A Pilot Study: Favorable Effects of Clostridium butyricum on Intestinal Microbiota for Adjuvant Therapy of Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
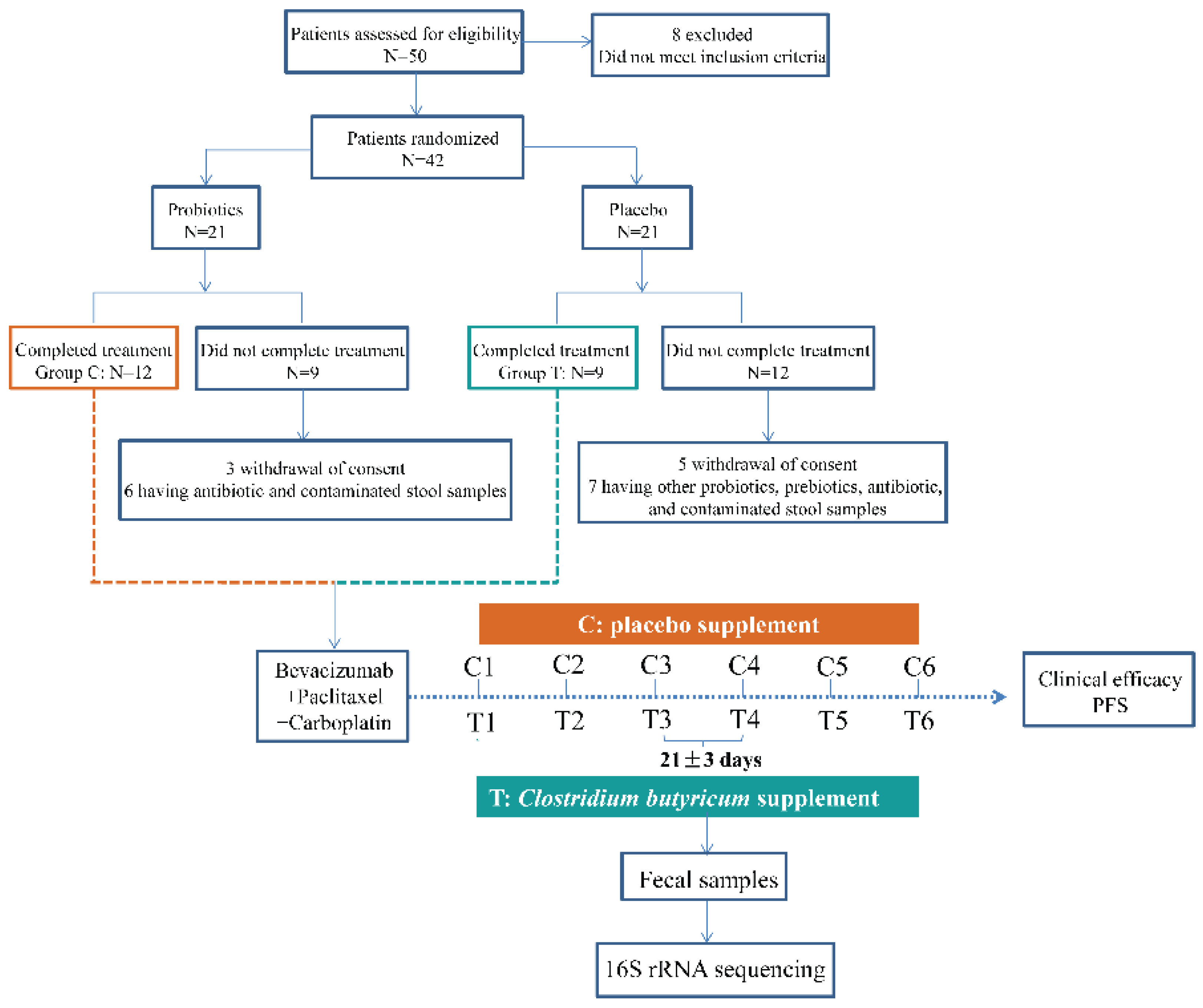
2.1. Patients and Medications
2.2. Fecal Samples Collection
2.3. DNA Extraction and Gene Amplicon Sequencing
2.4. Sequencing Processing
2.5. Network Analysis
2.6. Statistical Analysis
3. Results
3.1. Dynamic Taxonomical Composition Changes of Intestinal Microbiota
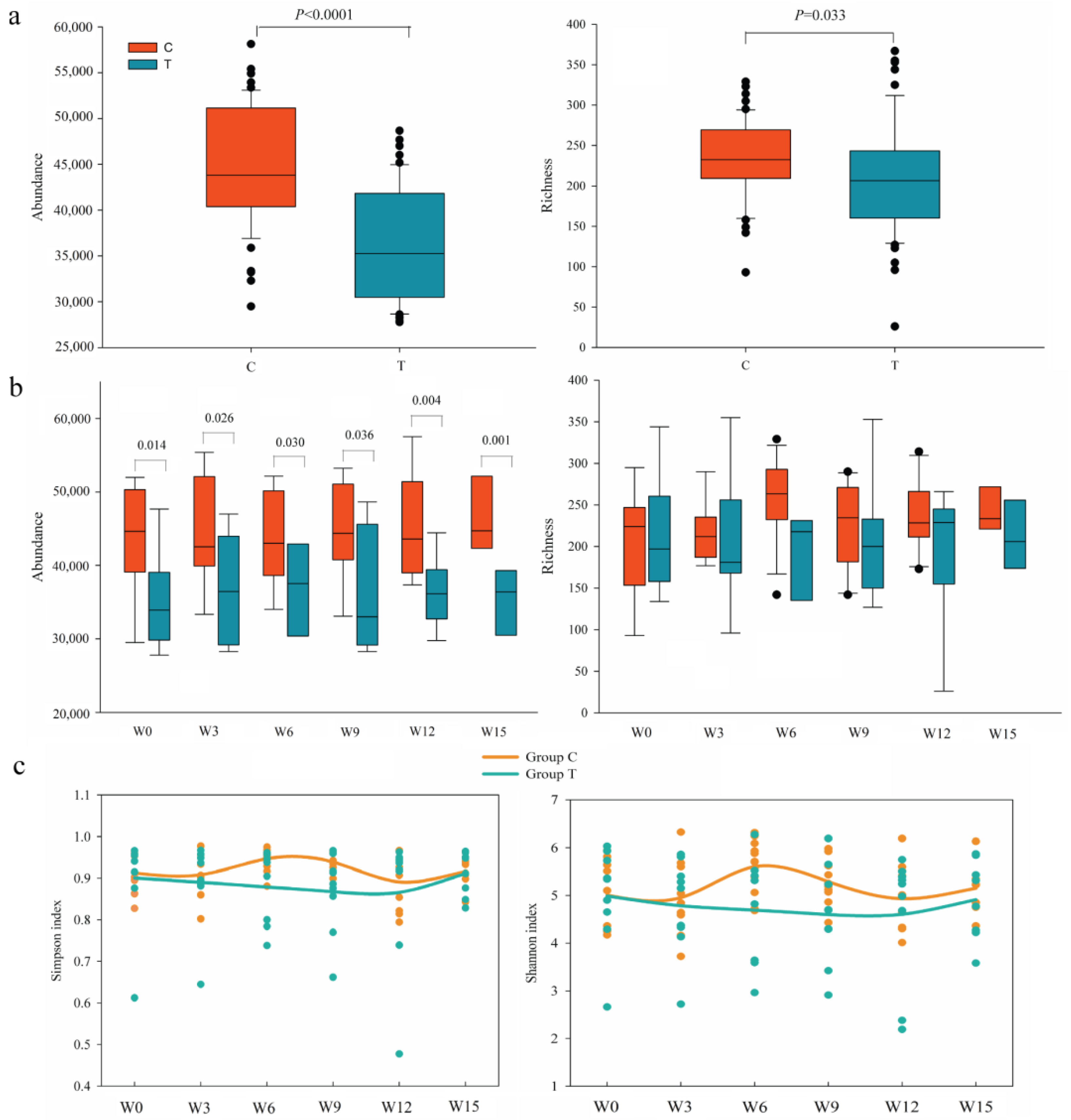
3.2. Dynamic Diversity Changes of Intestinal Microbiota
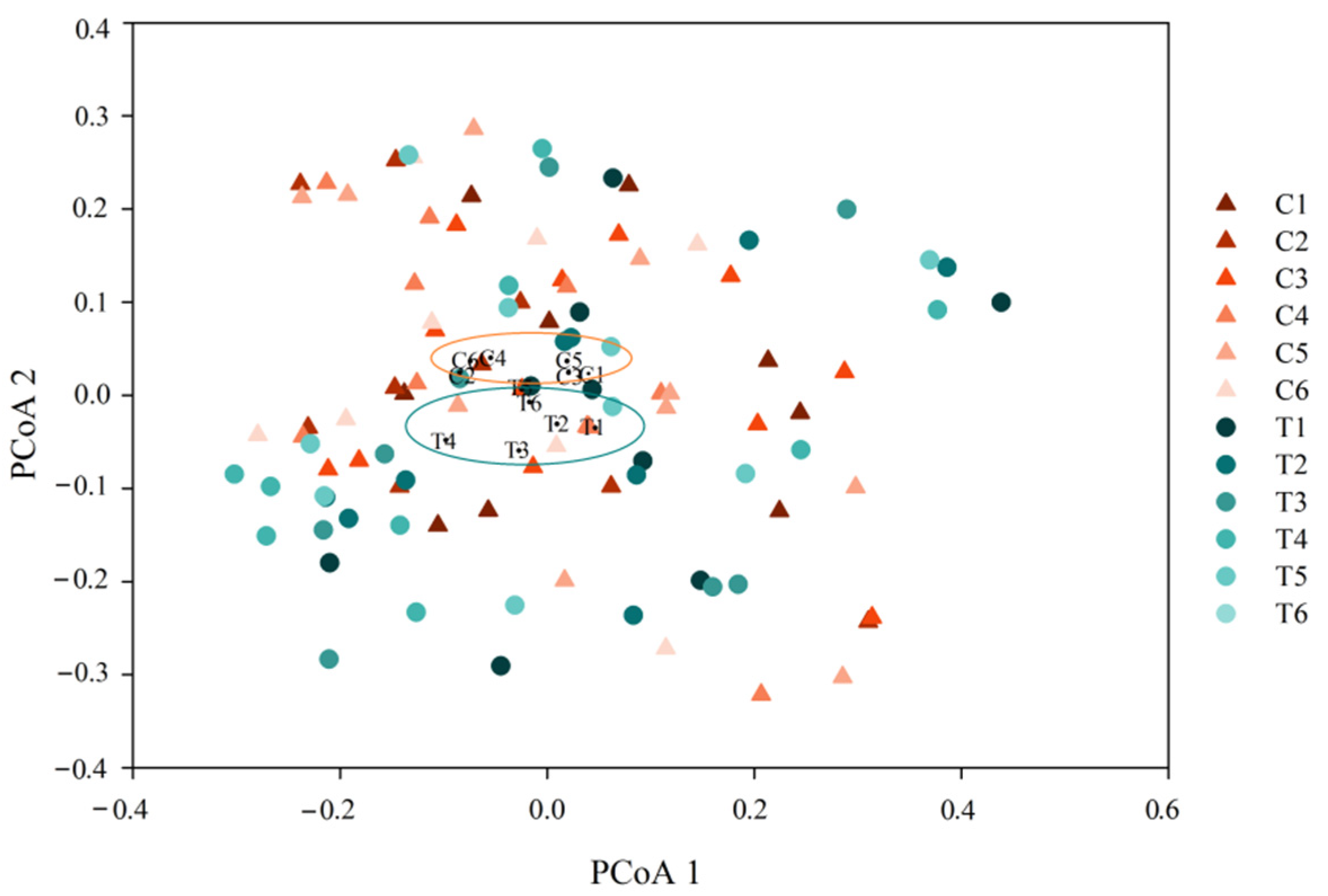
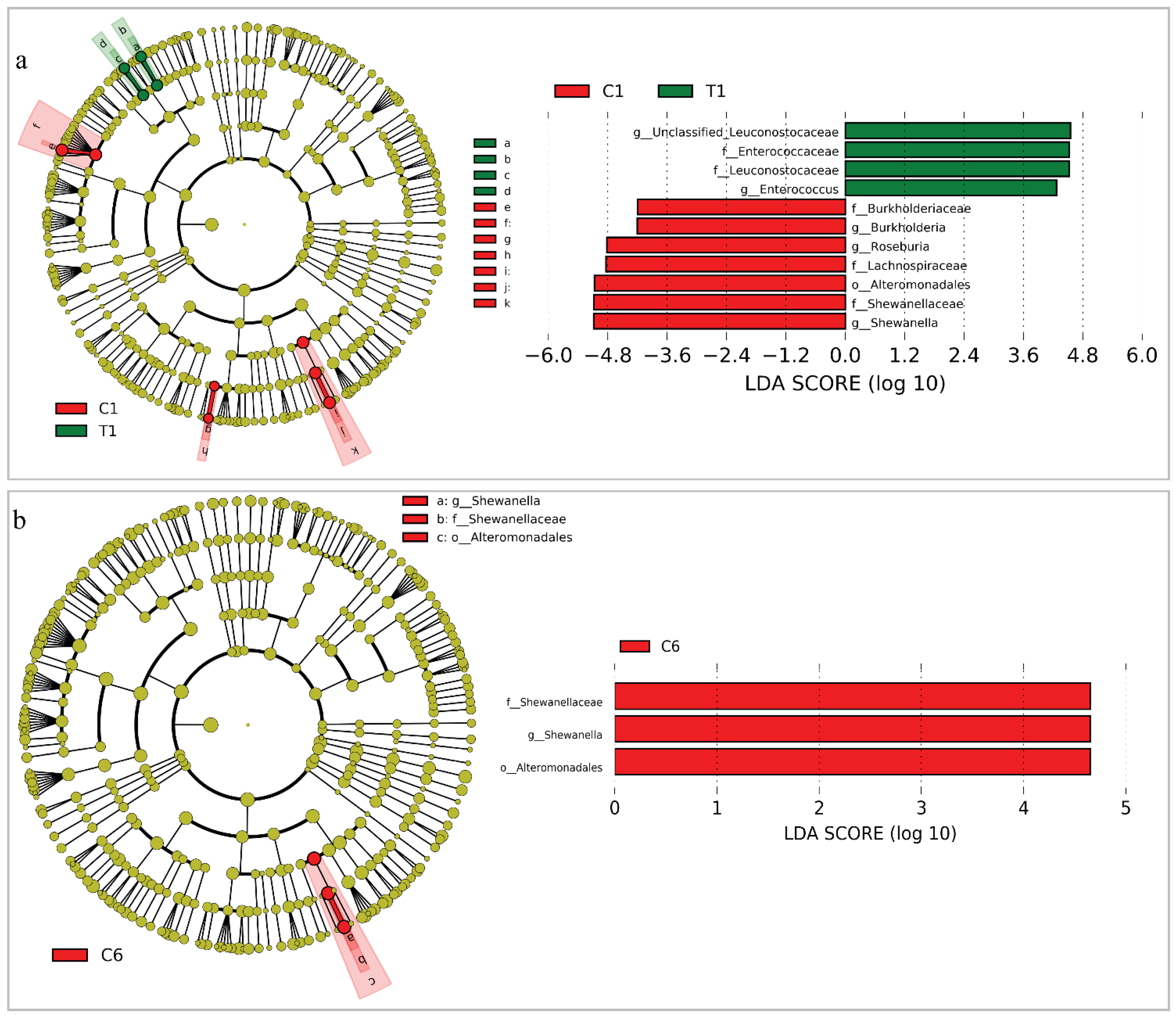
3.3. Changes of the Whole Community Structure and Potential Biomarkers of Intestinal Microbiota
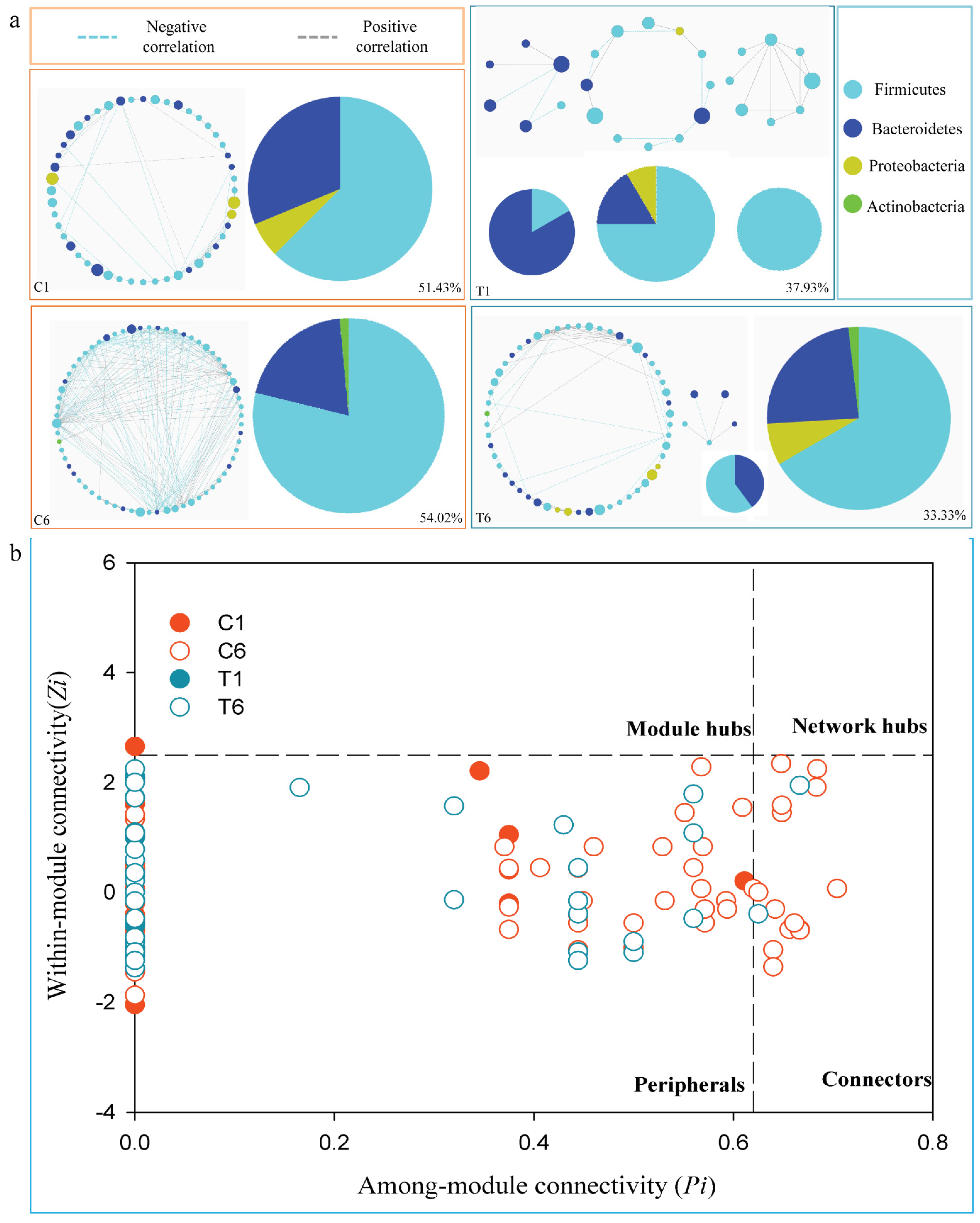
3.4. Dynamic Changes of Phylogenetic Interactional Network of Intestinal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.; Chirieac, L.R.; D’Amico, T.A.; DeCamp, M.M.; Dilling, T.J.; Dobelbower, M.; et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer NE 2017, 15, 504–535. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wu, Y.L.; Chen, G.; Liu, X.; Zhu, Y.; Lu, S.; Feng, J.; He, J.; Han, B.; Wang, J.; et al. BEYOND: A Randomized, Double-Blind, Placebo-Controlled, Multicenter, Phase III Study of First-Line Carboplatin/Paclitaxel Plus Bevacizumab or Placebo in Chinese Patients with Advanced or Recurrent Nonsquamous Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2015, 33, 2197–2204. [Google Scholar] [CrossRef]
- Sandler, A.; Gray, R.; Perry, M.C.; Brahmer, J.; Schiller, J.H.; Dowlati, A.; Lilenbaum, R.; Johnson, D.H. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N. Engl. J. Med. 2006, 355, 2542–2550. [Google Scholar] [CrossRef] [Green Version]
- Reck, M.; von Pawel, J.; Zatloukal, P.; Ramlau, R.; Gorbounova, V.; Hirsh, V.; Leighl, N.; Mezger, J.; Archer, V.; Moore, N.; et al. Overall survival with cisplatin-gemcitabine and bevacizumab or placebo as first-line therapy for nonsquamous non-small-cell lung cancer: Results from a randomised phase III trial (AVAiL). Ann. Oncol. 2010, 21, 1804–1809. [Google Scholar] [CrossRef]
- Crinò, L.; Dansin, E.; Garrido, P.; Griesinger, F.; Laskin, J.; Pavlakis, N.; Stroiakovski, D.; Thatcher, N.; Tsai, C.M.; Wu, Y.L.; et al. Safety and efficacy of first-line bevacizumab-based therapy in advanced non-squamous non-small-cell lung cancer (SAiL, MO19390): A phase 4 study. Lancet Oncol. 2010, 11, 733–740. [Google Scholar] [CrossRef]
- Sandler, A.B.; Kong, G.; Strickland, D. Treatment outcomes by tumor histology in Eastern Cooperative Group (ECOG) Study E4599 of bevacizumab (BV) with paclitaxel/carboplatin (PC) for advanced non-small cell lung cancer (NSCLC). J. Thorac. Oncol. 2008, 3, S283. [Google Scholar]
- Liu, K.J.; Ding, L.Y.; Wu, H.Y. Bevacizumab in combination with anticancer drugs for previously treated advanced non-small cell lung cancer. Tumour Biol. 2015, 36, 1323–1327. [Google Scholar] [CrossRef] [PubMed]
- Pakkala, S.; Ramalingam, S.S. Combined inhibition of vascular endothelial growth factor and epidermal growth factor signaling in non-small-cell lung cancer therapy. Clin. Lung Cancer 2009, 10, S17–S23. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Ichiki, M.; Takahashi, K.; Hisamatsu, Y.; Takeoka, H.; Azuma, K.; Shukuya, T.; Nishikawa, K.; Tokito, T.; Ishii, H.; et al. A multicenter phase II trial of S-1 combined with bevacizumab after platinum-based chemotherapy in patients with advanced non-squamous non-small cell lung cancer. Cancer Chemother. Pharmacol. 2016, 78, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [Green Version]
- Rajpoot, M.; Sharma, A.K.; Sharma, A.; Gupta, G.K. Understanding the microbiome: Emerging biomarkers for exploiting the microbiota for personalized medicine against cancer. Semin. Cancer Biol. 2018, 52, 1–8. [Google Scholar] [CrossRef]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.M.; Serino, M.; Planas-Fèlix, M. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Santacroce, L.; Charitos, I.A.; Bottalico, L. A successful history: Probiotics and their potential as antimicrobials. Expert Rev. Anti Infect. Ther. 2019, 17, 635–645. [Google Scholar] [CrossRef]
- Ghoddusi, H.B.; Sherburn, R. Preliminary study on the isolation of Clostridium butyricum strains from natural sources in the UK and screening the isolates for presence of the type E botulinal toxin gene. Int. J. Food Microbiol. 2010, 142, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Yamashita, R.; Matsumoto, A.; Mori, T.; Inagaki, T.; Nonogaki, T.; Kuroki, Y.; Higashi, S.; Oka, K.; Takahashi, M.; et al. The impact of probiotic Clostridium butyricum MIYAIRI 588 on murine gut metabolic alterations. J. Infect. Chemother. 2019, 25, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Yamashita, R.; Matsumoto, A.; Mori, T.; Kuroki, Y.; Kudo, H.; Oka, K.; Takahashi, M.; Nonogaki, T.; Yamagishi, Y.; et al. The impact of Clostridium butyricum MIYAIRI 588 on the murine gut microbiome and colonic tissue. Anaerobe 2018, 54, 8–18. [Google Scholar] [CrossRef]
- Jia, L.; Shan, K.; Pan, L.L.; Feng, N.; Lv, Z.; Sun, Y.; Li, J.; Wu, C.; Zhang, H.; Chen, W.; et al. Clostridium butyricum CGMCC0313.1 Protects against Autoimmune Diabetes by Modulating Intestinal Immune Homeostasis and Inducing Pancreatic Regulatory T Cells. Front. Immunol. 2017, 8, 1345. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.Y.; Li, M.; Li, Y.Y.; Li, L.X.; Zhai, W.Z.; Wang, P.; Yang, X.X.; Gu, X.; Song, L.J.; Li, Z.; et al. The effect of Clostridium butyricum on symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: A randomized, double-blind, placebo-controlled trial. Sci. Rep. 2018, 8, 2964. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020, 469, 456–467. [Google Scholar] [CrossRef]
- Pan, L.L.; Niu, W.; Fang, X.; Liang, W.; Li, H.; Chen, W.; Zhang, H.; Bhatia, M.; Sun, J. Clostridium butyricum Strains Suppress Experimental Acute Pancreatitis by Maintaining Intestinal Homeostasis. Mol. Nutr. Food Res. 2019, 63, e1801419. [Google Scholar] [CrossRef]
- Tomita, Y.; Ikeda, T.; Sakata, S.; Saruwatari, K.; Sato, R.; Iyama, S.; Jodai, T.; Akaike, K.; Ishizuka, S.; Saeki, S.; et al. Association of Probiotic Clostridium butyricum Therapy with Survival and Response to Immune Checkpoint Blockade in Patients with Lung Cancer. Cancer Immunol. Res. 2020, 8, 1236–1242. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, H.; Chen, J.; Shang, S.; Wei, Q.; Yan, J.; Tu, X. Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3-V4 region of the 16S rRNA gene. Appl. Microbiol. Biot. 2016, 100, 3577–3586. [Google Scholar] [CrossRef]
- Wu, L.; Wen, C.; Qin, Y.; Yin, H.; Tu, Q.; Van Nostrand, J.D.; Yuan, T.; Yuan, M.; Deng, Y.; Zhou, J. Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 2015, 15, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhou, X.; Hale, L. Climate warming accelerates temporal scaling of grassland soil microbial biodiversity. Nat. Ecol. Evol. 2019, 3, 612–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, J.; Zhu, H.; Liu, D.; Li, T.; Zhang, C.; Zhu, J.; Lv, H.; Liu, K.; Hao, C.; Tian, Z.; et al. A Pilot Study: Changes of Gut Microbiota in Post-surgery Colorectal Cancer Patients. Front. Microbiol. 2018, 9, 2777. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011, 98, 152–153. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Cong, J.; Zhu, J.; Zhang, C.; Li, T.; Liu, K.; Liu, D.; Zhou, N.; Jiang, M.; Hou, H.; Zhang, X. Chemotherapy Alters the Phylogenetic Molecular Ecological Networks of Intestinal Microbial Communities. Front. Microbiol. 2019, 10, 1008. [Google Scholar] [CrossRef] [Green Version]
- Guimerà, R.; Nunes Amaral, L.A. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Wang, H.; Zhang, G.; Yan, X.; Ma, Z. Efficacy and Safety of Bevacizumab Combined with Chemotherapy as Second-line or Later-line Treatment in Advanced Nonsquamous Non-small Cell Lung Cancer. Chin. J. Lung Cancer 2018, 21, 513–518. [Google Scholar]
- Besse, B.; Le Moulec, S.; Mazières, J.; Senellart, H.; Barlesi, F.; Chouaid, C.; Dansin, E.; Bérard, H.; Falchero, L.; Gervais, R.; et al. Bevacizumab in Patients with Nonsquamous Non-Small Cell Lung Cancer and Asymptomatic, Untreated Brain Metastases (BRAIN): A Nonrandomized, Phase II Study. Clin. Cancer Res. 2015, 21, 1896–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, H.; Meng, Q.; Li, M.; Shi, L.; Tang, J.; Liu, Z. Outcomes of bevacizumab combined with chemotherapy in lung adenocarcinoma-induced malignant pleural effusion. Thorac. Cancer 2018, 9, 298–304. [Google Scholar] [CrossRef] [Green Version]
- Quan, R.; Huang, J.; Chen, N.; Fang, W.; Hu, Z.; Zhan, J.; Zhou, T.; Zhang, L.; Zhang, H. A retrospective analysis of efficacy and safety of adding bevacizumab to chemotherapy as first- and second-line therapy in advanced non-small-cell lung cancer (NSCLC). Tumour Biol. 2016, 37, 11479–11484. [Google Scholar] [CrossRef]
- Hakozaki, T.; Richard, C.; Elkrief, A.; Hosomi, Y.; Benlaïfaoui, M.; Mimpen, I.; Terrisse, S.; Derosa, L. The Gut Microbiome Associates with Immune Checkpoint Inhibition Outcomes in Patients with Advanced Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 1243–1250. [Google Scholar] [CrossRef]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [Green Version]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Guan, Y.; Lou, Y.; Chen, H.; Xu, M.; Deng, D.; Chen, J.; Ni, B.; Zhao, L.; et al. Dysbiosis of the Gut Microbiome is associated with Tumor Biomarkers in Lung Cancer. Int. J. Biol. Sci. 2019, 15, 2381–2392. [Google Scholar] [CrossRef]
- Tiago, S.; Vipul, Y.; Vanessa, Z.; Anders, B.; Bertil, A.; Basit, A.W. On the colonic bacterial metabolism of azo-bonded prodrugsof 5-aminosalicylic acid. J. Pharm. Sci. 2014, 103, 3171–3175. [Google Scholar]
- Haiser, H.J.; Gootenberg, D.B.; Chatman, K.; Sirasani, G.; Balskus, E.P.; Turnbaugh, P.J. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 2013, 341, 295–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, M.; Zimmermann-Kogadeeva, M. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 2019, 363, eaat9931. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, Y.; Fang, K.; Mao, K.; Dou, J.; Fan, H.; Zhou, C.; Wang, H. Lactobacillus acidophilus and Clostridium butyricum ameliorate colitis in murine by strengthening the gut barrier function and decreasing inflammatory factors. Benef. Microbes 2018, 9, 775–787. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, A.; Sato, T.; Kamada, N.; Mikami, Y.; Matsuoka, K.; Hisamatsu, T.; Hibi, T.; Roers, A.; Yagita, H.; Ohteki, T.; et al. A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe 2013, 13, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated with Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients with NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, T.; Tu, X.; Huang, Y.; Zhang, H.; Tan, D.; Jiang, W.; Cai, S.; Zhao, P.; Song, R.; et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 193. [Google Scholar] [CrossRef] [Green Version]
- Hagihara, M.; Kuroki, Y.; Ariyoshi, T.; Higashi, S.; Fukuda, K.; Yamashita, R.; Matsumoto, A.; Mori, T.; Mimura, K.; Yamaguchi, N.; et al. Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. iScience 2020, 23, 100772. [Google Scholar] [CrossRef] [Green Version]
- Airhart, S.; Cade, W.T.; Jiang, H.; Coggan, A.R.; Racette, S.B.; Korenblat, K.; Spearie, C.A.; Waller, S.; O’Connor, R.; Bashir, A.; et al. A Diet Rich in Medium-Chain Fatty Acids Improves Systolic Function and Alters the Lipidomic Profile in Patients with Type 2 Diabetes: A Pilot Study. J. Clin. Endocrinol. Metab. 2016, 101, 504–512. [Google Scholar] [CrossRef] [Green Version]
- Mehta, R.S.; Abu-Ali, G.S.; Drew, D.A.; Lloyd-Price, J.; Subramanian, A.; Lochhead, P.; Joshi, A.D.; Ivey, K.L.; Khalili, H.; Brown, G.T.; et al. Stability of the human faecal microbiome in a cohort of adult men. Nat. Microbiol. 2018, 3, 347–355. [Google Scholar] [CrossRef]
- Scholz, F.; Badgley, B.D.; Sadowsky, M.J.; Kaplan, D.H. Immune mediated shaping of microflora community composition depends on barrier site. PLoS ONE 2014, 9, e84019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Cont. Hosp. EP 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, H.; Cheng, L.; Wang, Y.; Zhang, Y.K.; Zhao, M.F.; Liang, G.D.; Zhang, M.C.; Li, Y.G.; Zhao, J.B.; Gao, Y.N.; et al. Dysbiosis of the Gut Microbiome in Lung Cancer. Front. Cell. Infect. Microbiol. 2019, 9, 112. [Google Scholar] [CrossRef]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Rhodes, K.A.; Schweizer, H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updates 2016, 28, 82–90. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Fang, Y.; Pang, F.; Liang, S.; Lu, X.; Kan, B.; Xu, J.; Zhao, J.; Du, Y.; Wang, D. Rare Shewanella spp. associated with pulmonary and bloodstream infections of cancer patients, China: A case report. BMC Infect. Dis. 2018, 18, 454. [Google Scholar] [CrossRef]
- Yousfi, K.; Bekal, S.; Usongo, V.; Touati, A. Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1353–1362. [Google Scholar] [CrossRef]
| Clinical Parameter | Group T | Group C | p Value |
|---|---|---|---|
| Age | 0.203 | ||
| ≥65 | 2 | 6 | |
| <65 | 7 | 6 | |
| Gender | 0.204 | ||
| Man | 5 | 10 | |
| Woman | 4 | 2 | |
| BMI | |||
| ≥24 | 3 | 4 | 0.596 |
| <24 | 6 | 8 | |
| Smoking | 0.204 | ||
| Yes | 5 | 10 | |
| No | 4 | 2 | |
| Stage | 0.448 | ||
| Ⅲ | 1 | 3 | |
| Ⅳ | 8 | 9 |
| Clinical Parameter | Group T | Group C | p Value | |
|---|---|---|---|---|
| Efficacy | Progression-free survival (days) | 174 ± 30 | 187 ± 38 | 0.799 |
| Overall survival (days) | 415 ± 50 | 421 ± 60 | 0.941 | |
| Safety | Frequency of adverse events | 9 ± 1 | 29 ± 4 | 0.001 |
| Type of adverse events | 7 ± 1 | 16 ± 1 | <0.001 | |
| Serious adverse events | 1/9 (11.1%) | 2/12 (16.7%) | 0.735 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, J.; Zhang, C.; Zhou, S.; Zhu, J.; Liang, C. A Pilot Study: Favorable Effects of Clostridium butyricum on Intestinal Microbiota for Adjuvant Therapy of Lung Cancer. Cancers 2022, 14, 3599. https://doi.org/10.3390/cancers14153599
Cong J, Zhang C, Zhou S, Zhu J, Liang C. A Pilot Study: Favorable Effects of Clostridium butyricum on Intestinal Microbiota for Adjuvant Therapy of Lung Cancer. Cancers. 2022; 14(15):3599. https://doi.org/10.3390/cancers14153599
Chicago/Turabian StyleCong, Jing, Chuantao Zhang, Siyu Zhou, Jingjuan Zhu, and Chengwei Liang. 2022. "A Pilot Study: Favorable Effects of Clostridium butyricum on Intestinal Microbiota for Adjuvant Therapy of Lung Cancer" Cancers 14, no. 15: 3599. https://doi.org/10.3390/cancers14153599






