Li–Fraumeni Syndrome: Mutation of TP53 Is a Biomarker of Hereditary Predisposition to Tumor: New Insights and Advances in the Treatment
Abstract
Simple Summary
Abstract
1. Introduction
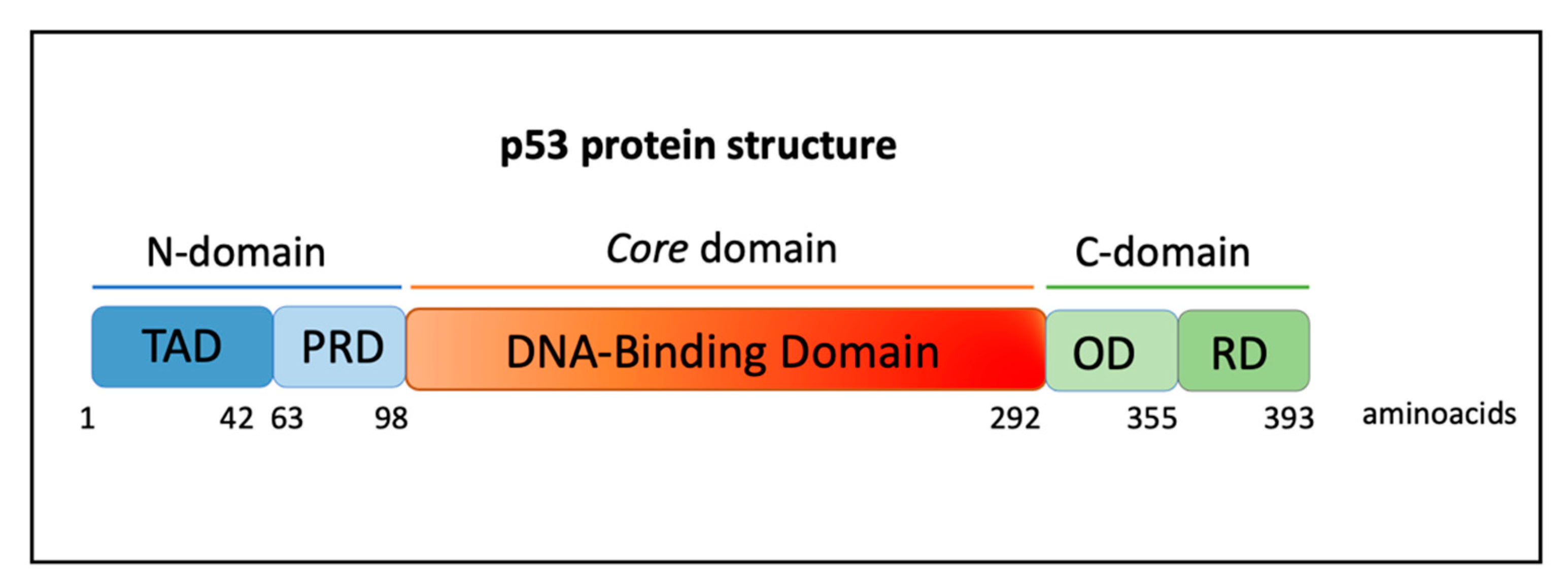
2. The TP53 Gene and Role of Mutant p53 Proteins in Cancer
3. Mutational Landscape of TP53 in LFS
4. Role of Non-Coding RNAs in LFS: Novel Mechanisms and Hypothesis
5. Tumor Prevention and Treatments
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, F.P. Soft-Tissue Sarcomas, Breast Cancer, and Other Neoplasms. Ann. Intern. Med. 1969, 71, 747. [Google Scholar] [CrossRef]
- Valdez, J.M.; Nichols, K.E.; Kesserwan, C. Li-Fraumeni syndrome: A paradigm for the understanding of hereditary cancer predisposition. Br. J. Haematol. 2017, 176, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Zelley, K.; Nichols, K.E.; Garber, J. Li-Fraumeni Syndrome; National Library of Medicine: Bethesda, MD, USA, 1993. [Google Scholar]
- Malkin, D.; Li, F.P.; Strong, L.C.; Fraumeni, J.F.; Nelson, C.E.; Kim, D.H.; Kassel, J.; Gryka, M.A.; Bischoff, F.Z.; Tainsky, M.A.; et al. Germ Line p53 Mutations in a Familial Syndrome of Breast Cancer, Sarcomas, and Other Neoplasms. Science 1990, 250, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Kamihara, J.; Rana, H.Q.; Garber, J.E. Germline TP53 Mutations and the Changing Landscape of Li-Fraumeni Syndrome. Hum. Mutat. 2014, 35, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Mai, P.L.; Best, A.F.; Peters, J.A.; DeCastro, R.M.; Khincha, P.P.; Loud, J.T.; Bremer, R.C.; Rosenberg, P.S.; Savage, S.A. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer 2016, 122, 3673–3681. [Google Scholar] [CrossRef]
- Strano, S.; Dell’Orso, S.; di Agostino, S.; Fontemaggi, G.; Sacchi, A.; Blandino, G. Mutant p53: An oncogenic transcription factor. Oncogene 2007, 26, 2212–2219. [Google Scholar] [CrossRef]
- Levine, A.J.; Chan, C.S.; Dudgeon, C.; Puzio-Kuter, A.; Hainaut, P. The Evolution of Tumors in Mice and Humans with Germline p53 Mutations. Cold Spring Harb. Symp. Quant. Biol. 2015, 80, 139–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kennedy, M.C.; Lowe, S.W. Mutant p53: It’s not all one and the same. Cell Death Differ. 2022, 29, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Amadou, A.; Achatz, M.I.W.; Hainaut, P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: Temporal phases of Li–Fraumeni syndrome. Curr. Opin. Oncol. 2018, 30, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.J.; Dodd-Eaton, E.B.; Peng, G.; Bojadzieva, J.; Chen, J.; Amos, C.I.; Frone, M.N.; Khincha, P.P.; Mai, P.L.; Savage, S.A.; et al. Penetrance of Different Cancer Types in Families with Li-Fraumeni Syndrome: A Validation Study Using Multicenter Cohorts. Cancer Res. 2020, 80, 354–360. [Google Scholar] [CrossRef]
- Kurian, A.W.; Hare, E.E.; Mills, M.A.; Kingham, K.E.; McPherson, L.; Whittemore, A.S.; McGuire, V.; Ladabaum, U.; Kobayashi, Y.; Lincoln, S.E.; et al. Clinical Evaluation of a Multiple-Gene Sequencing Panel for Hereditary Cancer Risk Assessment. J. Clin. Oncol. 2014, 32, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.; Ellisen, L.; et al. Frequency of mutations in individuals with breast cancer referred for BRCA 1 and BRCA 2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015, 121, 25–33. [Google Scholar] [CrossRef]
- Rana, H.Q.; Gelman, R.; LaDuca, H.; McFarland, R.; Dalton, E.; Thompson, J.; Speare, V.; Dolinsky, J.S.; Chao, E.C.; Garber, J.E. Differences in TP53 Mutation Carrier Phenotypes Emerge From Panel-Based Testing. JNCI J. Natl. Cancer Inst. 2018, 110, 863–870. [Google Scholar] [CrossRef]
- Kratz, C.P.; Freycon, C.; Maxwell, K.N.; Nichols, K.E.; Schiffman, J.D.; Evans, D.G.; Achatz, M.I.; Savage, S.A.; Weitzel, J.N.; Garber, J.E.; et al. Analysis of the Li-Fraumeni Spectrum Based on an International Germline TP53 Variant Data Set. JAMA Oncol. 2021, 7, 1800. [Google Scholar] [CrossRef]
- Chompret, A.; Abel, A.; Stoppa-lyonnet, D.; Brugieres, L.; Pages, S.; Feunteun, J.; Bonaiti-pellie, C. Sensitivity and predictive value of criteria for p53germline mutation screening. J. Med. Genet. 2001, 38, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Tinat, J.; Bougeard, G.; Baert-Desurmont, S.; Vasseur, S.; Martin, C.; Bouvignies, E.; Caron, O.; Paillerets, B.B.; Berthet, P.; Dugast, C.; et al. 2009 Version of the Chompret Criteria for Li Fraumeni Syndrome. J. Clin. Oncol. 2009, 27, e108–e109. [Google Scholar] [CrossRef] [PubMed]
- Nichols, K.E.; Malkin, D. Genotype Versus Phenotype: The Yin and Yang of Germline TP53 Mutations in Li-Fraumeni Syndrome. J. Clin. Oncol. 2015, 33, 2331–2333. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Axell, O.; van Leeuwen, T.; Konrat, R.; Kharaziha, P.; Larsson, C.; Wright, A.P.H.; Bajalica-Lagercrantz, S. Association between Predicted Effects of TP53 Missense Variants on Protein Conformation and Their Phenotypic Presentation as Li-Fraumeni Syndrome or Hereditary Breast Cancer. Int. J. Mol. Sci. 2021, 22, 6345. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.M.; Brondani, V.B.; Helena, V.P.; Charchar, H.L.S.; Zerbini, M.C.N.; Leite, L.A.S.; Hoff, A.O.; Latronico, A.C.; Mendonca, B.B.; Diz, M.D.P.E.; et al. Clinical spectrum of Li-Fraumeni syndrome/Li-Fraumeni-like syndrome in Brazilian individuals with the TP53 p.R337H mutation. J. Steroid Biochem. Mol. Biol. 2019, 190, 250–255. [Google Scholar] [CrossRef]
- Sandoval, R.L.; Polidorio, N.; Leite, A.C.R.; Cartaxo, M.; Pisani, J.P.; Quirino, C.V.; Cezana, L.; Pereira, N.G.; Pereira, A.A.L.; Rossi, B.M.; et al. Breast Cancer Phenotype Associated With Li-Fraumeni Syndrome: A Brazilian Cohort Enriched by TP53 p.R337H Carriers. Front. Oncol. 2022, 12, 826. [Google Scholar] [CrossRef]
- Lane, D.P.; Crawford, L.V. T antigen is bound to a host protein in SY40-transformed cells. Nature 1979, 278, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Linzer, D.I.H.; Levine, A.J. Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979, 17, 43–52. [Google Scholar] [CrossRef]
- Soussi, T. The history of p53. EMBO Rep. 2010, 11, 822–826. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Strasser, A.; Kelly, G.L. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026062. [Google Scholar] [CrossRef] [PubMed]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef]
- Boutelle, A.M.; Attardi, L.D. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021, 31, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [CrossRef]
- Hainaut, P.; Hollstein, M. p53 and Human Cancer: The First Ten Thousand Mutations. Adv. Cancer Res. 1999, 77, 81–137. [Google Scholar]
- Freed-Pastor, W.A.; Prives, C. Mutant p53: One name, many proteins. Genes Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef]
- Stein, Y.; Aloni-Grinstein, R.; Rotter, V. Mutant p53 oncogenicity: Dominant-negative or gain-of-function? Carcinogenesis 2020, 41, 1635–1647. [Google Scholar] [CrossRef]
- Overholtzer, M.; Rao, P.H.; Favis, R.; Lu, X.-Y.; Elowitz, M.B.; Barany, F.; Ladanyi, M.; Gorlick, R.; Levine, A.J. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc. Natl. Acad. Sci. USA 2003, 100, 11547–11552. [Google Scholar] [CrossRef]
- Cordani, M.; Oppici, E.; Dando, I.; Butturini, E.; Pozza, E.D.; Nadal-Serrano, M.; Oliver, J.; Roca, P.; Mariotto, S.; Cellini, B.; et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol. Oncol. 2016, 10, 1008–1029. [Google Scholar] [CrossRef] [PubMed]
- Ingallina, E.; Sorrentino, G.; Bertolio, R.; Lisek, K.; Zannini, A.; Azzolin, L.; Severino, L.U.; Scaini, D.; Mano, M.; Mantovani, F.; et al. Mechanical cues control mutant p53 stability through a mevalonate–RhoA axis. Nat. Cell Biol. 2018, 20, 28–35. [Google Scholar] [CrossRef]
- Blandino, G.; Valenti, F.; Sacconi, A.; di Agostino, S. Wild type- and mutant p53 proteins in mitochondrial dysfunction: Emerging insights in cancer disease. Semin. Cell Dev. Biol. 2020, 98, 105–117. [Google Scholar] [CrossRef]
- Butturini, E.; Butera, G.; Pacchiana, R.; de Prati, A.C.; Mariotto, S.; Donadelli, M. Redox Sensitive Cysteine Residues as Crucial Regulators of Wild-Type and Mutant p53 Isoforms. Cells 2021, 10, 3149. [Google Scholar] [CrossRef] [PubMed]
- di Agostino, S.; Strano, S.; Emiliozzi, V.; Zerbini, V.; Mottolese, M.; Sacchi, A.; Blandino, G.; Piaggio, G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006, 10, 191–202. [Google Scholar] [CrossRef]
- Aschauer, L.; Muller, P.A.J. Novel targets and interaction partners of mutant p53 Gain-Of-Function. Biochem. Soc. Trans. 2016, 44, 460–466. [Google Scholar] [CrossRef] [PubMed]
- di Agostino, S.; Sorrentino, G.; Ingallina, E.; Valenti, F.; Ferraiuolo, M.; Bicciato, S.; Piazza, S.; Strano, S.; del Sal, G.; Blandino, G. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 2016, 17, 188–201. [Google Scholar] [CrossRef]
- Pruszko, M.; Milano, E.; Forcato, M.; Donzelli, S.; Ganci, F.; di Agostino, S.; de Panfilis, S.; Fazi, F.; Bates, D.O.; Bicciato, S.; et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep 2017, 18, 1331–1351. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.P.; Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 2018, 25, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Pilley, S.; Rodriguez, T.A.; Vousden, K.H. Mutant p53 in cell-cell interactions. Genes Dev. 2021, 35, 433–448. [Google Scholar] [CrossRef]
- Freed-Pastor, W.A.; Mizuno, H.; Zhao, X.; Langerød, A.; Moon, S.-H.; Rodriguez-Barrueco, R.; Barsotti, A.; Chicas, A.; Li, W.; Polotskaia, A.; et al. Mutant p53 Disrupts Mammary Tissue Architecture via the Mevalonate Pathway. Cell 2012, 148, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.J.; Trinidad, A.G.; Timpson, P.; Morton, J.P.; Zanivan, S.; van den Berghe, P.V.E.; Nixon, C.; Karim, S.A.; Caswell, P.T.; Noll, J.E.; et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 2013, 32, 1252–1265. [Google Scholar] [CrossRef]
- Lozano, G. The Enigma of p53. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 37–40. [Google Scholar] [CrossRef][Green Version]
- Hassin, O.; Nataraj, N.B.; Shreberk-Shaked, M.; Aylon, Y.; Yaeger, R.; Fontemaggi, G.; Mukherjee, S.; Maddalena, M.; Avioz, A.; Iancu, O.; et al. Different hotspot p53 mutants exert distinct phenotypes and predict outcome of colorectal cancer patients. Nat. Commun. 2022, 13, 2800. [Google Scholar] [CrossRef]
- Lozano, G. Mouse Models of p53 Functions. Cold Spring Harb. Perspect. Biol. 2010, 2, a001115. [Google Scholar] [CrossRef]
- Kim, M.P.; Zhang, Y.; Lozano, G. Mutant p53: Multiple Mechanisms Define Biologic Activity in Cancer. Front. Oncol. 2015, 5, 249. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Janic, A.; Chen, Y.; Chang, C.; Lieschke, E.C.; Diepstraten, S.T.; Kueh, A.J.; Bernardini, J.P.; Dewson, G.; O’Reilly, L.A.; et al. Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes Dev. 2018, 32, 1420–1429. [Google Scholar] [CrossRef]
- Souza, L.C.D.E.; Faletti, A.; Veríssimo, C.P.; Stelling, M.P.; Borges, H.L. p53 Signaling on Microenvironment and Its Contribution to Tissue Chemoresistance. Membranes 2022, 12, 202. [Google Scholar] [CrossRef]
- Jackson, J.G.; Lozano, G. The mutant p53 mouse as a pre-clinical model. Oncogene 2013, 32, 4325–4330. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.N.; Eriksson, S.E.; Bianchi, J.; Wiman, K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 2018, 18, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Blandino, G.; di Agostino, S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J. Exp. Clin. Cancer Res. 2018, 37, 30. [Google Scholar] [CrossRef] [PubMed]
- Sallman, D.A.; DeZern, A.E.; Garcia-Manero, G.; Steensma, D.P.; Roboz, G.J.; Sekeres, M.A.; Cluzeau, T.; Sweet, K.L.; McLemore, A.; McGraw, K.L.; et al. Eprenetapopt (APR-246) and Azacitidine in TP53 -Mutant Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1584–1594. [Google Scholar] [CrossRef]
- Ceder, S.; Eriksson, S.E.; Cheteh, E.H.; Dawar, S.; Benitez, M.C.; Bykov, V.J.N.; Fujihara, K.M.; Grandin, M.; Li, X.; Ramm, S.; et al. A thiol-bound drug reservoir enhances APR-246-induced mutant p53 tumor cell death. EMBO Mol. Med. 2021, 13, e10852. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Fortuno, C.; Lee, K.; Olivier, M.; Pesaran, T.; Mai, P.L.; Andrade, K.C.; Attardi, L.D.; Crowley, S.; Evans, D.G.; Feng, B.; et al. Specifications of the ACMG/AMP variant interpretation guidelines for germline TP53 variants. Hum. Mutat. 2021, 42, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Wasserman, J.D.; Novokmet, A.; Eichler-Jonsson, C.; Ribeiro, R.C.; Rodriguez-Galindo, C.; Zambetti, G.P.; Malkin, D. Prevalence and Functional Consequence of TP53 Mutations in Pediatric Adrenocortical Carcinoma: A Children’s Oncology Group Study. J. Clin. Oncol. 2015, 33, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Fortuno, C.; Pesaran, T.; Dolinsky, J.; Yussuf, A.; McGoldrick, K.; Kho, P.F.; James, P.A.; Spurdle, A.B. p53 major hotspot variants are associated with poorer prognostic features in hereditary cancer patients. Cancer Genet. 2019, 236, 21–27. [Google Scholar] [CrossRef]
- Bougeard, G.; Renaux-Petel, M.; Flaman, J.-M.; Charbonnier, C.; Fermey, P.; Belotti, M.; Gauthier-Villars, M.; Stoppa-Lyonnet, D.; Consolino, E.; Brugières, L.; et al. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J. Clin. Oncology. 2015, 33, 2345–2352. [Google Scholar] [CrossRef]
- Paduano, F.; Fabiani, F.; Colao, E.; Trapasso, F.; Perrotti, N.; Barbieri, V.; Baudi, F.; Iuliano, R. Case Report: Identification of a Novel Pathogenic Germline TP53 Variant in a Family With Li–Fraumeni Syndrome. Front. Genet. 2021, 12, 1541. [Google Scholar] [CrossRef]
- Kharaziha, P.; Ceder, S.; Axell, O.; Krall, M.; Fotouhi, O.; Böhm, S.; Lain, S.; Borg, Å.; Larsson, C.; Wiman, K.G.; et al. Functional characterization of novel germline TP53 variants in Swedish families. Clin. Genet. 2019, 96, 216–225. [Google Scholar] [CrossRef]
- Fortuno, C.; Pesaran, T.; Mester, J.; Dolinsky, J.; Yussuf, A.; McGoldrick, K.; James, P.A.; Spurdle, A.B. Genotype-phenotype correlations among TP53 carriers: Literature review and analysis of probands undergoing multi-gene panel testing and single-gene testing. Cancer Genet. 2020, 248, 11–17. [Google Scholar] [CrossRef]
- de Andrade, K.C.; Khincha, P.P.; Hatton, J.N.; Frone, M.N.; Wegman-Ostrosky, T.; Mai, P.L.; Best, A.F.; Savage, S.A. Cancer incidence, patterns, and genotype–phenotype associations in individuals with pathogenic or likely pathogenic germline TP53 variants: An observational cohort study. Lancet Oncol. 2021, 22, 1787–1798. [Google Scholar] [CrossRef]
- Giacomelli, A.O.; Yang, X.; Lintner, R.E.; McFarland, J.M.; Duby, M.; Kim, J.; Howard, T.P.; Takeda, D.Y.; Ly, S.H.; Kim, E.; et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat. Genet. 2018, 50, 1381–1387. [Google Scholar] [CrossRef]
- Ribeiro, R.C.; Michalkiewicz, E.L.; Figueiredo, B.C.; DeLacerda, L.; Sandrini, F.; Pianovsky, M.D.; Sampaio, G.; Sandrini, R. Adrenocortical tumors in children. Braz. J. Med. Biol. Res. 2000, 33, 1225–1234. [Google Scholar] [CrossRef]
- Pinto, E.M.; Chen, X.; Easton, J.; Finkelstein, D.; Liu, Z.; Pounds, S.; Rodriguez-Galindo, C.; Lund, T.C.; Mardis, E.R.; Wilson, R.K.; et al. Genomic landscape of paediatric adrenocortical tumours. Nat. Commun. 2015, 6, 6302. [Google Scholar] [CrossRef]
- Pinto, E.M.; Figueiredo, B.C.; Chen, W.; Galvao, H.C.R.; Formiga, M.N.; Fragoso, M.C.B.V.; Ashton-Prolla, P.; Ribeiro, E.M.S.F.; Felix, G.; Costa, T.E.B.; et al. XAF1 as a modifier of p53 function and cancer susceptibility. Sci. Adv. 2020, 6, eaba3231. [Google Scholar] [CrossRef]
- Powers, J.; Pinto, E.M.; Barnoud, T.; Leung, J.C.; Martynyuk, T.; Kossenkov, A.v.; Philips, A.H.; Desai, H.; Hausler, R.; Kelly, G.; et al. A Rare TP53 Mutation Predominant in Ashkenazi Jews Confers Risk of Multiple Cancers. Cancer Res. 2020, 80, 3732–3744. [Google Scholar] [CrossRef] [PubMed]
- Kratz, C.P.; Achatz, M.I.; Brugières, L.; Frebourg, T.; Garber, J.E.; Greer, M.-L.C.; Hansford, J.R.; Janeway, K.A.; Kohlmann, W.K.; McGee, R.; et al. Cancer Screening Recommendations for Individuals with Li-Fraumeni Syndrome. Clin. Cancer Res. 2017, 23, e38–e45. [Google Scholar] [CrossRef]
- Bougeard, G. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J. Med. Genet. 2006, 43, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Bond, G.L.; Hu, W.; Bond, E.E.; Robins, H.; Lutzker, S.G.; Arva, N.C.; Bargonetti, J.; Bartel, F.; Taubert, H.; Wuerl, P.; et al. A Single Nucleotide Polymorphism in the MDM2 Promoter Attenuates the p53 Tumor Suppressor Pathway and Accelerates Tumor Formation in Humans. Cell 2004, 119, 591–602. [Google Scholar] [CrossRef]
- Renaux-Petel, M.; Sesboüé, R.; Baert-Desurmont, S.; Vasseur, S.; Fourneaux, S.; Bessenay, E.; Frébourg, T.; Bougeard, G. The MDM2 285G–309G haplotype is associated with an earlier age of tumour onset in patients with Li-Fraumeni syndrome. Fam. Cancer 2014, 13, 127–130. [Google Scholar] [CrossRef]
- Said, B.I.; Malkin, D. A functional variant in miR-605 modifies the age of onset in Li-Fraumeni syndrome. Cancer Genet. 2015, 208, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Bandeira, I.C.; Vieira, I.A.; Andreis, T.F.; Reis, L.B.; Macedo, G.S.; Vianna, F.S.L.; Santos-Silva, P.; Palmero, E.I.; Galvão, H.d.R.; Ramos, C.R.N.; et al. MIR605 rs2043556 is associated with the occurrence of multiple primary tumors in TP53 p.(Arg337His) mutation carriers. Cancer Genet. 2020, 240, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Tabori, U.; Nanda, S.; Druker, H.; Lees, J.; Malkin, D. Younger Age of Cancer Initiation Is Associated with Shorter Telomere Length in Li-Fraumeni Syndrome. Cancer Res. 2007, 67, 1415–1418. [Google Scholar] [CrossRef]
- Shlien, A.; Tabori, U.; Marshall, C.R.; Pienkowska, M.; Feuk, L.; Novokmet, A.; Nanda, S.; Druker, H.; Scherer, S.W.; Malkin, D. Excessive genomic DNA copy number variation in the Li–Fraumeni cancer predisposition syndrome. Proc. Natl. Acad. Sci. USA 2008, 105, 11264–11269. [Google Scholar] [CrossRef]
- di Agostino, S. The Impact of Mutant p53 in the Non-Coding RNA World. Biomolecules 2020, 10, 472. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K. Emerging roles of long non-coding RNAs in the p53 network. RNA Biol. 2020, 17, 1648–1656. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Thorne, R.F.; Zhang, X.D.; Wu, M.; Liu, L. Non-coding RNAs, guardians of the p53 galaxy. Semin. Cancer Biol. 2021, 75, 72–83. [Google Scholar] [CrossRef]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar]
- Palcau, A.C.; Canu, V.; Donzelli, S.; Strano, S.; Pulito, C.; Blandino, G. CircPVT1: A pivotal circular node intersecting Long Non-Coding-PVT1 and c-MYC oncogenic signals. Mol. Cancer 2022, 21, 33. [Google Scholar] [CrossRef]
- Xiao, J.; Lin, H.; Luo, X.; Luo, X.; Wang, Z. miR-605 joins p53 network to form a p53: miR-605: Mdm2 positive feedback loop in response to stress. EMBO J. 2011, 30, 524–532. [Google Scholar] [CrossRef]
- Zhang, M.W.; Jin, M.J.; Zhang, S.C. Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol. Carcinog. 2012, 51, E21–E31. [Google Scholar] [PubMed]
- Capaci, V.; Bascetta, L.; Fantuz, M.; Beznoussenko, G.V.; Sommaggio, R.; Cancila, V.; Bisso, A.; Campaner, E.; Mironov, A.A.; Wiśniewski, J.R.; et al. Mutant p53 induces Golgi tubulo-vesiculation driving a prometastatic secretome. Nat. Commun. 2020, 11, 3945. [Google Scholar] [CrossRef]
- Madrigal, T.; Hernández-Monge, J.; Herrera, L.A.; la Rosa, C.H.G.; Domínguez-Gómez, G.; Candelaria, M.; Luna-Maldonado, F.; González, K.G.C.; Díaz-Chávez, J. Regulation of miRNAs Expression by Mutant p53 Gain of Function in Cancer. Front. Cell Dev. Biol. 2021, 9, 695723. [Google Scholar] [CrossRef] [PubMed]
- Vahabi, M.; Blandino, G.; di Agostino, S. MicroRNAs in head and neck squamous cell carcinoma: A possible challenge as biomarkers, determinants for the choice of therapy and targets for personalized molecular therapies. Transl. Cancer Res. 2021, 10, 3090–3110. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef]
- Li-Fraumeni Syndrome Study, (n.d.). Available online: https://lfs.cancer.gov/ (accessed on 16 June 2022).
- Frebourg, T.; Lagercrantz, S.B.; Oliveira, C.; Magenheim, R.; Evans, D.G. Guidelines for the Li–Fraumeni and heritable TP53-related cancer syndromes. Eur. J. Hum. Genet. 2020, 28, 1379–1386. [Google Scholar] [CrossRef]
- Daly, M.B.; Pilarski, R.; Yurgelun, M.B.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Garber, J.E.; et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J. Natl. Compr. Cancer Netw. 2020, 18, 380–391. [Google Scholar] [CrossRef]
- Birch, J.M.; Hartley, A.L.; Tricker, K.J.; Prosser, J.; Condie, A.; Kelsey, A.M.; Harris, M.; Jones, P.H.; Binchy, A.; Crowther, D. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res. 1994, 54, 1298–1304. [Google Scholar]
- Eeles, R.A. Germline mutations in the TP53 gene. Cancer Surv 1995, 25, 101–124. [Google Scholar] [PubMed]
- Villani, A.; Shore, A.; Wasserman, J.D.; Stephens, D.; Kim, R.H.; Druker, H.; Gallinger, B.; Naumer, A.; Kohlmann, W.; Novokmet, A.; et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016, 17, 1295–1305. [Google Scholar] [CrossRef]
- Ballinger, M.L.; Best, A.; Mai, P.L.; Khincha, P.P.; Loud, J.T.; Peters, J.A.; Achatz, M.I.; Chojniak, R.; da Costa, A.B.; Santiago, K.M.; et al. Baseline Surveillance in Li-Fraumeni Syndrome Using Whole-Body Magnetic Resonance Imaging. JAMA Oncol. 2017, 3, 1634. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, T.; Yamazaki, F.; Nakano, Y.; Tamura, C.; Tashiro, S.; Hattori, H.; Nakagawara, A.; Tsunematsu, Y. Medical guidelines for Li–Fraumeni syndrome 2019, version 1.1. Int. J. Clin. Oncol. 2021, 26, 2161–2178. [Google Scholar] [CrossRef] [PubMed]
- Omran, M.; Tham, E.; Brandberg, Y.; Ahlström, H.; Lundgren, C.; Paulsson-Karlsson, Y.; Kuchinskaya, E.; Silander, G.; Rosén, A.; Persson, F.; et al. Whole-Body MRI Surveillance—Baseline Findings in the Swedish Multicentre Hereditary TP53-Related Cancer Syndrome Study (SWEP53). Cancers 2022, 14, 380. [Google Scholar] [CrossRef]
- Polotskaia, A.; Xiao, G.; Reynoso, K.; Martin, C.; Qiu, W.-G.; Hendrickson, R.C.; Bargonetti, J. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc. Natl. Acad. Sci. USA 2015, 112, E1220–E1229. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, Z.; Myers, J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell. Biochem. 2016, 117, 2682–2692. [Google Scholar] [CrossRef]
- Silwal-Pandit, L.; Langerød, A.; Børresen-Dale, A.-L. TP53 Mutations in Breast and Ovarian Cancer. Cold Spring Harb Perspect Med. 2017, 7, a026252. [Google Scholar] [CrossRef]
- Tuna, M.; Ju, Z.; Yoshihara, K.; Amos, C.I.; Tanyi, J.L.; Mills, G.B. Clinical relevance of TP53 hotspot mutations in high-grade serous ovarian cancers. Br. J. Cancer 2020, 122, 405–412. [Google Scholar] [CrossRef]
- Le, A.N.; Harton, J.; Desai, H.; Powers, J.; Zelley, K.; Bradbury, A.R.; Nathanson, K.L.; Shah, P.D.; Doucette, A.; Freedman, G.M.; et al. Frequency of radiation-induced malignancies post-adjuvant radiotherapy for breast cancer in patients with Li-Fraumeni syndrome. Breast Cancer Res. Treat. 2020, 181, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, P.G.; Luo, Y.; Kohlmann, W.; Schiffman, J.; Maese, L.; Bishop, A.J.; Lloyd, S.; Kokeny, K.E.; Hitchcock, Y.J.; Poppe, M.M.; et al. Radiation therapy and secondary malignancy in Li-Fraumeni syndrome: A hereditary cancer registry study. Cancer Med. 2020, 9, 7954–7963. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, J.; Walcott, F.L.; Kang, J.-G.; Starost, M.F.; Talagala, S.L.; Zhuang, J.; Park, J.-H.; Huffstutler, R.D.; Bryla, C.M.; et al. Inhibiting mitochondrial respiration prevents cancer in a mouse model of Li-Fraumeni syndrome. J. Clin. Investig. 2016, 127, 132–136. [Google Scholar] [CrossRef]
- Walcott, F.L.; Wang, P.-Y.; Bryla, C.M.; Huffstutler, R.D.; Singh, N.; Pollak, M.N.; Khincha, P.P.; Savage, S.A.; Mai, P.L.; Dodd, K.W.; et al. Pilot Study Assessing Tolerability and Metabolic Effects of Metformin in Patients with Li-Fraumeni Syndrome. JNCI Cancer Spectr. 2020, 4, pkaa063. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.R.; Morris, A.D. Metformin in Cancer Treatment and Prevention. Annu. Rev. Med. 2015, 66, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.S.O.; Golovoy, M.; Abdullah, Y.; Ahmed, R.S.I.; Dou, Q.P. Repurposing of Metformin for Cancer Therapy: Updated Patent and Literature Review. Recent Pat. Anti-Cancer Drug Discov. 2021, 16, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Fleshner, N. The role of metformin, statins and diet in men on active surveillance for prostate cancer. World J. Urol. 2022, 40, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, N.; Iannantuoni, F.; Gruevska, A.; Muntane, J.; Rocha, M.; Victor, V.M. Mechanisms of action of metformin in type 2 diabetes: Effects on mitochondria and leukocyte-endothelium interactions. Redox Biol. 2020, 34, 101517. [Google Scholar] [CrossRef]
- Beaubier, N.; Bontrager, M.; Huether, R.; Igartua, C.; Lau, D.; Tell, R.; Bobe, A.M.; Bush, S.; Chang, A.L.; Hoskinson, D.C.; et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat. Biotechnol. 2019, 37, 1351–1360. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Gray, R.; Chen, A.; Li, S.; Patton, D.; Hamilton, S.R.; Williams, P.M.; Mitchell, E.P.; Iafrate, A.J.; Sklar, J.; et al. The Molecular Analysis for Therapy Choice (NCI-MATCH) Trial: Lessons for Genomic Trial Design. JNCI J. Natl. Cancer Inst. 2020, 112, 1021–1029. [Google Scholar] [CrossRef]
- Reed, M.R.; Lyle, A.G.; de Loose, A.; Maddukuri, L.; Learned, K.; Beale, H.C.; Kephart, E.T.; Cheney, A.; van den Bout, A.; Lee, M.P.; et al. A Functional Precision Medicine Pipeline Combines Comparative Transcriptomics and Tumor Organoid Modeling to Identify Bespoke Treatment Strategies for Glioblastoma. Cells 2021, 10, 3400. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocca, V.; Blandino, G.; D’Antona, L.; Iuliano, R.; Di Agostino, S. Li–Fraumeni Syndrome: Mutation of TP53 Is a Biomarker of Hereditary Predisposition to Tumor: New Insights and Advances in the Treatment. Cancers 2022, 14, 3664. https://doi.org/10.3390/cancers14153664
Rocca V, Blandino G, D’Antona L, Iuliano R, Di Agostino S. Li–Fraumeni Syndrome: Mutation of TP53 Is a Biomarker of Hereditary Predisposition to Tumor: New Insights and Advances in the Treatment. Cancers. 2022; 14(15):3664. https://doi.org/10.3390/cancers14153664
Chicago/Turabian StyleRocca, Valentina, Giovanni Blandino, Lucia D’Antona, Rodolfo Iuliano, and Silvia Di Agostino. 2022. "Li–Fraumeni Syndrome: Mutation of TP53 Is a Biomarker of Hereditary Predisposition to Tumor: New Insights and Advances in the Treatment" Cancers 14, no. 15: 3664. https://doi.org/10.3390/cancers14153664
APA StyleRocca, V., Blandino, G., D’Antona, L., Iuliano, R., & Di Agostino, S. (2022). Li–Fraumeni Syndrome: Mutation of TP53 Is a Biomarker of Hereditary Predisposition to Tumor: New Insights and Advances in the Treatment. Cancers, 14(15), 3664. https://doi.org/10.3390/cancers14153664








