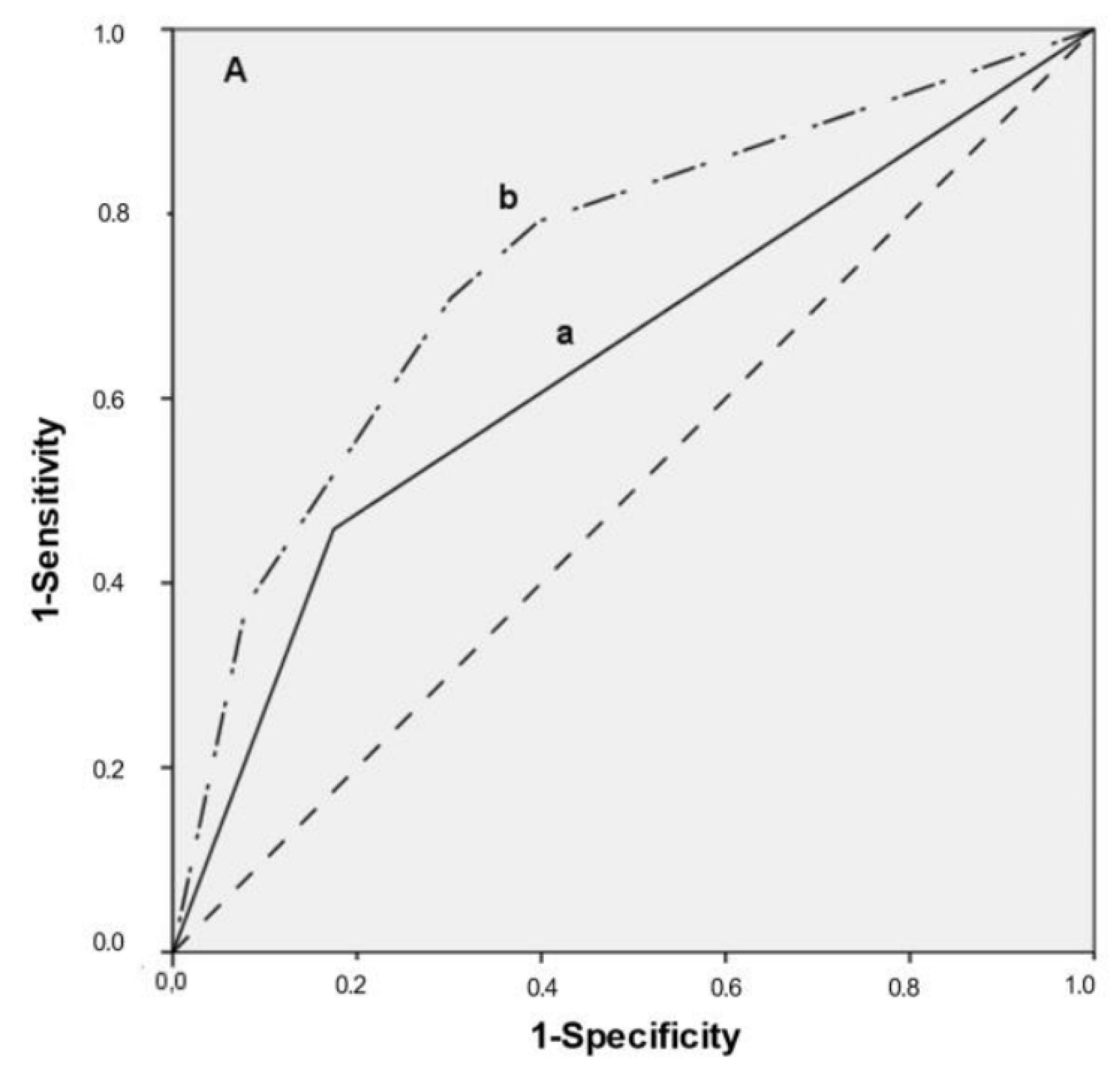
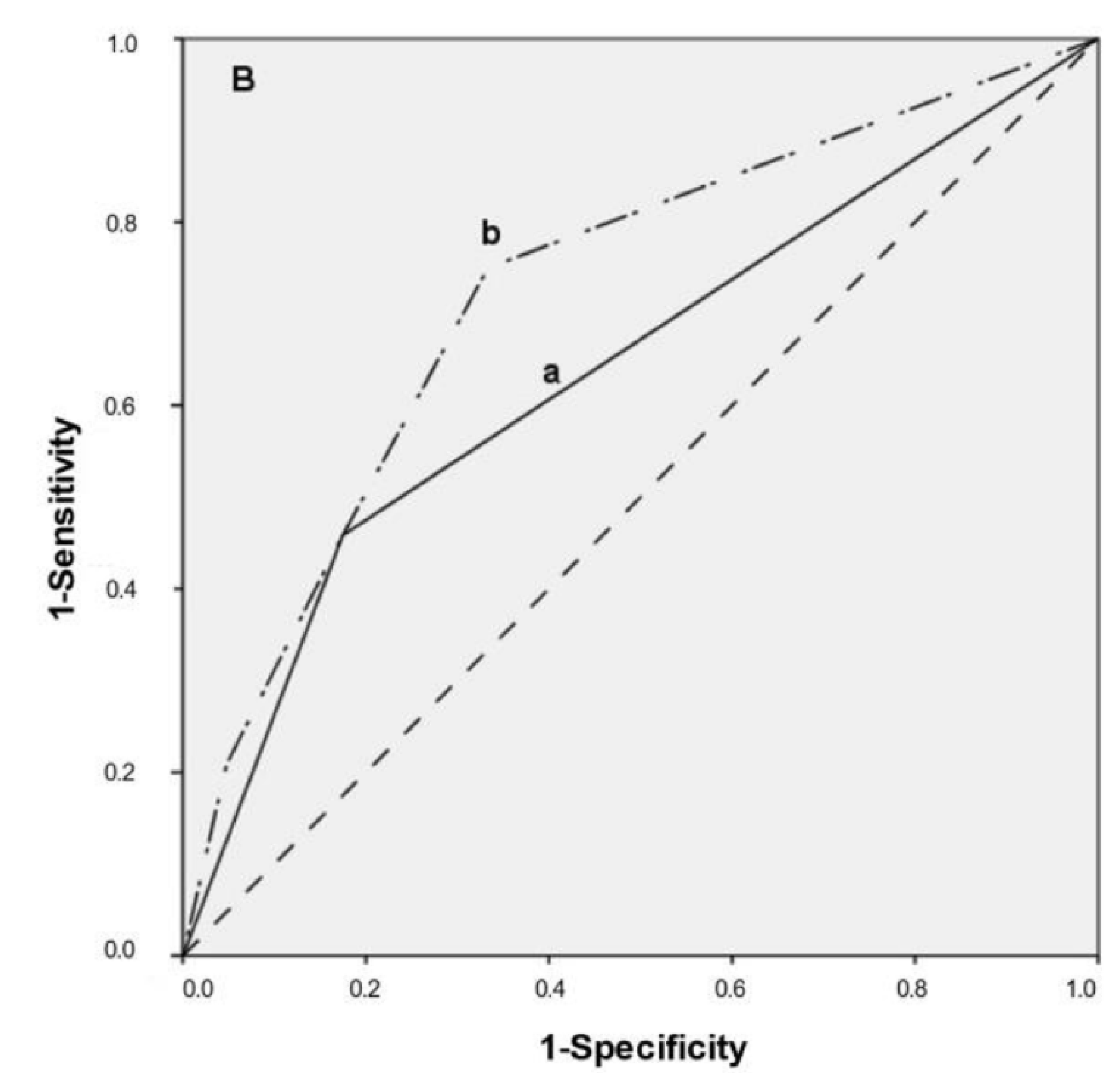
3.4. Multivariate Predictive Model, ROC Curves
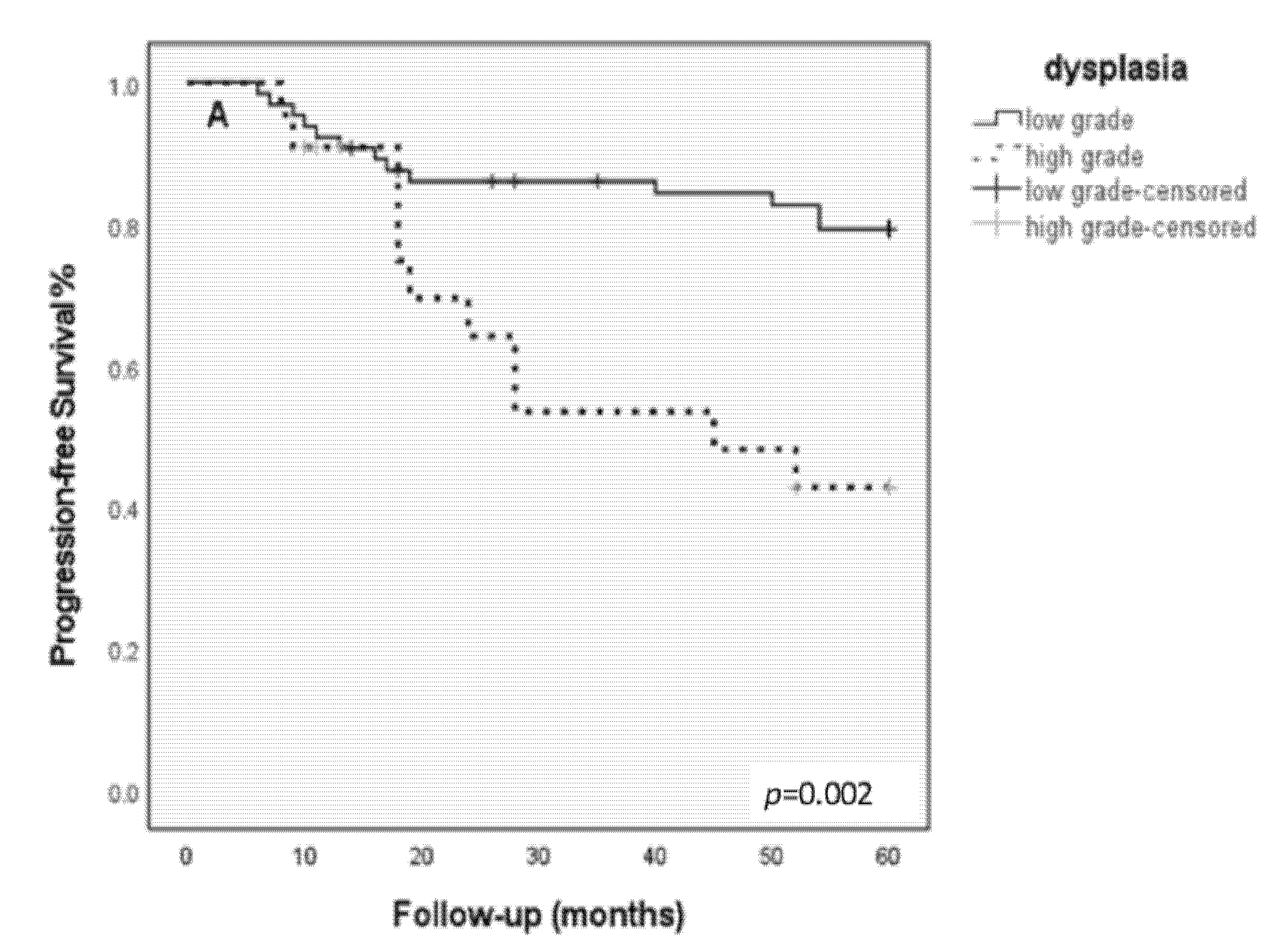
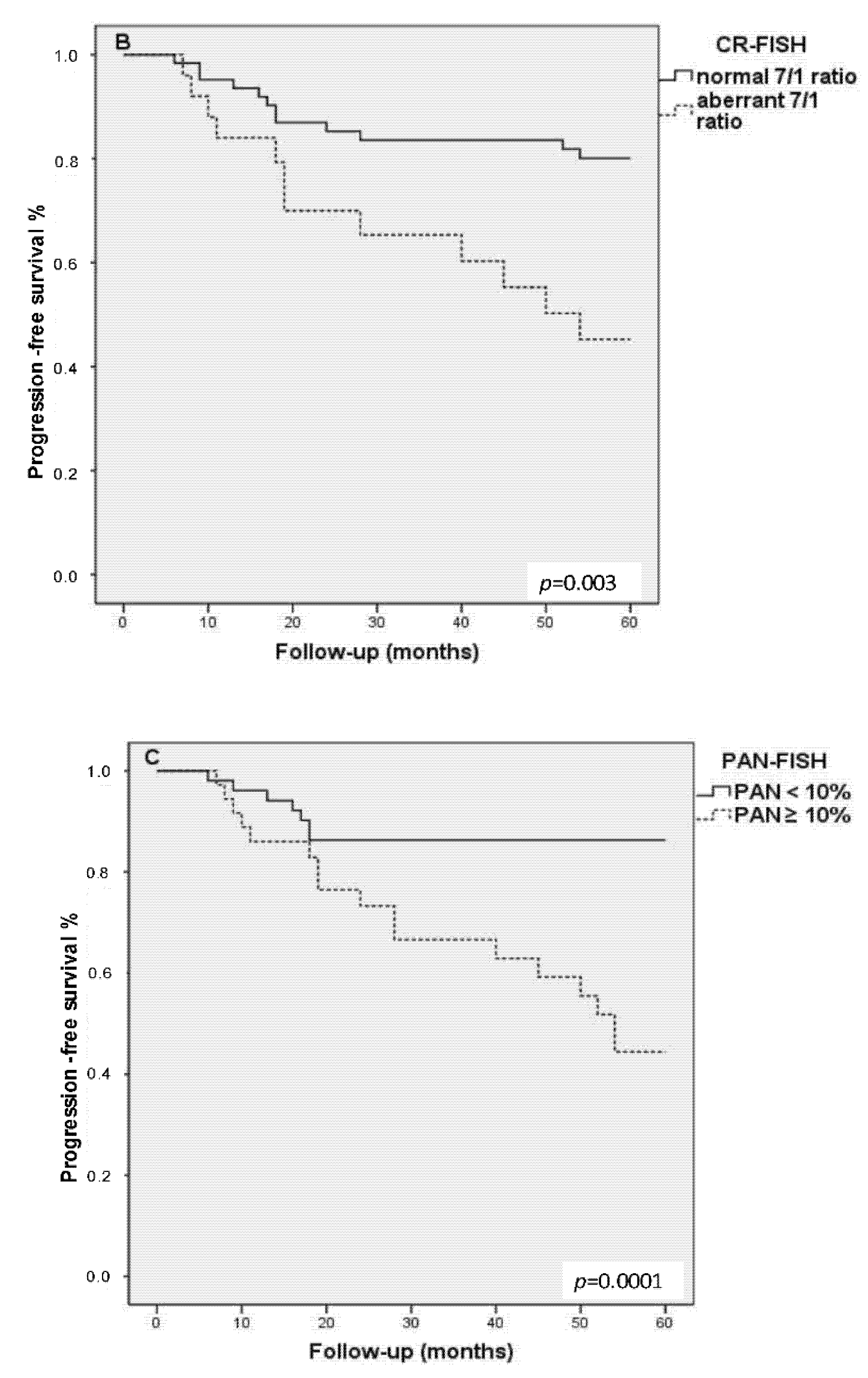
3.5. Kaplan–Meier Progression-Free Survival Analysis
4. Discussion
4.1. PAN-FISH Is More Favorable Than CR-FISH for CI Detection
4.2. FISH Not Always Correctly Predicts Outcome
4.3. The Role of Histopathological Diagnosis and the Combination with PAN-FISH
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hashibe, M.; Brennan, P.; Chuang, S.C.; Boccia, S.; Castellsague, X.; Chen, C.; Curado, M.P.; Dal Maso, L.; Daudt, A.W.; Fabianova, E.; et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol. Biomark. Prev. 2009, 18, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, J.H.; Grandis, J.R.; Ferris, R.L. HPV-Associated Head and Neck Cancer: Unique Features of Epidemiology and Clinical Management. Annu. Rev. Med. 2016, 67, 91–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafkamp, H.C.; Mooren, J.J.; Claessen, S.M.; Klingenberg, B.; Voogd, A.C.; Bot, F.J.; Klussmann, J.P.; Hopman, A.H.; Manni, J.J.; Kremer, B.; et al. P21 Cip1/WAF1 expression is strongly associated with HPV-positive tonsillar carcinoma and a favorable prognosis. Mod. Pathol. 2009, 22, 686–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, K.D.; Parkinson, E.K.; Harrison, P.R. Profiling early head and neck cancer. Nat. Rev. 2005, 5, 127–135. [Google Scholar] [CrossRef]
- Pezzuto, F.; Buonaguro, L.; Caponigro, F.; Ionna, F.; Starita, N.; Annunziata, C.; Buonaguro, F.M.; Tornesello, M.L. Update on Head and Neck Cancer: Current Knowledge on Epidemiology, Risk Factors, Molecular Features and Novel Therapies. Oncology 2015, 89, 125–136. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. Reply. N. Engl. J. Med. 2020, 382, e57. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef]
- Bergshoeff, V.E.; Hopman, A.H.; Zwijnenberg, I.R.; Ramaekers, F.C.; Bot, F.J.; Kremer, B.; Manni, J.J.; Speel, E.J. Chromosome instability in resection margins predicts recurrence of oral squamous cell carcinoma. J. Pathol. 2008, 215, 347–348. [Google Scholar] [CrossRef]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef] [Green Version]
- Mao, L.; Hong, W.K.; Papadimitrakopoulou, V.A. Focus on head and neck cancer. Cancer Cell 2004, 5, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Fleskens, S.; Slootweg, P. Grading systems in head and neck dysplasia: Their prognostic value, weaknesses and utility. Head Neck Oncol. 2009, 1, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleskens, S.A.; Bergshoeff, V.E.; Voogd, A.C.; van Velthuysen, M.L.; Bot, F.J.; Speel, E.J.; Kremer, B.; Takes, R.; Slootweg, P. Interobserver variability of laryngeal mucosal premalignant lesions: A histopathological evaluation. Mod. Pathol. 2011, 24, 892–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gale, N.; Poljak, M.; Zidar, N. Update from the 4th Edition of the World Health Organization Classification of Head and Neck Tumours: What is New in the 2017 WHO Blue Book for Tumours of the Hypopharynx, Larynx, Trachea and Parapharyngeal Space. Head Neck Pathol. 2017, 11, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Garnis, C.; Chari, R.; Buys, T.P.; Zhang, L.; Ng, R.T.; Rosin, M.P.; Lam, W.L. Genomic imbalances in precancerous tissues signal oral cancer risk. Mol. Cancer 2009, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Ai, H.; Barrera, J.E.; Meyers, A.D.; Shroyer, K.R.; Varella-Garcia, M. Chromosomal aneuploidy precedes morphological changes and supports multifocality in head and neck lesions. Laryngoscope 2001, 111, 1853–1858. [Google Scholar] [CrossRef]
- Veltman, J.A.; Bot, F.J.; Huynen, F.C.; Ramaekers, F.C.; Manni, J.J.; Hopman, A.H. Chromosome instability as an indicator of malignant progression in laryngeal mucosa. J. Clin. Oncol. 2000, 18, 1644–1651. [Google Scholar] [CrossRef] [Green Version]
- Veltman, J.A.; van Weert, I.; Aubele, M.; Bot, F.J.; Ramaekers, F.C.; Manni, J.J.; Hopman, A.H. Specific steps in aneuploidization correlate with loss of heterozygosity of 9p21, 17p13 and 18q21 in the progression of pre-malignant laryngeal lesions. Int. J. Cancer 2001, 91, 193–199. [Google Scholar] [CrossRef]
- Bergshoeff, V.E.; Van der Heijden, S.J.; Haesevoets, A.; Litjens, S.G.; Bot, F.J.; Voogd, A.C.; Chenault, M.N.; Hopman, A.H.; Schuuring, E.; Van der Wal, J.M.; et al. Chromosome instability predicts progression of premalignant lesions of the larynx. Pathology 2014, 46, 216–224. [Google Scholar] [CrossRef]
- Siebers, T.J.; Bergshoeff, V.E.; Otte-Holler, I.; Kremer, B.; Speel, E.J.; van der Laak, J.A.; Merkx, M.A.; Slootweg, P.J. Chromosome instability predicts the progression of premalignant oral lesions. Oral Oncol. 2013, 49, 1121–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soder, A.I.; Hopman, A.H.; Ramaekers, F.C.; Conradt, C.; Bosch, F.X. Distinct nonrandom patterns of chromosomal aberrations in the progression of squamous cell carcinomas of the head and neck. Cancer Res. 1995, 55, 5030–5037. [Google Scholar] [PubMed]
- Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Sethi, N.; MacLennan, K.; Wood, H.M.; Rabbitts, P. Past and future impact of next-generation sequencing in head and neck cancer. Head Neck 2016, 38, E2395–E2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Poh, C.F.; Williams, M.; Laronde, D.M.; Berean, K.; Gardner, P.J.; Jiang, H.; Wu, L.; Lee, J.J.; Rosin, M.P. Loss of heterozygosity (LOH) profiles--validated risk predictors for progression to oral cancer. Cancer Prev. Res. 2012, 5, 1081–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleskens, S.J.; Takes, R.P.; Otte-Holler, I.; van Doesburg, L.; Smeets, A.; Speel, E.J.; Slootweg, P.J.; van der Laak, J.A. Simultaneous assessment of DNA ploidy and biomarker expression in paraffin-embedded tissue sections. Histopathology 2010, 57, 14–26. [Google Scholar] [CrossRef]
- Abou-Elhamd, K.-E.; Habib, T.N. The flow cytometric analysis of premalignant and malignant lesions in head and neck squamous cell carcinoma. Oral Oncol. 2007, 43, 366–372. [Google Scholar] [CrossRef]
- Saintigny, P.; Zhang, L.; Fan, Y.H.; El-Naggar, A.K.; Papadimitrakopoulou, V.A.; Feng, L.; Lee, J.J.; Kim, E.S.; Ki Hong, W.; Mao, L. Gene expression profiling predicts the development of oral cancer. Cancer Prev. Res. 2011, 4, 218–229. [Google Scholar] [CrossRef] [Green Version]
- Herbergs, J.; Speel, E.J.; Ramaekers, F.C.; de Bruine, A.P.; Arends, J.W.; Hopman, A.H. Combination of lamin immunocytochemistry and in situ hybridization for the analysis of chromosome copy numbers in tumor cell areas with high nuclear density. Cytometry 1996, 23, 1–7. [Google Scholar] [CrossRef]
- Jurmeister, P.; Lenze, D.; Berg, E.; Mende, S.; Schaper, F.; Kellner, U.; Herbst, H.; Sers, C.; Budczies, J.; Dietel, M.; et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015, 87, 122–129. [Google Scholar] [CrossRef]
- Thunnissen, E.; Bubendorf, L.; Dietel, M.; Elmberger, G.; Kerr, K.; Lopez-Rios, F.; Moch, H.; Olszewski, W.; Pauwels, P.; Penault-Llorca, F.; et al. EML4-ALK testing in non-small cell carcinomas of the lung: A review with recommendations. Virchows Arch. 2012, 461, 245–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, H.; Allmanritter, J.; Doglioni, C.; Marx, A.; Muller, J.; Gattenlohner, S.; Staiger, A.M.; Rosenwald, A.; Ott, G.; Ott, M.M. Fluorescence in situ analysis of soft tissue tumor associated genetic alterations in formalin-fixed paraffin-embedded tissue. Pathol. Res. Pract. 2014, 210, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Horn, H.; Ziepert, M.; Becher, C.; Barth, T.F.; Bernd, H.W.; Feller, A.C.; Klapper, W.; Hummel, M.; Stein, H.; Hansmann, M.L.; et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 2013, 121, 2253–2263. [Google Scholar] [CrossRef] [Green Version]
- Bergethon, K.; Shaw, A.T.; Ou, S.H.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radonic, T.; Geurts-Giele, W.R.R.; Samsom, K.G.; Roemen, G.; von der Thusen, J.H.; Thunnissen, E.; Meijssen, I.C.; Sleddens, H.; Dinjens, W.N.M.; Boelens, M.C.; et al. RET Fluorescence In Situ Hybridization Analysis Is a Sensitive but Highly Unspecific Screening Method for RET Fusions in Lung Cancer. J. Thorac. Oncol. 2021, 16, 798–806. [Google Scholar] [CrossRef]
- Bubendorf, L.; Dafni, U.; Schobel, M.; Finn, S.P.; Tischler, V.; Sejda, A.; Marchetti, A.; Thunnissen, E.; Verbeken, E.K.; Warth, A.; et al. Prevalence and clinical association of MET gene overexpression and amplification in patients with NSCLC: Results from the European Thoracic Oncology Platform (ETOP) Lungscape project. Lung Cancer 2017, 111, 143–149. [Google Scholar] [CrossRef]
- Creytens, D.; van Gorp, J.; Ferdinande, L.; Speel, E.J.; Libbrecht, L. Detection of MDM2/CDK4 amplification in lipomatous soft tissue tumors from formalin-fixed, paraffin-embedded tissue: Comparison of multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridization (FISH). Appl. Immunohistochem. Mol. Morphol. 2015, 23, 126–133. [Google Scholar] [CrossRef]
- van den Bent, M.J.; Looijenga, L.H.; Langenberg, K.; Dinjens, W.; Graveland, W.; Uytdewilligen, L.; Sillevis Smitt, P.A.; Jenkins, R.B.; Kros, J.M. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer 2003, 97, 1276–1284. [Google Scholar] [CrossRef]
- Tsuta, K.; Kohno, T.; Yoshida, A.; Shimada, Y.; Asamura, H.; Furuta, K.; Kushima, R. RET-rearranged non-small-cell lung carcinoma: A clinicopathological and molecular analysis. Br. J. Cancer 2014, 110, 1571–1578. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerami, P.; Jewell, S.S.; Morrison, L.E.; Blondin, B.; Schulz, J.; Ruffalo, T.; Matushek, P.T.; Legator, M.; Jacobson, K.; Dalton, S.R.; et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am. J. Surg. Pathol. 2009, 33, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Huysentruyt, C.J.; Baldewijns, M.M.; Ruland, A.M.; Tonk, R.J.; Vervoort, P.S.; Smits, K.M.; van de Beek, C.; Speel, E.J. Modified UroVysion scoring criteria increase the urothelial carcinoma detection rate in cases of equivocal urinary cytology. Histopathology 2011, 58, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Coindre, J.M.; Pedeutour, F.; Aurias, A. Well-differentiated and dedifferentiated liposarcomas. Virchows Arch. 2010, 456, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Califano, J.; van der Riet, P.; Westra, W.; Nawroz, H.; Clayman, G.; Piantadosi, S.; Corio, R.; Lee, D.; Greenberg, B.; Koch, W.; et al. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996, 56, 2488–2492. [Google Scholar] [CrossRef]
- Hall, G.L.; Shaw, R.J.; Field, E.A.; Rogers, S.N.; Sutton, D.N.; Woolgar, J.A.; Lowe, D.; Liloglou, T.; Field, J.K.; Risk, J.M. p16 Promoter methylation is a potential predictor of malignant transformation in oral epithelial dysplasia. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2174–2179. [Google Scholar] [CrossRef] [Green Version]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nature Rev. 2010, 10, 878–889. [Google Scholar] [CrossRef] [Green Version]
- Pierssens, D.; Borgemeester, M.C.; van der Heijden, S.J.H.; Peutz-Kootstra, C.J.; Ruland, A.M.; Haesevoets, A.M.; Kessler, P.; Kremer, B.; Speel, E.M. Chromosome instability in tumor resection margins of primary OSCC is a predictor of local recurrence. Oral Oncol. 2017, 66, 14–21. [Google Scholar] [CrossRef]

| Tumor Entity | DNA Probe(s) for | Tissue | Evaluation Criterium | Cut-Off Value for Positivity | N = Nuclei to Be Scored | References |
|---|---|---|---|---|---|---|
| Lung cancer | ALK | FFPE * cyto | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Jurmeister et al. [30] Thunnissen et al. [31] |
| ROS1 | FFPE * cyto | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Jurmeister et al. [30] Bergethon et al. [34] | |
| RET | FFPE * cyto | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Tsuta et al. [39] Radonic et al. [35] | |
| MET | FFPE * cyto | amplification | MET/CEP7 ǁ- ratio ≥2 | n = 50 | Bubendorf et al. [36] | |
| Different types of lymphomas | Bcl2 | FFPE * | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Horn et al. [32,33] |
| Bcl6 | FFPE * | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Horn et al. [32,33] | |
| Myc | FFPE * | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Horn et al. [32,33] | |
| Breast cancer | ERBB2/CEP17 | FFPE * cyto | amplification/CNG ‡ | ERBB2/CEP17 ǁ Copy Ratio ≥2.0 | n = 20 | ASCO/CAP guidelines 2018 [40,41] |
| Melanoma | 11q13/ 6q23/6p25/ CEP6 ǁ | FFPE * | loss/ CNG‡ | PAN † ≥ (29–55%)/ multiparameter model/ CR ¶ | n = 30 | Gerami et al. [42] |
| Bladder cancer | Urovysion (chrom. 3,7,9p21,17) | Urine cytology | CNV § | CNG ‡ for chromosomes 3,7,17 (≥4); 9p21 loss in ≥12 | n = 25 | Huysentruyt et al. [43] |
| Liposarcoma | MDM/CEP12 | FFPE * | amplification | MDM2/CEP12 ǁ- ratio ≥2; | n = 20 | Coindre et al., Creytens et al. [37,44] |
| Different sarcoma types | DDIT3, EWSR1, FOXO1, FUS SS18, MDM2 | FFPE * | rearrangement | PAN † ≥ 50% PAN † ≥ 15% | n = 50 n = 100 | Modified from Horn et al. [32] |
| Oligodendroglioma | 1p/19q | FFPE * | Loss/ CNG ‡ | 1p/1q and 19q/19p ratio ≤0.8 | n = 20 | Modified from Van den Bent et al. [38] |
| Patient Population | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Histopathology | CR | PAN | |||||||
| Low-Grade | High-Grade | p-Value | Normal | Aberrant | p-Value | <10% | ≥10% | p-Value | |
| Gender | p = NS | p = NS | p = NS | ||||||
| Male (n = 53) | 39 | 14 | 36 | 17 | 30 | 23 | |||
| Female (n = 34) | 26 | 8 | 25 | 9 | 24 | 10 | |||
| Age (mean, (SD)) | 57.7 (11.8) | 60.7 (11.9) | p = NS | 57.6 (11,18) | 61.35 (12.96) | p = NS | 57.8 (12.4) | 59.9 (10.8) | p = NS |
| Histopathology | p = NS | p = 0.02 * | |||||||
| Low-grade dysplasia (n = 65) | 48 (73%) | 17 (27%) | 43 (65%) | 22 (35%) | |||||
| High-grade dysplasia (n = 22) | 14 (58%) | 8 (42%) | 8 (33%) | 14 (67%) | |||||
| 5-year disease free survival | p = 0.002 | p = 0.003 | p = 0.0001 | ||||||
| Parameter | OR (95% Confidence Interval) | p-Value |
|---|---|---|
| Univariate | ||
| Histopathology | 4.0 (1.4–11.2) | 0.009 * |
| (low-grade vs. high-grade) | ||
| CR-FISH | 3.8 (1.4–10.5) | 0.009 * |
| PAN-FISH 0%) | 5.6 (2.0–15.8) | 0.001 * |
| Model I | ||
| Histopathology (low-grade vs. high-grade) | 3.9 (1.3–11.6) | 0.014 * |
| CR-FISH | 3.7 (1.3–10.8) | 0.014 * |
| Model II | ||
| Histopathology (low-grade vs. high-grade) | 2.9 (1.0–8.8) | 0.058 |
| PAN-FISH | 4.6 (1.6–13.4) | 0.005 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergshoeff, V.E.; Balkenhol, M.C.A.; Haesevoets, A.; Ruland, A.; Chenault, M.N.; Nelissen, R.C.; Peutz, C.J.; Clarijs, R.; Van der Laak, J.A.W.M.; Takes, R.P.; et al. Evaluation Criteria for Chromosome Instability Detection by FISH to Predict Malignant Progression in Premalignant Glottic Laryngeal Lesions. Cancers 2022, 14, 3260. https://doi.org/10.3390/cancers14133260
Bergshoeff VE, Balkenhol MCA, Haesevoets A, Ruland A, Chenault MN, Nelissen RC, Peutz CJ, Clarijs R, Van der Laak JAWM, Takes RP, et al. Evaluation Criteria for Chromosome Instability Detection by FISH to Predict Malignant Progression in Premalignant Glottic Laryngeal Lesions. Cancers. 2022; 14(13):3260. https://doi.org/10.3390/cancers14133260
Chicago/Turabian StyleBergshoeff, Verona E., Maschenka C. A. Balkenhol, Annick Haesevoets, Andrea Ruland, Michelene N. Chenault, Rik C. Nelissen, Carine J. Peutz, Ruud Clarijs, Jeroen A. W. M. Van der Laak, Robert P. Takes, and et al. 2022. "Evaluation Criteria for Chromosome Instability Detection by FISH to Predict Malignant Progression in Premalignant Glottic Laryngeal Lesions" Cancers 14, no. 13: 3260. https://doi.org/10.3390/cancers14133260






