The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
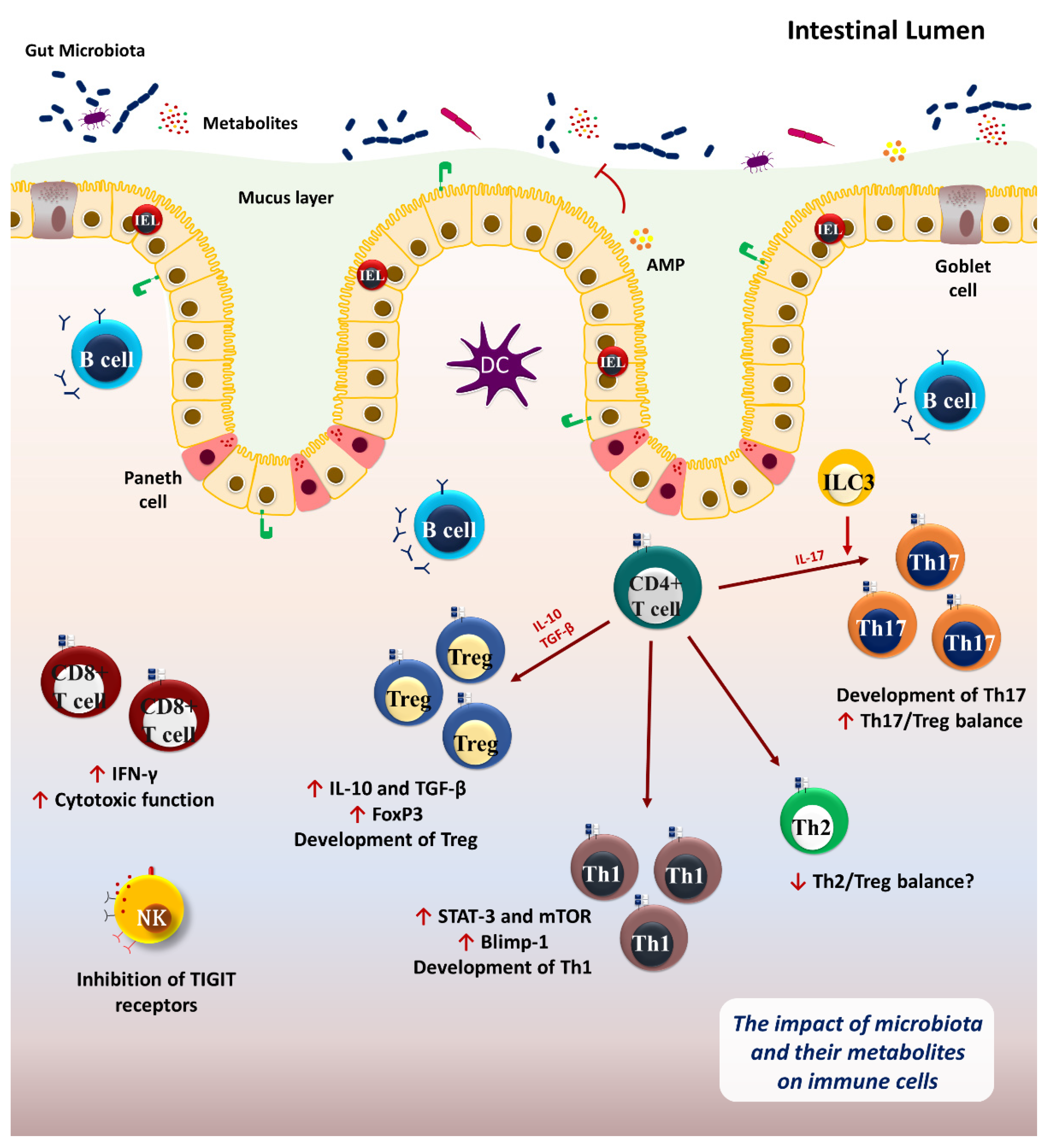
2. Microbiome and Immune System
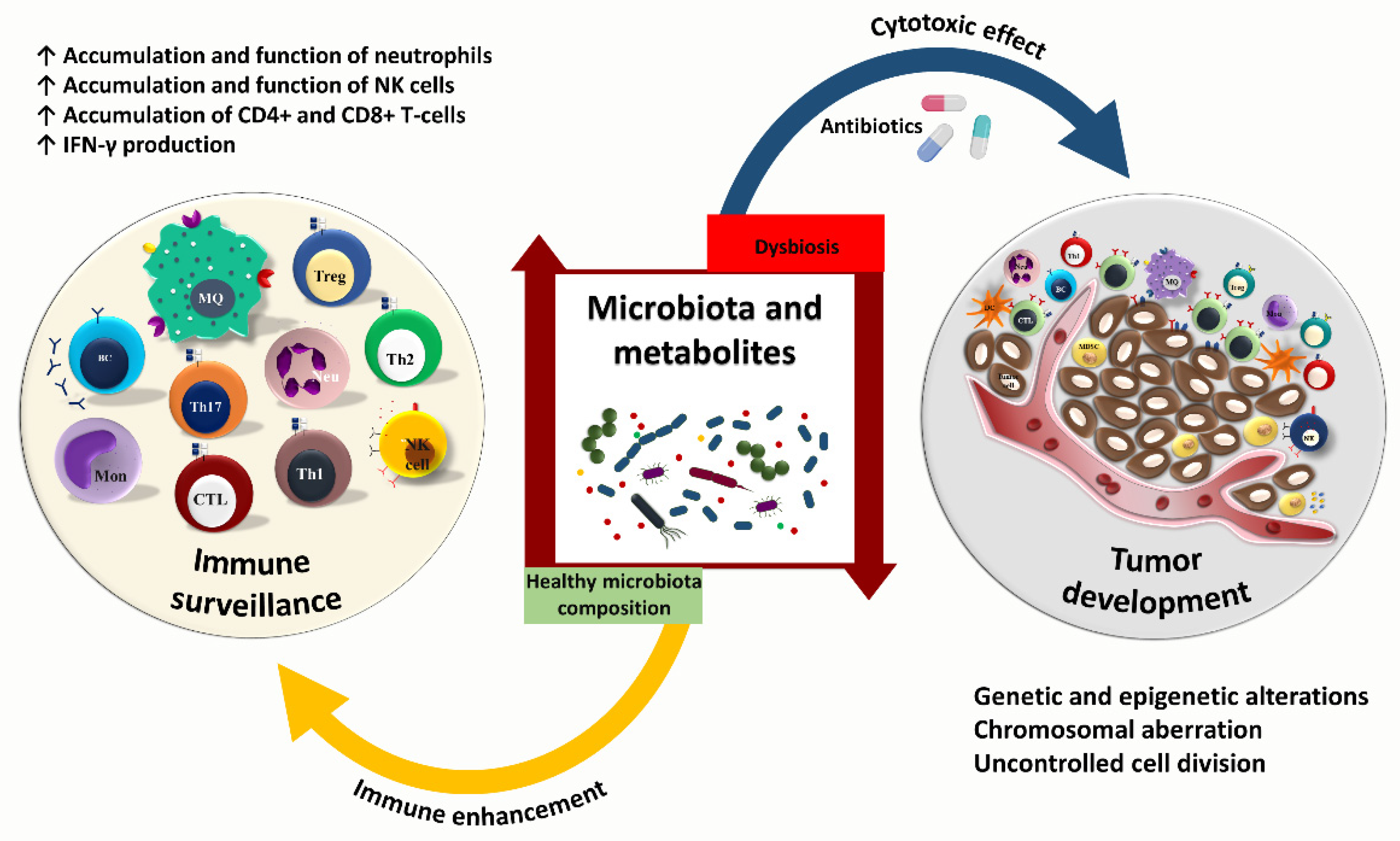
3. Cancer—Microbiome—Immune Axis
4. Gut Microbiota and Anti-Tumor Immunity
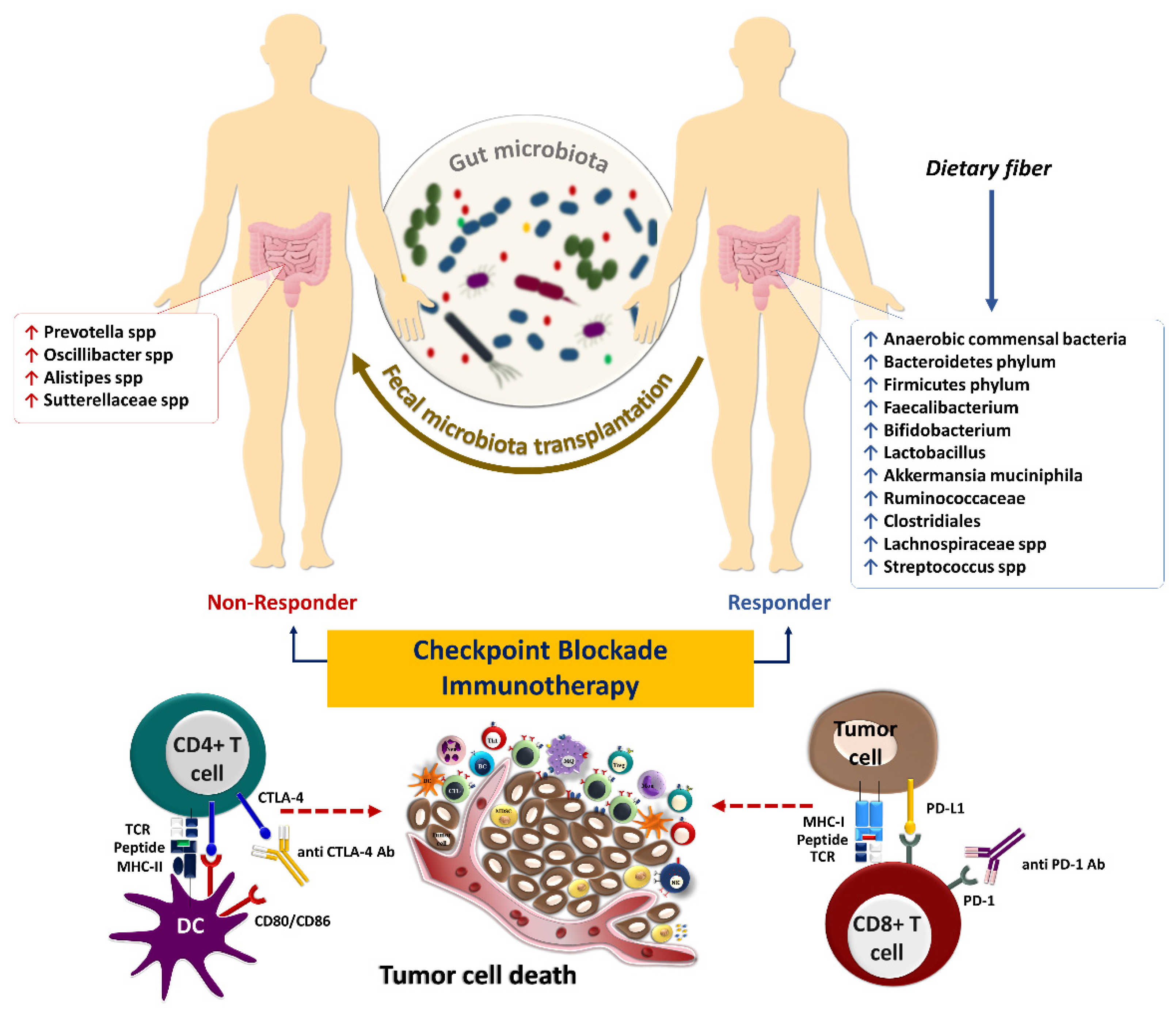
5. Gut Microbiota and Response to Systemic Therapy, including Immunotherapy
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lewandowska, A.M.; Rudzki, M.; Rudzki, S.; Lewandowski, T.; Laskowska, B. Environmental risk factors for cancer—Review paper. Ann. Agric. Environ. Med. 2019, 26, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Vinay, D.S.; Ryan, E.P.; Pawelec, G.; Talib, W.H.; Stagg, J.; Elkord, E.; Lichtor, T.; Decker, W.K.; Whelan, R.L.; Kumara, H.; et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015, 35, S185–S198. [Google Scholar] [CrossRef]
- Boland, P.; Pavlick, A.C.; Weber, J.; Sandigursky, S. Immunotherapy to treat malignancy in patients with pre-existing autoimmunity. J. Immunother. Cancer 2020, 8, 1–9. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Barbosa, A.M.; Gomes-Gonçalves, A.; Castro, A.G.; Torrado, E. Immune System Efficiency in Cancer and the Microbiota Influence. Pathobiology 2021, 88, 170–186. [Google Scholar] [CrossRef]
- Wu, J.; Wang, S.; Zheng, B.; Qiu, X.; Wang, H.; Chen, L. Modulation of Gut Microbiota to Enhance Effect of Checkpoint Inhibitor Immunotherapy. Front. Immunol. 2021, 12, 669150. [Google Scholar] [CrossRef]
- Boyaka, P.N.; Fujihashi, K. 20-Host Defenses at Mucosal Surfaces. In Clinical Immunology, 5th ed.; Rich, R.R., Fleisher, T.A., Shearer, W.T., Schroeder, H.W., Frew, A.J., Weyand, C.M., Eds.; Elsevier: London, UK, 2019; pp. 285–298.e281. [Google Scholar] [CrossRef]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef]
- Poggi, A.; Benelli, R.; Vene, R.; Costa, D.; Ferrari, N.; Tosetti, F.; Zocchi, M.R. Human Gut-Associated Natural Killer Cells in Health and Disease. Front. Immunol. 2019, 10, 961. [Google Scholar] [CrossRef] [Green Version]
- Elphick, D.A.; Mahida, Y.R. Paneth cells: Their role in innate immunity and inflammatory disease. Gut 2005, 54, 1802–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reboldi, A.; Cyster, J.G. Peyer’s patches: Organizing B-cell responses at the intestinal frontier. Immunol. Rev. 2016, 271, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Shale, M.; Schiering, C.; Powrie, F. CD4(+) T-cell subsets in intestinal inflammation. Immunol. Rev 2013, 252, 164–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konjar, Š.; Ferreira, C.; Blankenhaus, B.; Veldhoen, M. Intestinal Barrier Interactions with Specialized CD8 T Cells. Front. Immunol. 2017, 8, 1281. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Lee, N.; Kim, W.-U. Microbiota in T-cell homeostasis and inflammatory diseases. Exp. Mol. Med. 2017, 49, e340. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Bauer, H.; Horowitz, R.E.; Levenson, S.M.; Popper, H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 1963, 42, 471–483. [Google Scholar]
- Umesaki, Y.; Setoyama, H.; Matsumoto, S.; Okada, Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology 1993, 79, 32–37. [Google Scholar]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Frutos Rde, L.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, E.A.; King, K.Y.; Baldridge, M.T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Phys. 2018, 9, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cording, S.; Fleissner, D.; Heimesaat, M.M.; Bereswill, S.; Loddenkemper, C.; Uematsu, S.; Akira, S.; Hamann, A.; Huehn, J. Commensal microbiota drive proliferation of conventional and Foxp3(+) regulatory CD4(+) T cells in mesenteric lymph nodes and Peyer’s patches. Eur. J. Microbiol. Immunol. 2013, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Xue, H.; Ma, Q.; He, X.; Ma, L.; Shi, B.; Sun, S.; Yao, X. Heterogeneity of CD4(+)CD25(+)Foxp3(+)Treg TCR β CDR3 Repertoire Based on the Differences of Symbiotic Microorganisms in the Gut of Mice. Front. Cell Dev. Biol. 2020, 8, 576445. [Google Scholar] [CrossRef]
- Wan, Y.Y.; Flavell, R.A. Yin-Yang functions of transforming growth factor-beta and T regulatory cells in immune regulation. Immunol. Rev. 2007, 220, 199–213. [Google Scholar] [CrossRef]
- Hsu, P.; Santner-Nanan, B.; Hu, M.; Skarratt, K.; Lee, C.H.; Stormon, M.; Wong, M.; Fuller, S.J.; Nanan, R. IL-10 Potentiates Differentiation of Human Induced Regulatory T Cells via STAT3 and Foxo1. J. Immunol. 2015, 195, 3665. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef] [Green Version]
- Erturk-Hasdemir, D.; Oh Sungwhan, F.; Okan Nihal, A.; Stefanetti, G.; Gazzaniga Francesca, S.; Seeberger Peter, H.; Plevy Scott, E.; Kasper Dennis, L. Symbionts exploit complex signaling to educate the immune system. Proc. Nat. Acad. Sci. USA 2019, 116, 26157–26166. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Nat. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Vieira, R.S.; Castoldi, A.; Basso, P.J.; Hiyane, M.I.; Câmara, N.O.S.; Almeida, R.R. Butyrate Attenuates Lung Inflammation by Negatively Modulating Th9 Cells. Front. Immunol. 2019, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gallausiaux, C.; Beguet-Crespel, F.; Marinelli, L.; Jamet, A.; Ledue, F.; Blottiere, H.M.; Lapaque, N. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018, 8, 9742. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Chen, L.; Yang, W.; Huang, X.; Ma, C.; Chen, F.; Xiao, Y.; Zhao, Y.; Ma, C.; et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018, 9, 3555. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Sun, M.; Wu, W.; Yang, W.; Huang, X.; Xiao, Y.; Ma, C.; Xu, L.; Yao, S.; Liu, Z.; et al. Microbiota Metabolite Butyrate Differentially Regulates Th1 and Th17 Cells’ Differentiation and Function in Induction of Colitis. Inflamm. Bowel Dis. 2019, 25, 1450–1461. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, A.; Nakatani, A.; Hasegawa, S.; Irie, J.; Ozawa, K.; Tsujimoto, G.; Suganami, T.; Itoh, H.; Kimura, I. The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS ONE 2017, 12, e0179696. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Guan, X.; Chen, D.; Ma, W. The Th17/Treg Cell Balance: A Gut Microbiota-Modulated Story. Microorganisms 2019, 7, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Ma, X.; Liu, P.; Ge, W.; Hu, L.; Zuo, Z.; Xiao, H.; Liao, W. Treatment and mechanism of fecal microbiota transplantation in mice with experimentally induced ulcerative colitis. Exp. Biol. Med. 2021, 246, 1563–1575. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Levy, M.; Pevsner-Fishcer, M.; Elinav, E. Inflammasomes and intestinal inflammation. Mucosal Immunol. 2017, 10, 865–883. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Kern, L.; Elinav, E. The NLRP6 inflammasome. Immunology 2021, 162, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalova, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef] [Green Version]
- Pedicord, V.A.; Lockhart, A.A.K.; Rangan, K.J.; Craig, J.W.; Loschko, J.; Rogoz, A.; Hang, H.C.; Mucida, D. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci. Immunol. 2016, 1, eaai7732. [Google Scholar] [CrossRef] [Green Version]
- Flannigan, K.L.; Denning, T.L. Segmented filamentous bacteria-induced immune responses: A balancing act between host protection and autoimmunity. Immunology 2018, 154, 537–546. [Google Scholar] [CrossRef] [Green Version]
- Gorjifard, S.; Goldszmid, R.S. Microbiota-myeloid cell crosstalk beyond the gut. J. Leukoc. Biol. 2016, 100, 865–879. [Google Scholar] [CrossRef]
- Sankaran-Walters, S.; Hart, R.; Dills, C. Guardians of the Gut: Enteric Defensins. Front. Microbiol. 2017, 8, 647. [Google Scholar] [CrossRef] [Green Version]
- Majumdar, S.; Ruiz, D.Z.; Wu, W.J.; Orozco, S.; Bettini, M.; Diehl, G.; Bettini, M.L. Regulation of thymic T-cell development by intestinal microbiota. J. Immunol. 2020, 204, 84–85. [Google Scholar]
- Odamaki, T.; Kato, K.; Sugahara, H.; Hashikura, N.; Takahashi, S.; Xiao, J.Z.; Abe, F.; Osawa, R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016, 16, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Wang, Y.; Li, H.; Dai, Y.; Chen, D.; Wang, M.; Jiang, X.; Huang, Z.; Yu, H.; Huang, J.; et al. Altered Fecal Microbiota Composition in Older Adults With Frailty. Front. Cell Infect. Microbiol. 2021, 11, 696186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Li, S.; Gan, R.Y.; Zhou, T.; Xu, D.P.; Li, H.B. Impacts of gut bacteria on human health and diseases. Int J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, L.; Hu, D.; Zeng, S.; Han, Y.; Shen, H. The role of the tumor microbe microenvironment in the tumor immune microenvironment: Bystander, activator, or inhibitor? J. Exp. Clin. Cancer Res. CR 2021, 40, 327. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. New Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balachandran, V.P.; Łuksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef]
- Bessell, C.A.; Isser, A.; Havel, J.J.; Lee, S.; Bell, D.R.; Hickey, J.W.; Chaisawangwong, W.; Glick Bieler, J.; Srivastava, R.; Kuo, F.; et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. JCI Insight 2020, 5, 1–19. [Google Scholar] [CrossRef]
- Fluckiger, A.; Daillère, R.; Sassi, M.; Sixt, B.S.; Liu, P.; Loos, F.; Richard, C.; Rabu, C.; Alou, M.T.; Goubet, A.G.; et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 2020, 369, 936–942. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef]
- Lopès, A.; Billard, E.; Casse, A.H.; Villéger, R.; Veziant, J.; Roche, G.; Carrier, G.; Sauvanet, P.; Briat, A.; Pagès, F.; et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int. J. Cancer 2020, 146, 3147–3159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- do Vale, A.; Cabanes, D.; Sousa, S. Bacterial Toxins as Pathogen Weapons Against Phagocytes. Front. Microbiol. 2016, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Barth, H.; Fischer, S.; Möglich, A.; Förtsch, C. Clostridial C3 Toxins Target Monocytes/Macrophages and Modulate Their Functions. Front. Immunol. 2015, 6, 339. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Kato, I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Zhou, R.; Wang, H.; Li, W.; Pan, M.; Yao, X.; Zhan, W.; Yang, S.; Xu, L.; Ding, Y.; et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019, 26, 2447–2463. [Google Scholar] [CrossRef] [PubMed]
- Nakkarach, A.; Foo, H.L.; Song, A.A.-L.; Mutalib, N.E.A.; Nitisinprasert, S.; Withayagiat, U. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. Microb. Cell Fact. 2021, 20, 36. [Google Scholar] [CrossRef]
- Park, J.; Goergen, C.J.; HogenEsch, H.; Kim, C.H. Chronically Elevated Levels of Short-Chain Fatty Acids Induce T Cell-Mediated Ureteritis and Hydronephrosis. J. Immunol. 2016, 196, 2388–2400. [Google Scholar] [CrossRef] [Green Version]
- Ryu, T.Y.; Kim, K.; Han, T.S.; Lee, M.O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.J.; An, D.M.; Jung, C.R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022, 16, 1205–1221. [Google Scholar] [CrossRef]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef]
- Mikó, E.; Vida, A.; Kovács, T.; Ujlaki, G.; Trencsényi, G.; Márton, J.; Sári, Z.; Kovács, P.; Boratkó, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta. Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J. Transl. Med. 2012, 10, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engevik, M.A.; Danhof, H.A.; Ruan, W.; Engevik, A.C.; Chang-Graham, A.L.; Engevik, K.A.; Shi, Z.; Zhao, Y.; Brand, C.K.; Krystofiak, E.S.; et al. Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. MBio 2021, 12, e02706-20. [Google Scholar] [CrossRef]
- Cuellar-Gómez, H.; Ocharán-Hernández, M.E.; Calzada-Mendoza, C.C.; Comoto-Santacruz, D.A. Association of Fusobacterium nucleatum infection and colorectal cancer: A Mexican study. Rev. Gastroenterol. Mex. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Merino, J.; Isla, B.; Combes, T.; Martinez-Estrada, F.; Maluquer De Motes, C. Beneficial bacteria activate type-I interferon production via the intracellular cytosolic sensors STING and MAVS. Gut Microb. 2020, 11, 771–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tikka, C.; Manthari, R.K.; Ommati, M.M.; Niu, R.; Sun, Z.; Zhang, J.; Wang, J. Immune disruption occurs through altered gut microbiome and NOD2 in arsenic induced mice: Correlation with colon cancer markers. Chemosphere 2020, 246, 125791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Gou, Y.; Liang, S.; Chen, N.; Liu, Y.; He, Q.; Zhang, J. Dysbiosis of Gut Microbiota Promotes Hepatocellular Carcinoma Progression by Regulating the Immune Response. J. Immunol. Res. 2021, 2021, 4973589. [Google Scholar] [CrossRef]
- Zhou, J.; Boutros, M. JNK-dependent intestinal barrier failure disrupts host-microbe homeostasis during tumorigenesis. Proc. Nat. Acad. Sci. USA 2020, 117, 9401–9412. [Google Scholar] [CrossRef] [Green Version]
- Szczyrek, M.; Bitkowska, P.; Chunowski, P.; Czuchryta, P.; Krawczyk, P.; Milanowski, J. Diet, Microbiome, and Cancer Immunotherapy-A Comprehensive Review. Nutrients 2021, 13, 2217. [Google Scholar] [CrossRef]
- Vieira, A.T.; Galvão, I.; Amaral, F.A.; Teixeira, M.M.; Nicoli, J.R.; Martins, F.S. Oral treatment with Bifidobacterium longum 51A reduced inflammation in a murine experimental model of gout. Benef. Microb. 2015, 6, 799–806. [Google Scholar] [CrossRef]
- Vieira, A.T.; Rocha, V.M.; Tavares, L.; Garcia, C.C.; Teixeira, M.M.; Oliveira, S.C.; Cassali, G.D.; Gamba, C.; Martins, F.S.; Nicoli, J.R. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 5(1A). Microb. Infect. 2016, 18, 180–189. [Google Scholar] [CrossRef]
- Lakritz, J.R.; Poutahidis, T.; Levkovich, T.; Varian, B.J.; Ibrahim, Y.M.; Chatzigiagkos, A.; Mirabal, S.; Alm, E.J.; Erdman, S.E. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. Int. J. Cancer 2014, 135, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.Y.; Xu, J.Y.; Shi, X.Y.; Huang, W.; Ruan, T.Y.; Xie, P.; Ding, J.L. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. 2013, 93, 844–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.G.; Udayanga, K.G.; Totsuka, N.; Weinberg, J.B.; Núñez, G.; Shibuya, A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE₂. Cell Host Microbe 2014, 15, 95–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, Y.; Ji, Q. Tumor-Associated Macrophages Regulate PD-1/PD-L1 Immunosuppression. Front. Immunol. 2022, 13, 874589. [Google Scholar] [CrossRef] [PubMed]
- Hezaveh, K.; Shinde, R.S.; Klötgen, A.; Halaby, M.J.; Lamorte, S.; Ciudad, M.T.; Quevedo, R.; Neufeld, L.; Liu, Z.Q.; Jin, R.; et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 2022, 55, 324–340.e328. [Google Scholar] [CrossRef]
- Thiele Orberg, E.; Fan, H.; Tam, A.J.; Dejea, C.M.; Destefano Shields, C.E.; Wu, S.; Chung, L.; Finard, B.B.; Wu, X.; Fathi, P.; et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017, 10, 421–433. [Google Scholar] [CrossRef] [Green Version]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Lin, S.J.; Yan, D.C.; Lee, W.I.; Kuo, M.L.; Hsiao, H.S.; Lee, P.Y. Effect of azithromycin on natural killer cell function. Int. Immunopharmacol. 2012, 13, 8–14. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Xiao, X.; Ren, J.; Tang, Y.; Weng, H.; Yang, Q.; Wu, M.; Tang, W. Proteomic analysis of bladder cancer indicates Prx-I as a key molecule in BI-TK/GCV treatment system. PLoS ONE 2014, 9, e98764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.; Kim, G.; Jeon, B.N.; Fang, S.; Park, H. Bifidobacterium Strain-Specific Enhances the Efficacy of Cancer Therapeutics in Tumor-Bearing Mice. Cancers 2021, 13, 957. [Google Scholar] [CrossRef] [PubMed]
- Demin, N.N.; Karmanova, I.G.; Rubinskaya, N.L.; Khomutetskaya, O.E. Comparative neurochemical and physiological characteristics of catalepsy-type rest and sleep. Neurosci. Behav. Physiol. 1978, 9, 98–102. [Google Scholar] [CrossRef]
- Li, Y.; Tinoco, R.; Elmen, L.; Segota, I.; Xian, Y.; Fujita, Y.; Sahu, A.; Zarecki, R.; Marie, K.; Feng, Y.; et al. Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5(-/-) mice. Nat. Commun. 2019, 10, 1492. [Google Scholar] [CrossRef] [Green Version]
- Russell, B.L.; Sooklal, S.A.; Malindisa, S.T.; Daka, L.J.; Ntwasa, M. The Tumor Microenvironment Factors That Promote Resistance to Immune Checkpoint Blockade Therapy. Front. Oncol. 2021, 11, 641428. [Google Scholar] [CrossRef]
- Ghosh, C.; Luong, G.; Sun, Y. A snapshot of the PD-1/PD-L1 pathway. J. Cancer 2021, 12, 2735–2746. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Wojtukiewicz, M.Z.; Rek, M.M.; Karpowicz, K.; Górska, M.; Polityńska, B.; Wojtukiewicz, A.M.; Moniuszko, M.; Radziwon, P.; Tucker, S.C.; Honn, K.V. Inhibitors of immune checkpoints-PD-1, PD-L1, CTLA-4-new opportunities for cancer patients and a new challenge for internists and general practitioners. Cancer Metastasis Rev. 2021, 40, 949–982. [Google Scholar] [CrossRef]
- Dai, Z.; Zhang, J.; Wu, Q.; Fang, H.; Shi, C.; Li, Z.; Lin, C.; Tang, D.; Wang, D. Intestinal microbiota: A new force in cancer immunotherapy. Cell Commun. Signal. 2020, 18, 90. [Google Scholar] [CrossRef]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, T.; Liang, J.; Huang, Q.; Huang, J.D.; Ke, Y.; Sun, H. Current status of intratumour microbiome in cancer and engineered exogenous microbiota as a promising therapeutic strategy. Biomed. Pharmacother. 2022, 145, 112443. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalaora, S.; Lee, J.S.; Barnea, E.; Levy, R.; Greenberg, P.; Alon, M.; Yagel, G.; Bar Eli, G.; Oren, R.; Peri, A.; et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat. Commun. 2020, 11, 896. [Google Scholar] [CrossRef] [Green Version]
- Norton, J.E., Jr.; Kommineni, S.; Akrivoulis, P.; Gutierrez, D.A.; Hazuda, D.J.; Swaminathan, G. Primary Human Dendritic Cells and Whole-Blood Based Assays to Evaluate Immuno-Modulatory Properties of Heat-Killed Commensal Bacteria. Vaccines 2021, 9, 225. [Google Scholar] [CrossRef]
- Montalban-Arques, A.; Katkeviciute, E.; Busenhart, P.; Bircher, A.; Wirbel, J.; Zeller, G.; Morsy, Y.; Borsig, L.; Glaus Garzon, J.F.; Müller, A.; et al. Commensal Clostridiales strains mediate effective anti-cancer immune response against solid tumors. Cell Host Microbe 2021, 29, 1573–1588.e1577. [Google Scholar] [CrossRef]
- Inamura, K. Roles of microbiota in response to cancer immunotherapy. Semin. Cancer Biol. 2020, 65, 164–175. [Google Scholar] [CrossRef]
- Routy, B.; Gopalakrishnan, V.; Daillere, R.; Zitvogel, L.; Wargo, J.A.; Kroemer, G. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 2018, 15, 382–396. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Wu, N.; Yang, X.; Zhang, R.; Li, J.; Xiao, X.; Hu, Y.; Chen, Y.; Yang, F.; Lu, N.; Wang, Z.; et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013, 66, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCulloch, J.A.; Davar, D.; Rodrigues, R.R.; Badger, J.H.; Fang, J.R.; Cole, A.M.; Balaji, A.K.; Vetizou, M.; Prescott, S.M.; Fernandes, M.R.; et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 2022, 28, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Thomas, A.M.; Bolte, L.A.; Bjork, J.R.; de Ruijter, L.K.; Armanini, F.; Asnicar, F.; Blanco-Miguez, A.; Board, R.; Calbet-Llopart, N.; et al. Cross-cohort gut microbiome associations with immune checkpoint inhibitor response in advanced melanoma. Nat. Med. 2022, 28, 535–544. [Google Scholar] [CrossRef]
- Sims, T.T.; El Alam, M.B.; Karpinets, T.V.; Dorta-Estremera, S.; Hegde, V.L.; Nookala, S.; Yoshida-Court, K.; Wu, X.; Biegert, G.W.G.; Delgado Medrano, A.Y.; et al. Gut microbiome diversity is an independent predictor of survival in cervical cancer patients receiving chemoradiation. Commun. Biol. 2021, 4, 237. [Google Scholar] [CrossRef]
- Salgia, N.J.; Bergerot, P.G.; Maia, M.C.; Dizman, N.; Hsu, J.; Gillece, J.D.; Folkerts, M.; Reining, L.; Trent, J.; Highlander, S.K.; et al. Stool Microbiome Profiling of Patients with Metastatic Renal Cell Carcinoma Receiving Anti-PD-1 Immune Checkpoint Inhibitors. Eur. Urol. 2020, 78, 498–502. [Google Scholar] [CrossRef]
- Dizman, N.; Meza, L.; Bergerot, P.; Alcantara, M.; Dorff, T.; Lyou, Y.; Frankel, P.; Cui, Y.; Mira, V.; Llamas, M.; et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: A randomized phase 1 trial. Nat. Med. 2022, 28, 704–712. [Google Scholar] [CrossRef]
- Takada, K.; Shimokawa, M.; Takamori, S.; Shimamatsu, S.; Hirai, F.; Tagawa, T.; Okamoto, T.; Hamatake, M.; Tsuchiya-Kawano, Y.; Otsubo, K.; et al. Clinical impact of probiotics on the efficacy of anti-PD-1 monotherapy in patients with nonsmall cell lung cancer: A multicenter retrospective survival analysis study with inverse probability of treatment weighting. Int. J. Cancer 2021, 149, 473–482. [Google Scholar] [CrossRef]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Khan, M.A.W.; Zhang, X.; White, M.G.; Peterson, C.B.; et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Thomas, A.M.; Iebba, V.; Zalcman, G.; Friard, S.; Mazieres, J.; Audigier-Valette, C.; Moro-Sibilot, D.; Goldwasser, F.; et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 2022, 28, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Shen, P.; Shan, Y.; Yang, Y.; Deng, R.; Chen, W.; Lu, Y.; Wei, Z. Gut Microbiota-Mediated Modulation of Cancer Progression and Therapy Efficacy. Front. Cell Dev. Biol. 2021, 9, 626045. [Google Scholar] [CrossRef] [PubMed]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e712. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef] [Green Version]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Griffin, M.E.; Espinosa, J.; Becker, J.L.; Luo, J.D.; Carroll, T.S.; Jha, J.K.; Fanger, G.R.; Hang, H.C. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 2021, 373, 1040–1046. [Google Scholar] [CrossRef]
- Lee, K.A.; Luong, M.K.; Shaw, H.; Nathan, P.; Bataille, V.; Spector, T.D. The gut microbiome: What the oncologist ought to know. Br. J. Cancer 2021, 125, 1197–1209. [Google Scholar] [CrossRef]
- Cheng, W.Y.; Wu, C.Y.; Yu, J. The role of gut microbiota in cancer treatment: Friend or foe? Gut 2020, 69, 1867–1876. [Google Scholar] [CrossRef]
- Zackular, J.P.; Baxter, N.T.; Chen, G.Y.; Schloss, P.D. Manipulation of the Gut Microbiota Reveals Role in Colon Tumorigenesis. MSphere 2016, 1, e00001-15. [Google Scholar] [CrossRef] [Green Version]
- Imai, H.; Saijo, K.; Komine, K.; Yoshida, Y.; Sasaki, K.; Suzuki, A.; Ouchi, K.; Takahashi, M.; Takahashi, S.; Shirota, H.; et al. Antibiotics Improve the Treatment Efficacy of Oxaliplatin-Based but Not Irinotecan-Based Therapy in Advanced Colorectal Cancer Patients. J. Oncol. 2020, 2020, 1701326. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Eun, C.S.; Jo, S.V.; Lee, A.R.; Park, C.H.; Han, D.S. The impact of gut microbiota manipulation with antibiotics on colon tumorigenesis in a murine model. PLoS ONE 2019, 14, e0226907. [Google Scholar] [CrossRef]
- Patel, P.; Poudel, A.; Kafle, S.; Thapa Magar, M.; Cancarevic, I. Influence of Microbiome and Antibiotics on the Efficacy of Immune Checkpoint Inhibitors. Cureus 2021, 13, e16829. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhuang, T.; Shao, E.; Liu, Y.; He, H.; Shu, Z.; Huang, Y.; Yao, Y.; Lin, S.; Lin, S.; et al. Association of antibiotic exposure with the mortality in metastatic colorectal cancer patients treated with bevacizumab-containing chemotherapy: A hospital-based retrospective cohort study. PLoS ONE 2019, 14, e0221964. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Z.; Gao, P.; Song, Y.X.; Xu, Y.; Sun, J.X.; Chen, X.W.; Zhao, J.H.; Wang, Z.N. Antibiotic use and the efficacy of immune checkpoint inhibitors in cancer patients: A pooled analysis of 2740 cancer patients. Oncoimmunology 2019, 8, e1665973. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Liu, J.; Wu, S.; Xie, X. The impact of antibiotics on efficacy of immune checkpoint inhibitors in malignancies: A study based on 44 cohorts. Int. Immunopharmacol. 2021, 92, 107303. [Google Scholar] [CrossRef]
- Kim, H.; Lee, J.E.; Hong, S.H.; Lee, M.A.; Kang, J.H.; Kim, I.H. The effect of antibiotics on the clinical outcomes of patients with solid cancers undergoing immune checkpoint inhibitor treatment: A retrospective study. BMC Cancer 2019, 19, 1100. [Google Scholar] [CrossRef]
- Fessas, P.; Naeem, M.; Pinter, M.; Marron, T.U.; Szafron, D.; Balcar, L.; Saeed, A.; Jun, T.; Dharmapuri, S.; Gampa, A.; et al. Early Antibiotic Exposure Is Not Detrimental to Therapeutic Effect from Immunotherapy in Hepatocellular Carcinoma. Liver Cancer 2021, 10, 583–592. [Google Scholar] [CrossRef]
- Chen, L.; Zhai, Y.; Wang, Y.; Fearon, E.R.; Núñez, G.; Inohara, N.; Cho, K.R. Altering the Microbiome Inhibits Tumorigenesis in a Mouse Model of Oviductal High-Grade Serous Carcinoma. Cancer Res. 2021, 81, 3309–3318. [Google Scholar] [CrossRef]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A.; et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients with Cancer. JAMA Oncol. 2019, 5, 1774–1778. [Google Scholar] [CrossRef]
- Khan, U.; Ho, K.; Hwang, E.K.; Peña, C.; Brouwer, J.; Hoffman, K.; Betel, D.; Sonnenberg, G.F.; Faltas, B.; Saxena, A.; et al. Impact of Use of Antibiotics on Response to Immune Checkpoint Inhibitors and Tumor Microenvironment. Am. J. Clin. Oncol. 2021, 44, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Tsikala-Vafea, M.; Belani, N.; Vieira, K.; Khan, H.; Farmakiotis, D. Use of antibiotics is associated with worse clinical outcomes in patients with cancer treated with immune checkpoint inhibitors: A systematic review and meta-analysis. Int. J. Infect. Dis. 2021, 106, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and Anticancer Immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef] [Green Version]
- Bachem, A.; Makhlouf, C.; Binger, K.J.; de Souza, D.P.; Tull, D.; Hochheiser, K.; Whitney, P.G.; Fernandez-Ruiz, D.; Dähling, S.; Kastenmüller, W.; et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8(+) T Cells. Immunity 2019, 51, 285–297.e285. [Google Scholar] [CrossRef]
- Finlay, B.B.; Goldszmid, R.; Honda, K.; Trinchieri, G.; Wargo, J.; Zitvogel, L. Can we harness the microbiota to enhance the efficacy of cancer immunotherapy? Nat. Rev. Immunol. 2020, 20, 522–528. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aghamajidi, A.; Maleki Vareki, S. The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer. Cancers 2022, 14, 3563. https://doi.org/10.3390/cancers14153563
Aghamajidi A, Maleki Vareki S. The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer. Cancers. 2022; 14(15):3563. https://doi.org/10.3390/cancers14153563
Chicago/Turabian StyleAghamajidi, Azin, and Saman Maleki Vareki. 2022. "The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer" Cancers 14, no. 15: 3563. https://doi.org/10.3390/cancers14153563
APA StyleAghamajidi, A., & Maleki Vareki, S. (2022). The Effect of the Gut Microbiota on Systemic and Anti-Tumor Immunity and Response to Systemic Therapy against Cancer. Cancers, 14(15), 3563. https://doi.org/10.3390/cancers14153563





