Urine Cellular DNA Point Mutation and Methylation for Identifying Upper Tract Urinary Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
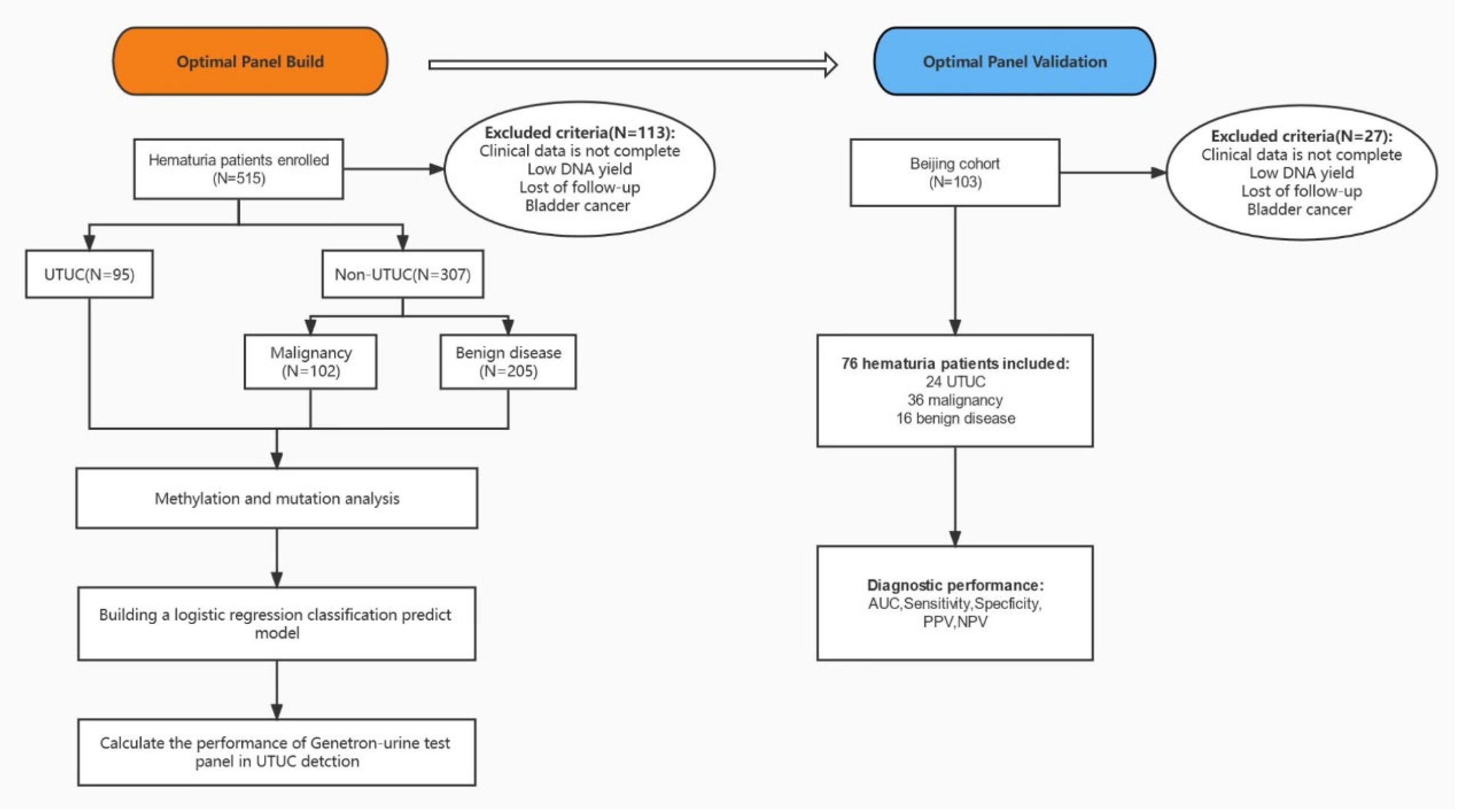
2.1. Patients and Samples
2.2. Sample Collection and Mutated Gene Detection
2.3. Methylation Analysis
2.4. Statistical Analysis and Logistic Regression Model
3. Results
3.1. The Baseline Characteristics of Included Patients
3.2. Univariate Logistic Regression of Significant Features
3.3. Multivariate Logistic Regression of Significant Features
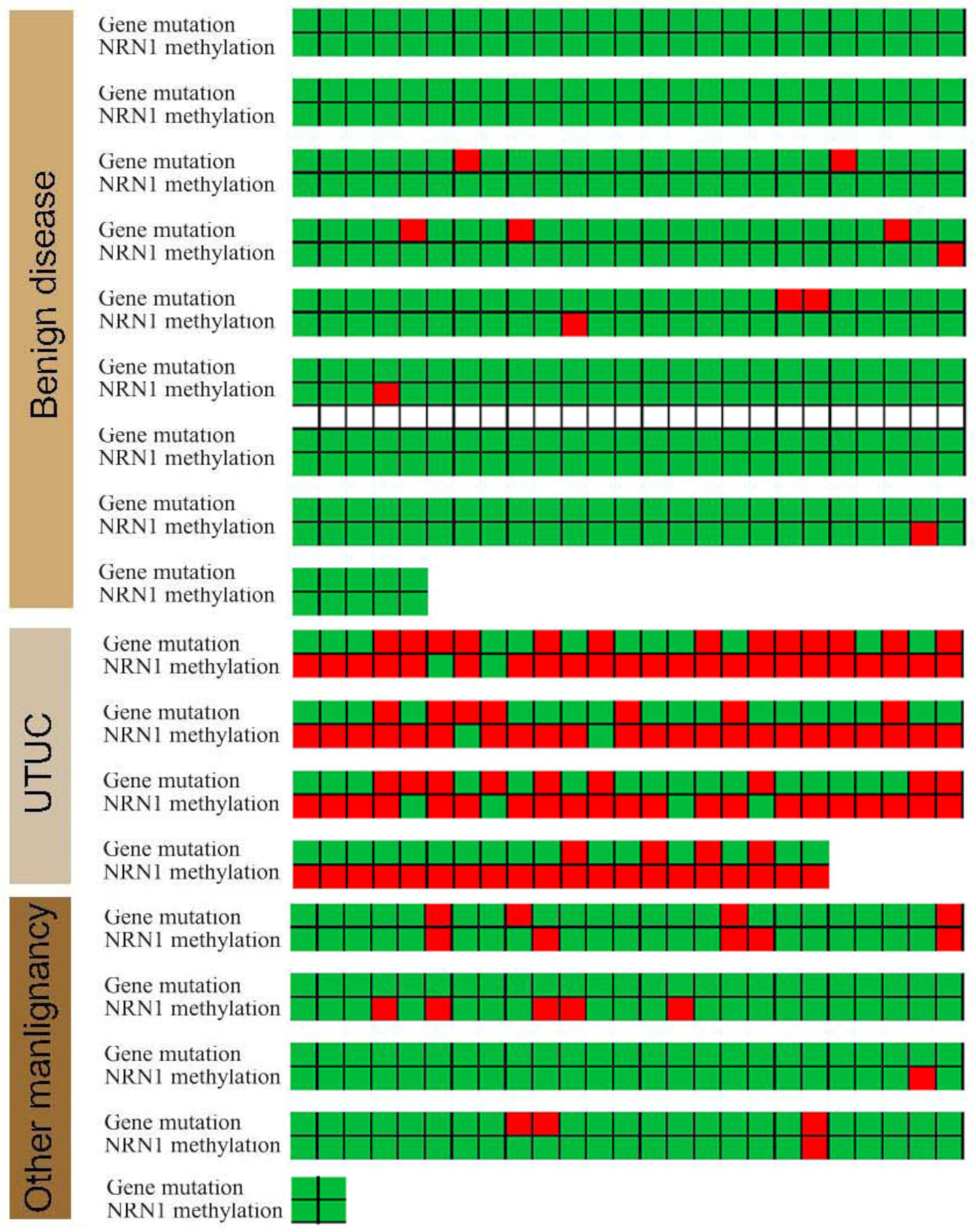
3.4. Gene Mutations and NRN1 Methylation Provided New Clinical Potential Applications
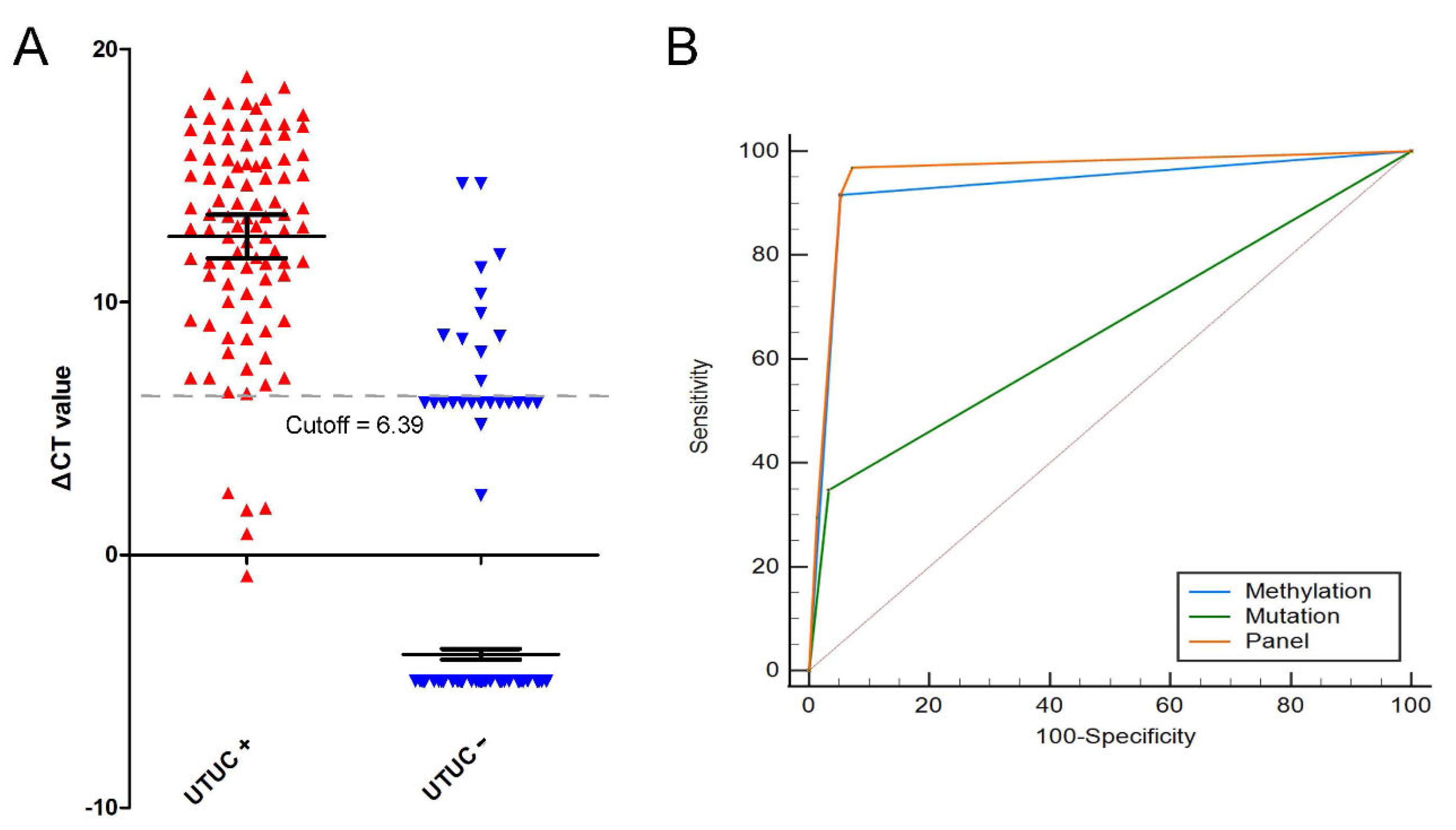
3.5. Gene Mutations and NRN1 Methylation as a Diagnostic Tool to Identify UTUC and Benign Disease
3.6. Gene Mutations and NRN1 Methylation as a Diagnostic Tool to Identify UTUC and Other Malignant Tumors
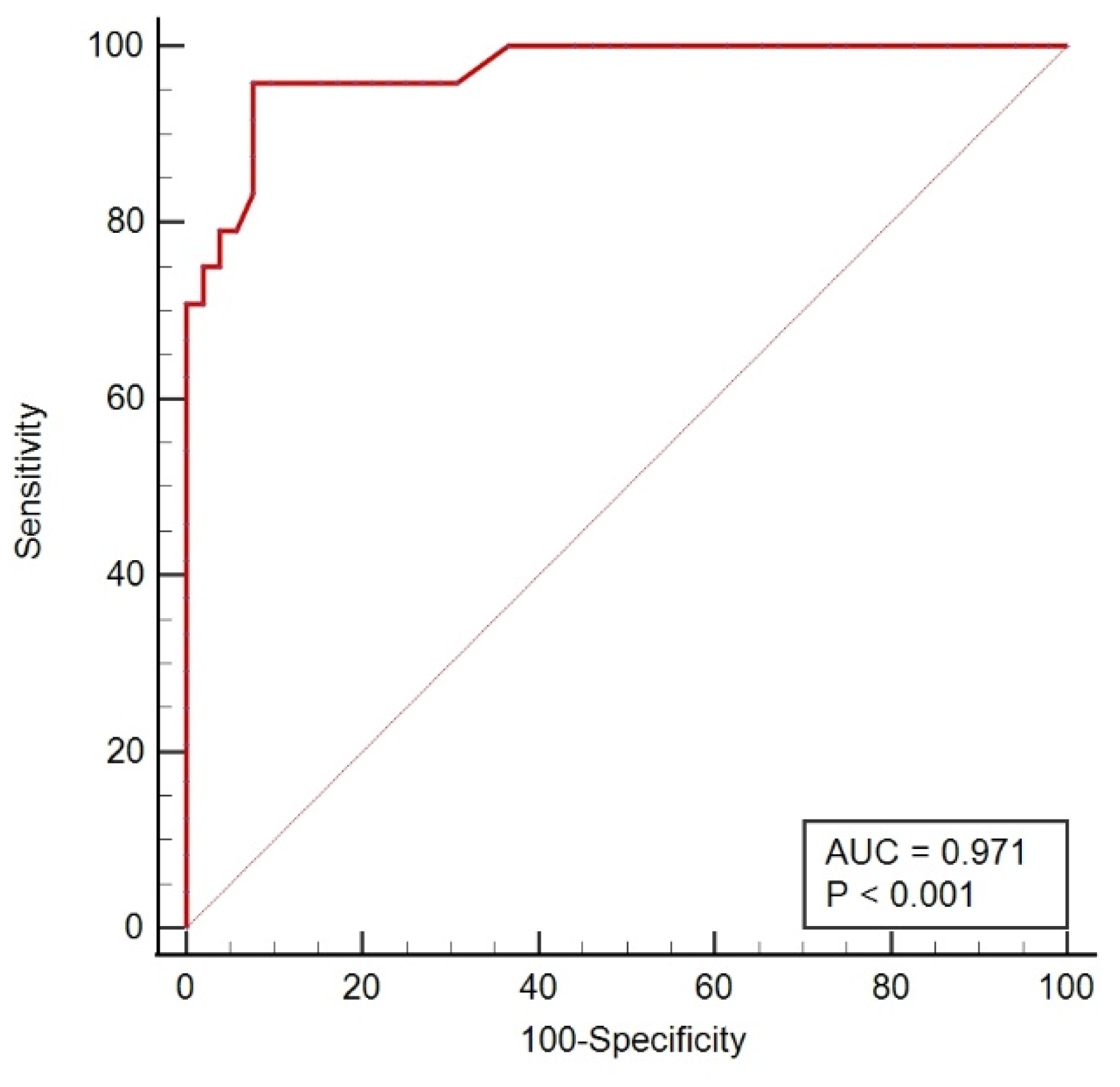
3.7. Panel Optimization
3.8. Novel Diagnostic Model, Cytology and FISH Comparison
3.9. The Performance of the Detector in an Independent Validation Data Set
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Roupret, M.; Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Comperat, E.M.; Cowan, N.C.; Dominguez-Escrig, J.L.; Gontero, P.; Hugh Mostafid, A.; et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2020 Update. Eur. Urol. 2021, 79, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Janisch, F.; Shariat, S.F.; Baltzer, P.; Fajkovic, H.; Kimura, S.; Iwata, T.; Korn, P.; Yang, L.; Glybochko, P.V.; Rink, M.; et al. Diagnostic performance of multidetector computed tomographic (MDCTU) in upper tract urothelial carcinoma (UTUC): A systematic review and meta-analysis. World J. Urol. 2020, 38, 1165–1175. [Google Scholar] [CrossRef]
- Rojas, C.P.; Castle, S.M.; Llanos, C.A.; Santos Cortes, J.A.; Bird, V.; Rodriguez, S.; Reis, I.M.; Zhao, W.; Gomez-Fernandez, C.; Leveillee, R.J.; et al. Low biopsy volume in ureteroscopy does not affect tumor biopsy grading in upper tract urothelial carcinoma. Urol. Oncol. 2013, 31, 1696–1700. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.Q.; Hong, P.; Xiong, G.Y.; Zhang, L.; Fang, D.; Li, X.S.; Zhang, K.; Zhou, L.Q. Impact of ureteroscopy before radical nephroureterectomy for upper tract urothelial carcinomas on oncological outcomes: A meta-analysis. BJU Int. 2018, 121, 184–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchioni, M.; Primiceri, G.; Cindolo, L.; Hampton, L.J.; Grob, M.B.; Guruli, G.; Schips, L.; Shariat, S.F.; Autorino, R. Impact of diagnostic ureteroscopy on intravesical recurrence in patients undergoing radical nephroureterectomy for upper tract urothelial cancer: A systematic review and meta-analysis. BJU Int. 2017, 120, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Malm, C.; Grahn, A.; Jaremko, G.; Tribukait, B.; Brehmer, M. Diagnostic accuracy of upper tract urothelial carcinoma: How samples are collected matters. Scand. J. Urol. 2017, 51, 137–145. [Google Scholar] [CrossRef]
- Audenet, F.; Isharwal, S.; Cha, E.K.; Donoghue, M.T.A.; Drill, E.N.; Ostrovnaya, I.; Pietzak, E.J.; Sfakianos, J.P.; Bagrodia, A.; Murugan, P.; et al. Clonal Relatedness and Mutational Differences between Upper Tract and Bladder Urothelial Carcinoma. Clin. Cancer Res. 2019, 25, 967–976. [Google Scholar] [CrossRef] [Green Version]
- Schmitz-Drager, B.J.; Droller, M.; Lokeshwar, V.B.; Lotan, Y.; Hudson, M.A.; van Rhijn, B.W.; Marberger, M.J.; Fradet, Y.; Hemstreet, G.P.; Malmstrom, P.U.; et al. Molecular markers for bladder cancer screening, early diagnosis, and surveillance: The WHO/ICUD consensus. Urol. Int. 2015, 94, 1–24. [Google Scholar] [CrossRef]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 2015, 51, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Ma, X.; Ai, X.; Gao, J.; Liang, Y.; Zhang, Q.; Ma, T.; Mao, K.; Zheng, Q.; Wang, S.; et al. A Urine-Based Liquid Biopsy Method for Detection of Upper Tract Urinary Carcinoma. Front. Oncol. 2020, 10, 597486. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Sato, Y.; Suzuki, H.; Kakiuchi, N.; Yoshizato, T.; Lenis, A.T.; Maekawa, S.; Yokoyama, A.; Takeuchi, Y.; Inoue, Y.; et al. Molecular classification and diagnostics of upper urinary tract urothelial carcinoma. Cancer Cell 2021, 39, 793–809.e798. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021, 37, 1012–1027. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Li, P.; Ning, J.; Luo, X.; Du, H.; Zhang, Q.; Zhou, G.; Du, Q.; Ou, Z.; Wang, L.; Wang, Y. New method to preserve the original proportion and integrity of urinary cell-free DNA. J. Clin. Lab. Anal. 2019, 33, e22668. [Google Scholar] [CrossRef]
- Zhao, C.; Pan, Y.; Wang, Y.; Li, Y.; Han, W.; Lu, L.; Tang, W.; Li, P.; Ou, Z.; Zhang, M.; et al. A novel cell-free single-molecule unique primer extension resequencing (cf-SUPER) technology for bladder cancer non-invasive detection in urine. Transl. Androl. Urol. 2020, 9, 1222–1231. [Google Scholar] [CrossRef]
- Ou, Z.; Li, K.; Yang, T.; Dai, Y.; Chandra, M.; Ning, J.; Wang, Y.; Xu, R.; Gao, T.; Xie, Y.; et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin. Transl. Med. 2020, 9, 4. [Google Scholar] [CrossRef] [Green Version]
- Guyatt, G.H.; Oxman, A.D.; Vist, G.E.; Kunz, R.; Falck-Ytter, Y.; Alonso-Coello, P.; Schunemann, H.J.; Group, G.W. GRADE: An emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008, 336, 924–926. [Google Scholar] [CrossRef] [Green Version]
- Szarvas, T.; Modos, O.; Horvath, A.; Nyirady, P. Why are upper tract urothelial carcinoma two different diseases? Transl. Androl. Urol. 2016, 5, 636–647. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liu, T.; Ge, N.; Liu, L.; Yuan, X.; Liu, J.; Kong, F.; Wang, C.; Ren, H.; Yan, K.; et al. TERT promoter mutations are associated with distant metastases in upper tract urothelial carcinomas and serve as urinary biomarkers detected by a sensitive castPCR. Oncotarget 2014, 5, 12428–12439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Liu, T.; Liu, L.; Liu, J.; Liu, C.; Wang, C.; Ge, N.; Ren, H.; Yan, K.; Hu, S.; et al. TERT promoter mutations in renal cell carcinomas and upper tract urothelial carcinomas. Oncotarget 2014, 5, 1829–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, Y.; Fujita, K.; Matsuzaki, K.; Matsushita, M.; Kawamura, N.; Koh, Y.; Nakano, K.; Wang, C.; Ishizuya, Y.; Yamamoto, Y.; et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci. 2019, 110, 1771–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Liu, T.; Liu, C.; Meng, Y.; Yuan, X.; Liu, L.; Ge, N.; Liu, J.; Wang, C.; Ren, H.; et al. TERT promoter mutations and TERT mRNA but not FGFR3 mutations are urinary biomarkers in Han Chinese patients with urothelial bladder cancer. Oncologist 2015, 20, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Hosen, M.I.; Sheikh, M.; Zvereva, M.; Scelo, G.; Forey, N.; Durand, G.; Voegele, C.; Poustchi, H.; Khoshnia, M.; Roshandel, G.; et al. Urinary TERT promoter mutations are detectable up to 10 years prior to clinical diagnosis of bladder cancer: Evidence from the Golestan Cohort Study. EBioMedicine 2020, 53, 102643. [Google Scholar] [CrossRef]
- Hurst, C.D.; Platt, F.M.; Knowles, M.A. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur. Urol. 2014, 65, 367–369. [Google Scholar] [CrossRef]
- Xing, X.; Yuan, X.; Liu, T.; Dai, M.; Fan, Y.; Liu, C.; Straat, K.; Bjorkholm, M.; Xu, D. Regulatory region mutations of TERT, PLEKHS1 and GPR126 genes as urinary biomarkers in upper tract urothelial carcinomas. J. Cancer 2021, 12, 3853–3861. [Google Scholar] [CrossRef]
- Berndl, F.; Hassler, M.R. Molecular intricacies of upper tract urothelial carcinoma and their relevance for therapy considerations. Curr. Opin. Urol. 2021, 32, 48–53. [Google Scholar] [CrossRef]
- Wisnieski, F.; Santos, L.C.; Calcagno, D.Q.; Geraldis, J.C.; Gigek, C.O.; Anauate, A.C.; Chen, E.S.; Rasmussen, L.T.; Payao, S.L.M.; Artigiani, R.; et al. The impact of DNA demethylation on the upregulation of the NRN1 and TNFAIP3 genes associated with advanced gastric cancer. J. Mol. Med. 2020, 98, 707–717. [Google Scholar] [CrossRef]
- Bosserhoff, A.K.; Schneider, N.; Ellmann, L.; Heinzerling, L.; Kuphal, S. The neurotrophin Neuritin1 (cpg15) is involved in melanoma migration, attachment independent growth, and vascular mimicry. Oncotarget 2017, 8, 1117–1131. [Google Scholar] [CrossRef] [Green Version]
- Sfakianos, J.P.; Cha, E.K.; Iyer, G.; Scott, S.N.; Zabor, E.C.; Shah, R.H.; Ren, Q.; Bagrodia, A.; Kim, P.H.; Hakimi, A.A.; et al. Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2015, 68, 970–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagrodia, A.; Cha, E.K.; Sfakianos, J.P.; Zabor, E.C.; Bochner, B.H.; Al-Ahmadie, H.A.; Solit, D.B.; Coleman, J.A.; Iyer, G.; Scott, S.N.; et al. Genomic Biomarkers for the Prediction of Stage and Prognosis of Upper Tract Urothelial Carcinoma. J. Urol. 2016, 195, 1684–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagrodia, A.; Audenet, F.; Pietzak, E.J.; Kim, K.; Murray, K.S.; Cha, E.K.; Sfakianos, J.P.; Iyer, G.; Singla, N.; Arcila, M.; et al. Genomic Profile of Urothelial Carcinoma of the Upper Tract from Ureteroscopic Biopsy: Feasibility and Validation Using Matched Radical Nephroureterectomy Specimens. Eur. Urol. Focus 2019, 5, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, K.; Sung, H.H.; Jeon, H.G.; Jeong, B.C.; Seo, S.I.; Jeon, S.S.; Lee, H.M.; Choi, H.Y.; Kwon, G.Y.; et al. Molecular Characterization of Urothelial Carcinoma of the Bladder and Upper Urinary Tract. Transl. Oncol. 2018, 11, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.; Pal, S.K.; Hahn, A.W.; Nussenzveig, R.H.; Pond, G.R.; Gupta, S.V.; Wang, J.; Bilen, M.A.; Naik, G.; Ghatalia, P.; et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer 2018, 124, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Moss, T.J.; Qi, Y.; Xi, L.; Peng, B.; Kim, T.B.; Ezzedine, N.E.; Mosqueda, M.E.; Guo, C.C.; Czerniak, B.A.; Ittmann, M.; et al. Comprehensive Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur. Urol. 2017, 72, 641–649. [Google Scholar] [CrossRef]
- Robinson, B.D.; Vlachostergios, P.J.; Bhinder, B.; Liu, W.; Li, K.; Moss, T.J.; Bareja, R.; Park, K.; Tavassoli, P.; Cyrta, J.; et al. Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat. Commun. 2019, 10, 2977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winters, B.R.; De Sarkar, N.; Arora, S.; Bolouri, H.; Jana, S.; Vakar-Lopez, F.; Cheng, H.H.; Schweizer, M.T.; Yu, E.Y.; Grivas, P.; et al. Genomic distinctions between metastatic lower and upper tract urothelial carcinoma revealed through rapid autopsy. JCI Insight 2019, 5, e128728. [Google Scholar] [CrossRef] [Green Version]
- Springer, S.U.; Chen, C.H.; Rodriguez Pena, M.D.C.; Li, L.; Douville, C.; Wang, Y.; Cohen, J.D.; Taheri, D.; Silliman, N.; Schaefer, J.; et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife 2018, 7, e32143. [Google Scholar] [CrossRef]
- McLaughlin, J.K.; Silverman, D.T.; Hsing, A.W.; Ross, R.K.; Schoenberg, J.B.; Yu, M.C.; Stemhagen, A.; Lynch, C.F.; Blot, W.J.; Fraumeni, J.F., Jr. Cigarette smoking and cancers of the renal pelvis and ureter. Cancer Res. 1992, 52, 254–257. [Google Scholar]
- Cosyns, J.P. Aristolochic acid and ‘Chinese herbs nephropathy’: A review of the evidence to date. Drug Saf. 2003, 26, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, T.A.; Grollman, A.P. Mutational signature of aristolochic acid: Clue to the recognition of a global disease. DNA Repair 2016, 44, 205–211. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | Number of UTUC Patients (n = 95) | Number of Non-UTUC Patients (n = 307) | p Value |
|---|---|---|---|
| Age, n (%) | 66.53 ± 11.14 | 58.93 ± 12.43 | <0.001 |
| Gender, n (%) | 0.480 | ||
| Male | 62 (65.26) | 188 (61.24) | |
| Female | 33 (34.74) | 119 (38.76) | |
| NRN1, n (%) | <0.001 | ||
| Positive | 87 (91.58) | 16 (5.21) | |
| Negative | 8 (8.42) | 291 (94.79) | |
| Gene Mutation, n (%) | <0.001 | ||
| Yes | 33 (34.74) | 10 (3.26) | |
| No | 62 (65.26) | 297 (96.74) | |
| Grade, n (%) | NA | NA | |
| Low grade | 17 (17.89) | ||
| High grade | 76 (80.00) | ||
| Gx | 2 (2.11) | ||
| Stage | NA | NA | |
| Ta-2 | 64 (67.36) | ||
| T3,4 | 29 (30.53) | ||
| Tx | 2 (2.11) |
| Variables | Test Performance | ||||
|---|---|---|---|---|---|
| AUC | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
| Gene mutations | 0.657 | 34.74 | 96.74 | 76.73 | 82.74 |
| NRN1 methylation | 0.932 | 91.58 | 94.79 | 84.58 | 97.36 |
| Panel | 0.958 | 91.58 | 94.79 | 84.58 | 97.36 |
| Variables of Models | Models with Different Features | |
|---|---|---|
| Panel | Panel + Age | |
| AUC | 0.958 (0.933–0.975) | 0.968 (0.945–0.983) |
| Sensitivity (%) | 91.58 (84.15–96.37) | 93.68 (86.83–97.65) |
| Specificity (%) | 94.79 (91.70–97.02) | 94.44 (92.55–97.57) |
| PPV (%) | 84.58 (77.14–89.88) | 86.46 (79.28–91.41) |
| NPV (%) | 97.36 (94.92–98.67) | 98.05 (95.73–99.11) |
| Urinary Cell-Free DNA | FISH | Cytology | ||||
|---|---|---|---|---|---|---|
| + | − | + | − | + | − | |
| UTUC+ | 89 | 6 | 81 | 14 | 39 | 56 |
| UTUC− | 17 | 290 | 26 | 281 | 42 | 265 |
| Sensitivity (%) | 93.68 | 85.26 | 41.05 | |||
| Specificity (%) | 94.44 | 91.53 | 86.32 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouyang, W.; Luo, L.; Zhang, J.; Xu, R.; Lu, Q.; Xu, Z.; Liu, J.; Li, P.; Zhang, Y.; Zhou, C.; et al. Urine Cellular DNA Point Mutation and Methylation for Identifying Upper Tract Urinary Carcinoma. Cancers 2022, 14, 3537. https://doi.org/10.3390/cancers14143537
Ouyang W, Luo L, Zhang J, Xu R, Lu Q, Xu Z, Liu J, Li P, Zhang Y, Zhou C, et al. Urine Cellular DNA Point Mutation and Methylation for Identifying Upper Tract Urinary Carcinoma. Cancers. 2022; 14(14):3537. https://doi.org/10.3390/cancers14143537
Chicago/Turabian StyleOuyang, Wei, Lufeng Luo, Junjie Zhang, Ran Xu, Qiang Lu, Zhenzhou Xu, Jianye Liu, Pei Li, Yaqun Zhang, Chuanchi Zhou, and et al. 2022. "Urine Cellular DNA Point Mutation and Methylation for Identifying Upper Tract Urinary Carcinoma" Cancers 14, no. 14: 3537. https://doi.org/10.3390/cancers14143537






