Overexpression of NNMT in Glioma Aggravates Tumor Cell Progression: An Emerging Therapeutic Target
Abstract
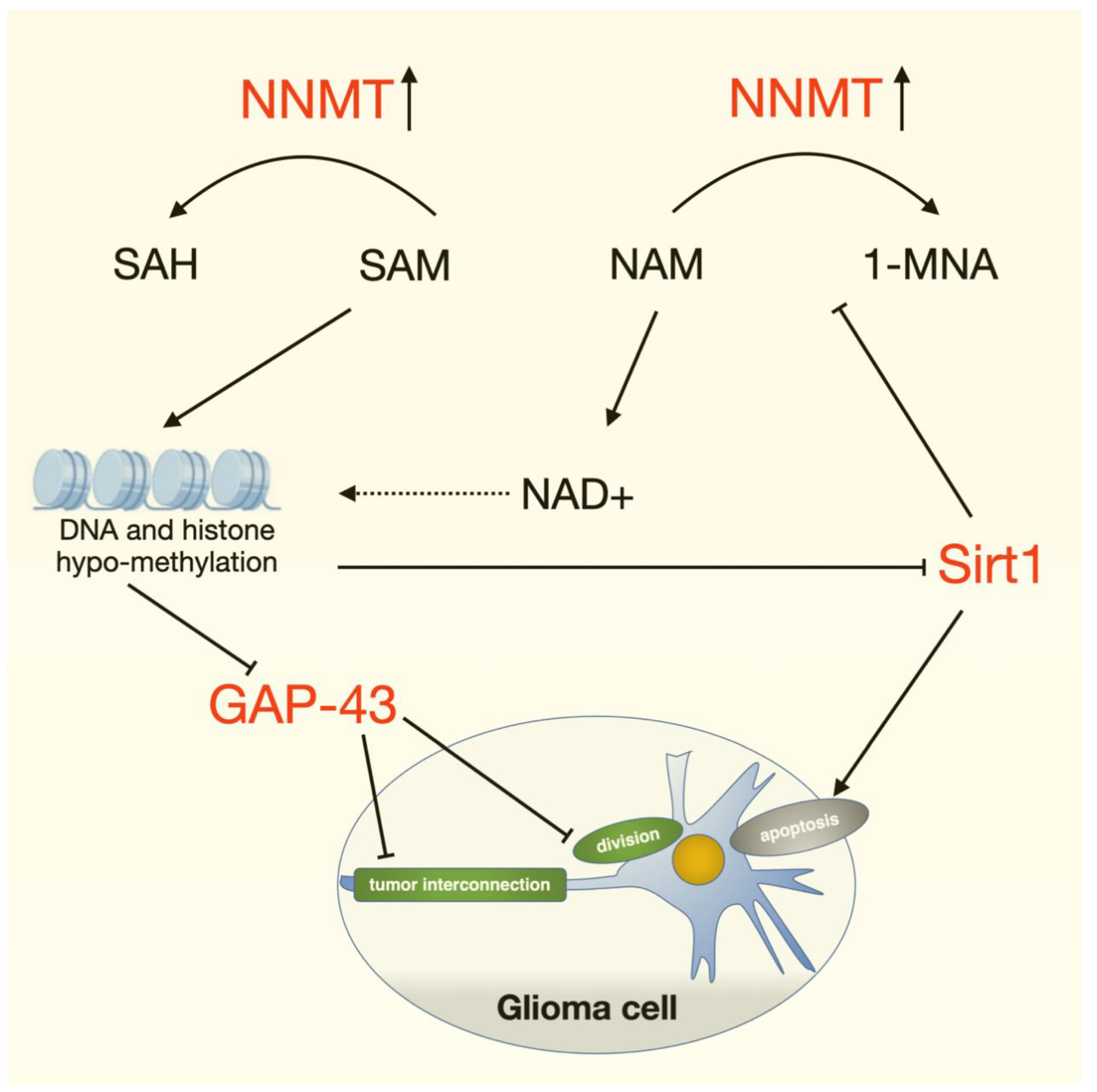
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
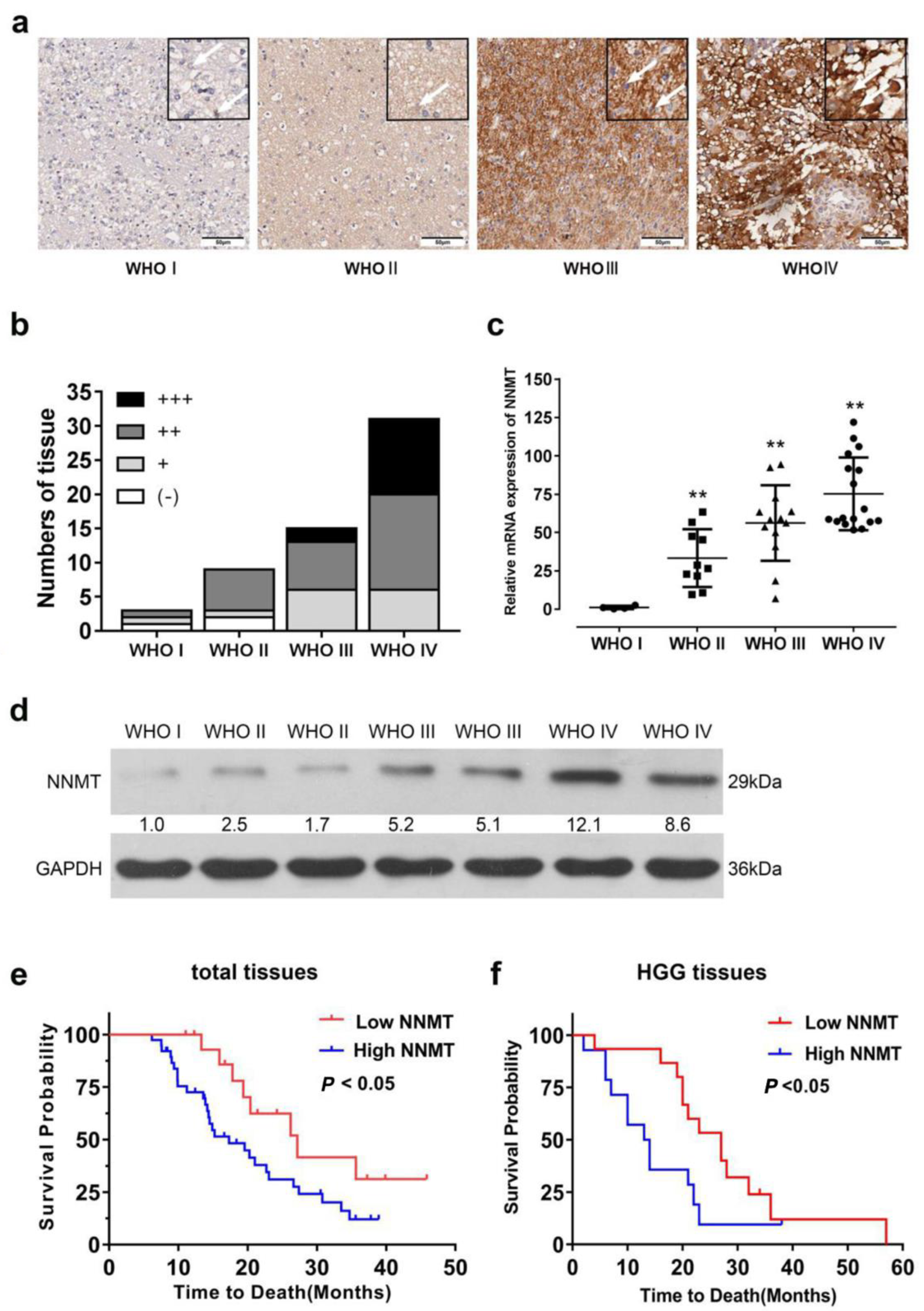
2.2. Immunohistochemical Analysis
2.3. Western Blot Assay
2.4. Quantitative Real-Time Polymerase Chain Reaction
2.5. Cell Cultures
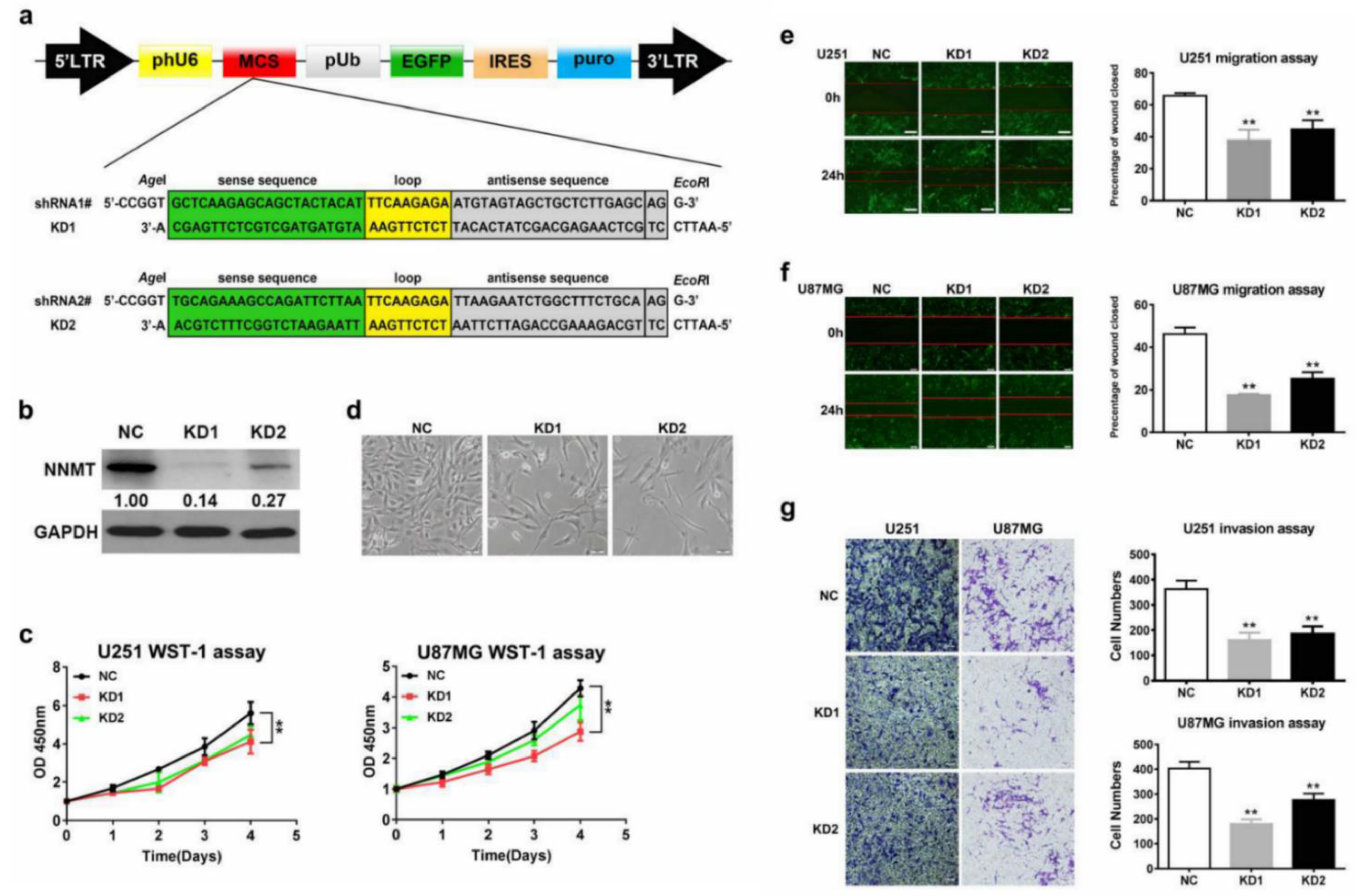
2.6. Cell Viability Assay
2.7. Wound Healing Assay
2.8. Transwell Migration and Invasion Assays
2.9. Determination of NAD/NADH Ratio
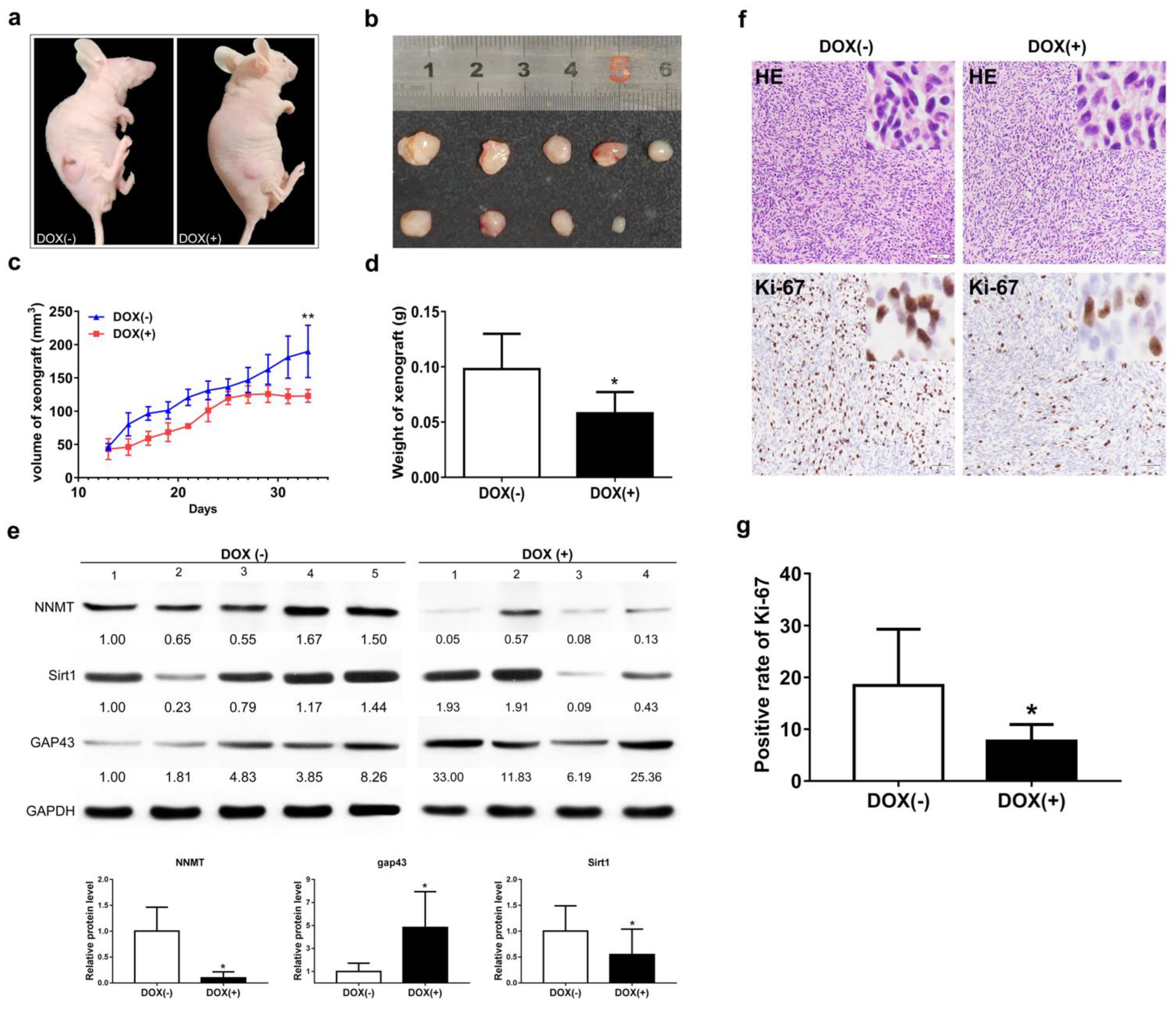
2.9.1. Experimental Animals
2.9.2. Hematoxylin and Eosin Staining
2.9.3. Statistical Analyses
3. Results
3.1. NNMT Expression Promotes Glioma Progression in Patients
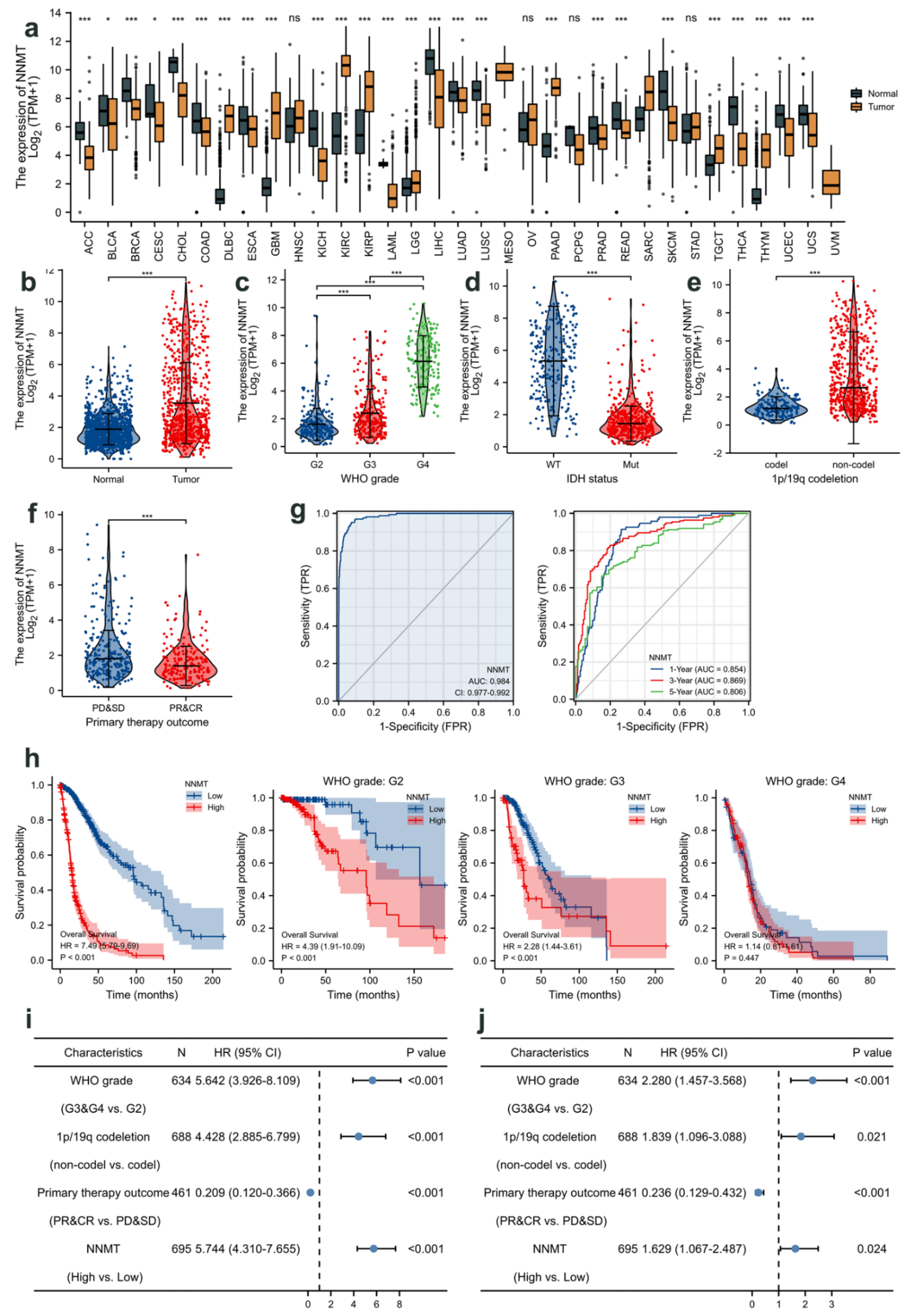
3.2. Relationship between NNMT Expression and Glioma Based on TCGA
3.3. NNMT Expression Promotes Proliferation, Migration, and Invasion of Glioma Cells
3.4. NNMT Gene Correlation Analysis
3.5. NNMT-Associated Downstream Proteins in Glioma Cells
3.6. NNMT Expression Is Closely Associated with the NAD/NADH Ratio
3.7. NNMT Knockdown Inhibits Tumor Growth by Promoting GAP43 Expression
3.8. Analysis of Differential Genes in Glioma Mice by DNA Methylation Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, R.; Cheng, L.; Yang, X.; Chen, X.; Miao, Y.; Qiu, Y.; Zhou, Z. Histone methyltransferase SUV39H2 regulates cell growth and chemosensitivity in glioma via regulation of hedgehog signaling. Cancer Cell Int. 2019, 19, 269. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhao, W.; Zhao, H.; Zhang, J. Diagnostic and prognostic value of microRNA-193b in patients with glioma and its effect on tumor progression. Oncol. Lett. 2019, 18, 4882–4890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Zhou, J.; Zou, D.; Hou, D.; Zhang, H.; Zhao, J.; Li, L.; Hu, J.; Zhang, Y.; Jing, Z. Overexpression of Limb-Bud and Heart (LBH) promotes angiogenesis in human glioma via VEGFA-mediated ERK signalling under hypoxia. eBioMedicine 2019, 48, 36–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Li, S.L.; Cao, Y.L.; Li, J.J.; Wang, Q.P. LKB1 suppresses glioma cell invasion via NF-κB/Snail signaling repression. OncoTargets Ther. 2019, 12, 2451–2463. [Google Scholar] [CrossRef] [Green Version]
- Malzkorn, B.; Reifenberger, G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr. Opin. Oncol. 2016, 28, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, H.; Chen, Z.; Fan, L.; Feng, S.; Cai, X.; Wang, H. Identification of 3 subpopulations of tumor-infiltrating immune cells for malignant transformation of low-grade glioma. Cancer Cell Int. 2019, 19, 265. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Xu, S.; Dai, G.; Xiao, Z.; Chen, L.; Liu, Z. Tumor treating fields for high-grade gliomas. Biomed. Pharmacother. 2020, 127, 110193. [Google Scholar] [CrossRef]
- Gao, Y.; Xuan, C.; Jin, M.; An, Q.; Zhuo, B.; Chen, X.; Wang, L.; Wang, Y.; Sun, Q.; Shi, Y. Ubiquitin ligase RNF5 serves an important role in the development of human glioma. Oncol. Lett. 2019, 18, 4659–4666. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Roila, F. Malignant glioma: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann. Oncol. 2009, 20 (Suppl. S4), 126–128. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cote, D.J.; Ascha, M.; Kruchko, C.; Barnholtz-Sloan, J.S. Adult Glioma Incidence and Survival by Race or Ethnicity in the United States from 2000 to 2014. JAMA Oncol. 2018, 4, 1254–1262. [Google Scholar] [CrossRef] [Green Version]
- Iyamu, I.D.; Huang, R. Development of fluorescence polarization-based competition assay for nicotinamide N-methyltransferase. Anal. Biochem. 2020, 604, 113833. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, C.W.; Yoon, G.; Hong, S.M.; Choi, K.Y. NNMT depletion contributes to liver cancer cell survival by enhancing autophagy under nutrient starvation. Oncogenesis 2018, 7, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neelakantan, H.; Wang, H.Y.; Vance, V.; Hommel, J.D.; McHardy, S.F.; Watowich, S.J. Structure-Activity Relationship for Small Molecule Inhibitors of Nicotinamide N-Methyltransferase. J. Med. Chem. 2017, 60, 5015–5028. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, J.; Wu, W.; Xie, S.; Yu, H.; Li, G.; Zhu, T.; Li, F.; Lu, J.; Wang, G.Y.; et al. Nicotinamide N-methyltransferase enhances chemoresistance in breast cancer through SIRT1 protein stabilization. Breast Cancer Res. 2019, 21, 64. [Google Scholar] [CrossRef] [Green Version]
- Mobley, A.; Zhang, S.; Bondaruk, J.; Wang, Y.; Majewski, T.; Caraway, N.P.; Huang, L.; Shoshan, E.; Velazquez-Torres, G.; Nitti, G.; et al. Aurora Kinase A is a Biomarker for Bladder Cancer Detection and Contributes to its Aggressive Behavior. Sci. Rep. 2017, 7, 40714. [Google Scholar] [CrossRef]
- Wu, M.; Hu, W.; Wang, G.; Yao, Y.; Yu, X.F. Nicotinamide N-Methyltransferase Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric Cancer. Front Genet. 2020, 11, 580299. [Google Scholar] [CrossRef]
- Song, M.; Li, Y.; Miao, M.; Zhang, F.; Yuan, H.; Cao, F.; Chang, W.; Shi, H.; Song, C. High stromal nicotinamide N-methyltransferase (NNMT) indicates poor prognosis in colorectal cancer. Cancer Med. 2020, 9, 2030–2038. [Google Scholar] [CrossRef]
- Harmankaya, İ.; Akar, S.; Uğraş, S.; Güler, A.H.; Ezveci, H.; Aydoğdu, M.; Çelik, Ç. Nicotinamide N-methyltransferase overexpression may be associated with poor prognosis in ovarian cancer. J. Obstet. Gynaecol. 2021, 41, 248–253. [Google Scholar] [CrossRef]
- Togni, L.; Mascitti, M.; Sartini, D.; Campagna, R.; Pozzi, V.; Salvolini, E.; Offidani, A.; Santarelli, A.; Emanuelli, M. Nicotinamide N-Methyltransferase in Head and Neck Tumors: A Comprehensive Review. Biomolecules 2021, 11, 1594. [Google Scholar] [CrossRef]
- Campagna, R.; Pozzi, V.; Sartini, D.; Salvolini, E.; Brisigotti, V.; Molinelli, E.; Campanati, A.; Offidani, A.; Emanuelli, M. Beyond Nicotinamide Metabolism: Potential Role of Nicotinamide N-Methyltransferase as a Biomarker in Skin Cancers. Cancers 2021, 13, 4943. [Google Scholar] [CrossRef]
- Eckert, M.A.; Coscia, F.; Chryplewicz, A.; Chang, J.W.; Hernandez, K.M.; Pan, S.; Tienda, S.M.; Nahotko, D.A.; Li, G.; Blaženović, I.; et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019, 569, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Campagna, R.; Salvolini, E.; Pompei, V.; Pozzi, V.; Salvucci, A.; Molinelli, E.; Brisigotti, V.; Sartini, D.; Campanati, A.; Offidani, A.; et al. Nicotinamide N-methyltransferase gene silencing enhances chemosensitivity of melanoma cell lines. Pigment Cell Melanoma Res. 2021, 34, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fang, S.; Shao, X.; Li, Y.; Tong, Q.; Kong, B.; Chen, L.; Wang, Y.; Yang, J.; Yu, H.; et al. Curcumin Reverses NNMT-Induced 5-Fluorouracil Resistance via Increasing ROS and Cell Cycle Arrest in Colorectal Cancer Cells. Biomolecules 2021, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
- Palanichamy, K.; Kanji, S.; Gordon, N.; Thirumoorthy, K.; Jacob, J.R.; Litzenberg, K.T.; Patel, D.; Chakravarti, A. NNMT Silencing Activates Tumor Suppressor PP2A, Inactivates Oncogenic STKs, and Inhibits Tumor Forming Ability. Clin. Cancer Res. 2017, 23, 2325–2334. [Google Scholar] [CrossRef] [Green Version]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [Green Version]
- Akar, S.; Harmankaya, İ.; Uğraş, S.; Çelik, Ç. Expression and Clinical Significance of Nicotinamide N-Methyltransferase in Cervical Squamous Cell Carcinoma. Int. J. Gynecol. Pathol. 2020, 39, 289–295. [Google Scholar] [CrossRef]
- Lu, X.M.; Long, H. Nicotinamide N-methyltransferase as a potential marker for cancer. Neoplasma 2018, 65, 656–663. [Google Scholar] [CrossRef]
- Shivarudrappa, A.H.; Ponesakki, G. Lutein reverses hyperglycemia-mediated blockage of Nrf2 translocation by modulating the activation of intracellular protein kinases in retinal pigment epithelial (ARPE-19) cells. J. Cell Commun. Signal. 2020, 14, 207–221. [Google Scholar] [CrossRef]
- Neelakantan, H.; Brightwell, C.R.; Graber, T.G.; Maroto, R.; Wang, H.L.; McHardy, S.F.; Papaconstantinou, J.; Fry, C.S.; Watowich, S.J. Small molecule nicotinamide N-methyltransferase inhibitor activates senescent muscle stem cells and improves regenerative capacity of aged skeletal muscle. Biochem. Pharmacol. 2019, 163, 481–492. [Google Scholar] [CrossRef]
- Xu, Y. MicroRNA-136-3p inhibits glioma tumorigenesis in vitro and in vivo by targeting KLF7. World J. Surg. Oncol. 2020, 18, 169. [Google Scholar] [CrossRef]
- Ding, C.; Wu, Z.; You, H.; Ge, H.; Zheng, S.; Lin, Y.; Wu, X.; Lin, Z.; Kang, D. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J. Exp. Clin. Cancer Res. 2019, 38, 506. [Google Scholar] [CrossRef] [Green Version]
- Blum, A.; Wang, P.; Zenklusen, J.C. SnapShot: TCGA-Analyzed Tumors. Cell 2018, 173, 530. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Liao, W.; Wos, F.; Johnston, A.D.; DeGrazia, J.; Ishii, J.; Bloom, T.; Zody, M.C.; Germer, S.; Greally, J.M. Whole-genome bisulfite sequencing with improved accuracy and cost. Genome Res. 2018, 28, 1364–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Hu, F.; Li, B.; Zhang, Y.; Chen, M.; Fan, T.; Wang, T. Whole genome bisulfite sequencing methylome analysis of mulberry (Morus alba) reveals epigenome modifications in response to drought stress. Sci. Rep. 2020, 10, 8013. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, Y. Integrated bioinformatics analysis of expression and gene regulation network of COL12A1 in colorectal cancer. Cancer Med. 2020, 9, 4743–4755. [Google Scholar] [CrossRef]
- Jackson, S.E.; Chester, J.D. Personalised cancer medicine. Int. J. Cancer 2015, 137, 262–266. [Google Scholar] [CrossRef]
- Vuong, H.G.; Altibi, A.M.A.; Duong, U.N.P.; Ngo, H.T.T.; Pham, T.Q.; Fung, K.M.; Hassell, L. BRAF Mutation is Associated with an Improved Survival in Glioma-a Systematic Review and Meta-analysis. Mol. Neurobiol. 2018, 55, 3718–3724. [Google Scholar] [CrossRef]
- Wu, D.; Hu, D.; Chen, H.; Shi, G.; Fetahu, I.S.; Wu, F.; Rabidou, K.; Fang, R.; Tan, L.; Xu, S.; et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 2018, 559, 637–641. [Google Scholar] [CrossRef]
- Ohkuma, T.; Peters, S.A.E.; Woodward, M. Sex differences in the association between diabetes and cancer: A systematic review and meta-analysis of 121 cohorts including 20 million individuals and one million events. Diabetologia 2018, 61, 2140–2154. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.Y.; Wu, Y.; Burke, S.P.; Gutmann, D.H. The 43000 growth-associated protein functions as a negative growth regulator in glioma. Cancer Res. 2003, 63, 2933–2939. [Google Scholar] [PubMed]
- Delsite, R.; Kachhap, S.; Anbazhagan, R.; Gabrielson, E.; Singh, K.K. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol. Cancer 2002, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, H.; Feng, J.; Li, Y.; Lv, J.; Shi, W.; Fan, W.; Xiao, L.; Sun, D.; Jiang, M.; et al. Transcriptome profiling unveils GAP43 regulates ABC transporters and EIF2 signaling in colorectal cancer cells. BMC Cancer 2021, 21, 24. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Quan, R.D.; Wu, C.Y.; Hu, J.; Lin, B.Y.; Dong, X.B.; Xia, E.J.; Bhandari, A.; Zhang, X.H.; Wang, O.C. Growth-associated protein 43 promotes thyroid cancer cell lines progression via epithelial-mesenchymal transition. J. Cell Mol. Med. 2019, 23, 7974–7984. [Google Scholar] [CrossRef]
- Xu, C.; Wang, L.; Fozouni, P.; Evjen, G.; Chandra, V.; Jiang, J.; Lu, C.; Nicastri, M.; Bretz, C.; Winkler, J.D.; et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat. Cell Biol. 2020, 22, 1170–1179. [Google Scholar] [CrossRef]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Bian, Y.; Zhang, Y.; Ren, G.; Li, G. Metformin activates AMPK/SIRT1/NF-κB pathway and induces mitochondrial dysfunction to drive caspase3/GSDME-mediated cancer cell pyroptosis. Cell Cycle 2020, 19, 1089–1104. [Google Scholar] [CrossRef]
- Nikas, I.P.; Paschou, S.A.; Ryu, H.S. The Role of Nicotinamide in Cancer Chemoprevention and Therapy. Biomolecules 2020, 10, 477. [Google Scholar] [CrossRef] [Green Version]
- Bell, E.H.; Zhang, P.; Fisher, B.J.; Macdonald, D.R.; McElroy, J.P.; Lesser, G.J.; Fleming, J.; Chakraborty, A.R.; Liu, Z.; Becker, A.P.; et al. Association of MGMT Promoter Methylation Status With Survival Outcomes in Patients With High-Risk Glioma Treated With Radiotherapy and Temozolomide: An Analysis From the NRG Oncology/RTOG 0424 Trial. JAMA Oncol. 2018, 4, 1405–1409. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; van Haren, M.J.; Buijs, N.; Innocenti, P.; Zhang, Y.; Sartini, D.; Campagna, R.; Emanuelli, M.; Parsons, R.B.; Jespers, W.; et al. Potent Inhibition of Nicotinamide N-Methyltransferase by Alkene-Linked Bisubstrate Mimics Bearing Electron Deficient Aromatics. J. Med. Chem. 2021, 64, 12938–12963. [Google Scholar] [CrossRef]
- van Haren, M.J.; Gao, Y.; Buijs, N.; Campagna, R.; Sartini, D.; Emanuelli, M.; Mateuszuk, L.; Kij, A.; Chlopicki, S.; Escudé Martinez de Castilla, P.; et al. Esterase-Sensitive Prodrugs of a Potent Bisubstrate Inhibitor of Nicotinamide N-Methyltransferase (NNMT) Display Cellular Activity. Biomolecules 2021, 11, 1357. [Google Scholar] [CrossRef] [PubMed]




| Primary Antibody | Manufacture | Dilution |
|---|---|---|
| NNMT | Abcam | 1:1000 |
| GAP43 | Proteintech | 1:1000 |
| Sirt1 | CST | 1:1000 |
| APOBEC3G | Proteintech | 1:1000 |
| CXCR7 | Proteintech | 1:1000 |
| NEGR1 | Abcam | 1:1000 |
| PAK3 | Abcam | 1:1000 |
| TROY | Abcam | 1:1000 |
| GAPDH | Proteintech | 1:5000 |
| Secondary antibody | ||
| Goat anti-rabbit HRP-IgG | Jackson | 1:2000 |
| Goat anti-mouse HRP-IgG | Jackson | 1:2000 |
| Sequences | Gene Name |
|---|---|
| 5′-ATATTCTGCCTAGACGGTGTGA-3′ | NNMT-F |
| 5′-TCAGTGACGACGATCTCCTTAAA-3′ | NNMT-R |
| 5′-AACACAACCGACTACCGAATC-3′ | SEMA3F-F |
| 5′-GGCTGCCCAGTGTATAATGAG-3′ | SEMA3F-R |
| 5′-CTGTGTCAGAAAAGAGACGGTC-3′ | APOBEC3G-F |
| 5′-GTACACGAACTTGCTCCAACA-3′ | APOBEC3G-R |
| 5′-TCTGCATCTCTTCGACTACTCA-3′ | CXCR7-F |
| 5′-GTAGAGCAGGACGCTTTTGTT-3′ | CXCR7-R |
| 5′-AGCCCATCCTTCGAGTACAAA-3′ | PAK3-F |
| 5′-TCTTGGTGCGATAACTGGTGG-3′ | PAK3-R |
| 5′-GGCCGCAACCAAAATTCAGG-3′ | GAP43-F |
| 5′-CGGCAGTAGTGGTGCCTTC-3′ | GAP43-R |
| 5′-GGGAGGTGATAAGTGGTCAGT-3′ | NEGR1-F |
| 5′-CTGGGTGTATGTTGAGTCTGAAC-3′ | NEGR1-R |
| 5′-CCAGCAAGGTCAACCTCGT-3′ | TORY-F |
| 5′-CAGAGCCGTTGTACTGAATGT-3′ | TORY-R |
| Characteristic | Levels | Low Expression of NNMT | High Expression of NNMT | p |
|---|---|---|---|---|
| n | 335 | 335 | ||
| WHO grade, n (%) | G2 | 173 (28.2%) | 43 (7%) | <0.001 |
| G3 | 130 (21.2%) | 107 (17.5%) | ||
| G4 | 0 (0%) | 160 (26.1%) | ||
| IDH status, n (%) | WT | 20 (3%) | 217 (32.8%) | <0.001 |
| Mut | 314 (47.5%) | 110 (16.6%) | ||
| 1p/19q codeletion, n (%) | codel | 145 (21.8%) | 23 (3.5%) | <0.001 |
| Noncodel | 190 (28.6%) | 306 (46.1%) | ||
| Primary therapy outcome, n (%) | PD | 49 (11%) | 54 (12.2%) | <0.001 |
| SD | 96 (21.6%) | 48 (10.8%) | ||
| PR | 39 (8.8%) | 23 (5.2%) | ||
| CR | 105 (23.6%) | 30 (6.8%) | ||
| Gender, n (%) | Female | 145 (21.6%) | 139 (20.7%) | 0.696 |
| Male | 190 (28.4%) | 196 (29.3%) | ||
| Race, n (%) | Asian | 7 (1.1%) | 6 (0.9%) | 0.519 |
| Black or African American | 13 (2%) | 19 (2.9%) | ||
| White | 311 (47.3%) | 302 (45.9%) | ||
| Age, n (%) | ≤60 | 305 (45.5%) | 226 (33.7%) | <0.001 |
| >60 | 30 (4.5%) | 109 (16.3%) | ||
| Histological type, n (%) | Astrocytoma | 99 (14.8%) | 93 (13.9%) | <0.001 |
| Glioblastoma | 0 (0%) | 160 (23.9%) | ||
| Oligoastrocytoma | 87 (13%) | 41 (6.1%) | ||
| Oligodendroglioma | 149 (22.2%) | 41 (6.1%) | ||
| Age, median (IQR) | 39 (32, 50) | 54 (40, 63) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Zou, Y.; Cai, Z.; Huang, J.; Hong, X.; Liang, Q.; Jin, W. Overexpression of NNMT in Glioma Aggravates Tumor Cell Progression: An Emerging Therapeutic Target. Cancers 2022, 14, 3538. https://doi.org/10.3390/cancers14143538
Sun W, Zou Y, Cai Z, Huang J, Hong X, Liang Q, Jin W. Overexpression of NNMT in Glioma Aggravates Tumor Cell Progression: An Emerging Therapeutic Target. Cancers. 2022; 14(14):3538. https://doi.org/10.3390/cancers14143538
Chicago/Turabian StyleSun, Wei, Yongxiang Zou, Zheng Cai, Jinxiang Huang, Xinjie Hong, Qiang Liang, and Weilin Jin. 2022. "Overexpression of NNMT in Glioma Aggravates Tumor Cell Progression: An Emerging Therapeutic Target" Cancers 14, no. 14: 3538. https://doi.org/10.3390/cancers14143538






