IKKε Inhibitor Amlexanox Promotes Olaparib Sensitivity through the C/EBP-β-Mediated Transcription of Rad51 in Castrate-Resistant Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Cloning, Viruses and Infections
2.3. Drugs
2.4. Cell Proliferation Assays
2.5. Drug Combination Analysis
2.6. Western Blot Analysis
2.7. Total RNA Extraction, Reverse Transcription and qPCR
2.8. Immunofluorescence
2.9. EdU (5-ethynyl-2′-deoxyuridine) Detection
2.10. Colony Formation
2.11. Luciferase Assay
2.12. Chromatin Immunoprecipitation
2.13. Murine Xenograft Model
2.14. Immunofluorescence on Xenograft Tissue
2.15. Statistical Analysis
3. Results
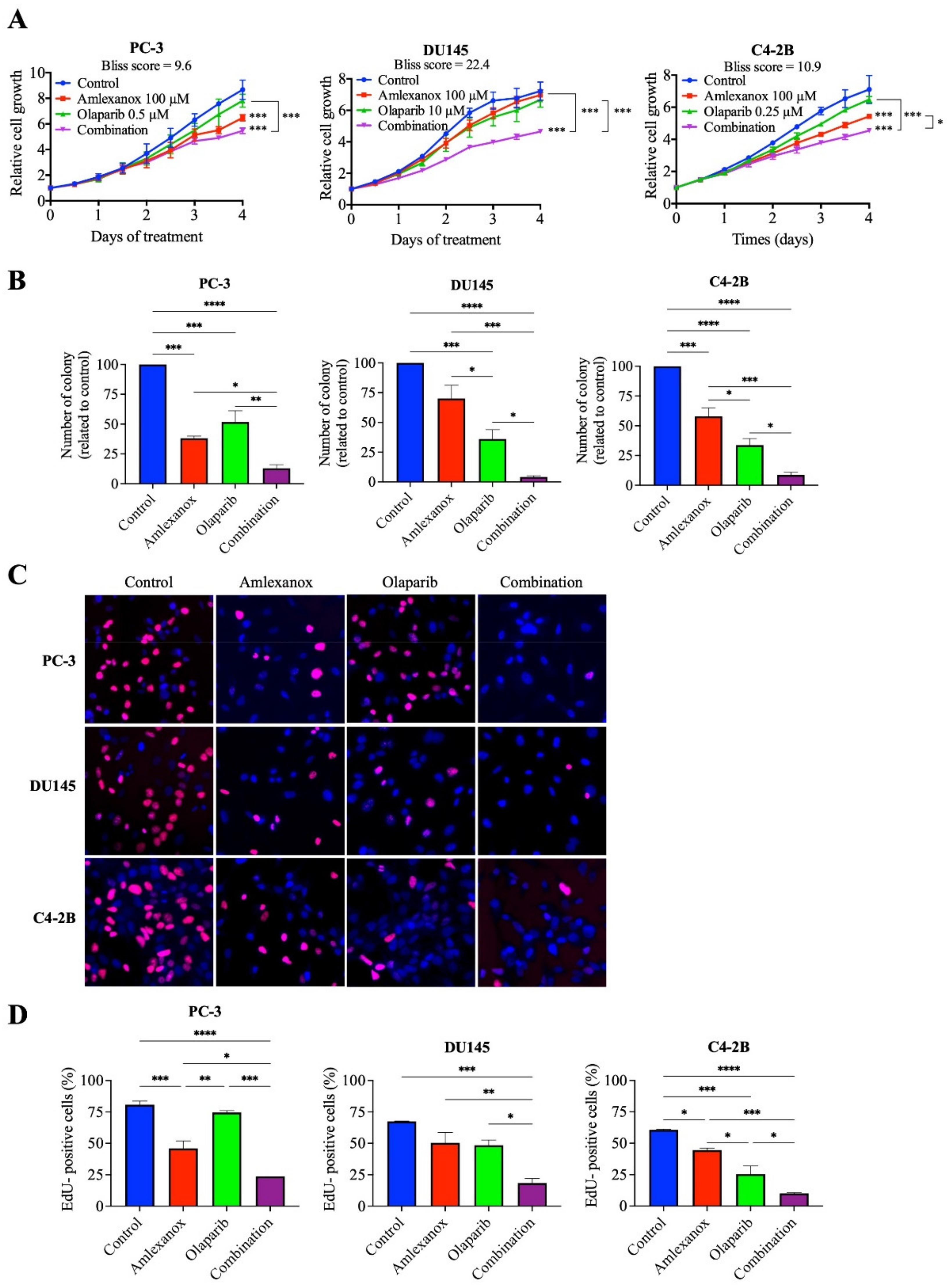
3.1. Effect of the Amlexanox-Olaparib Combination on CRPC Proliferation
3.2. Amlexanox-Olaparib Combination Increases DNA Damage
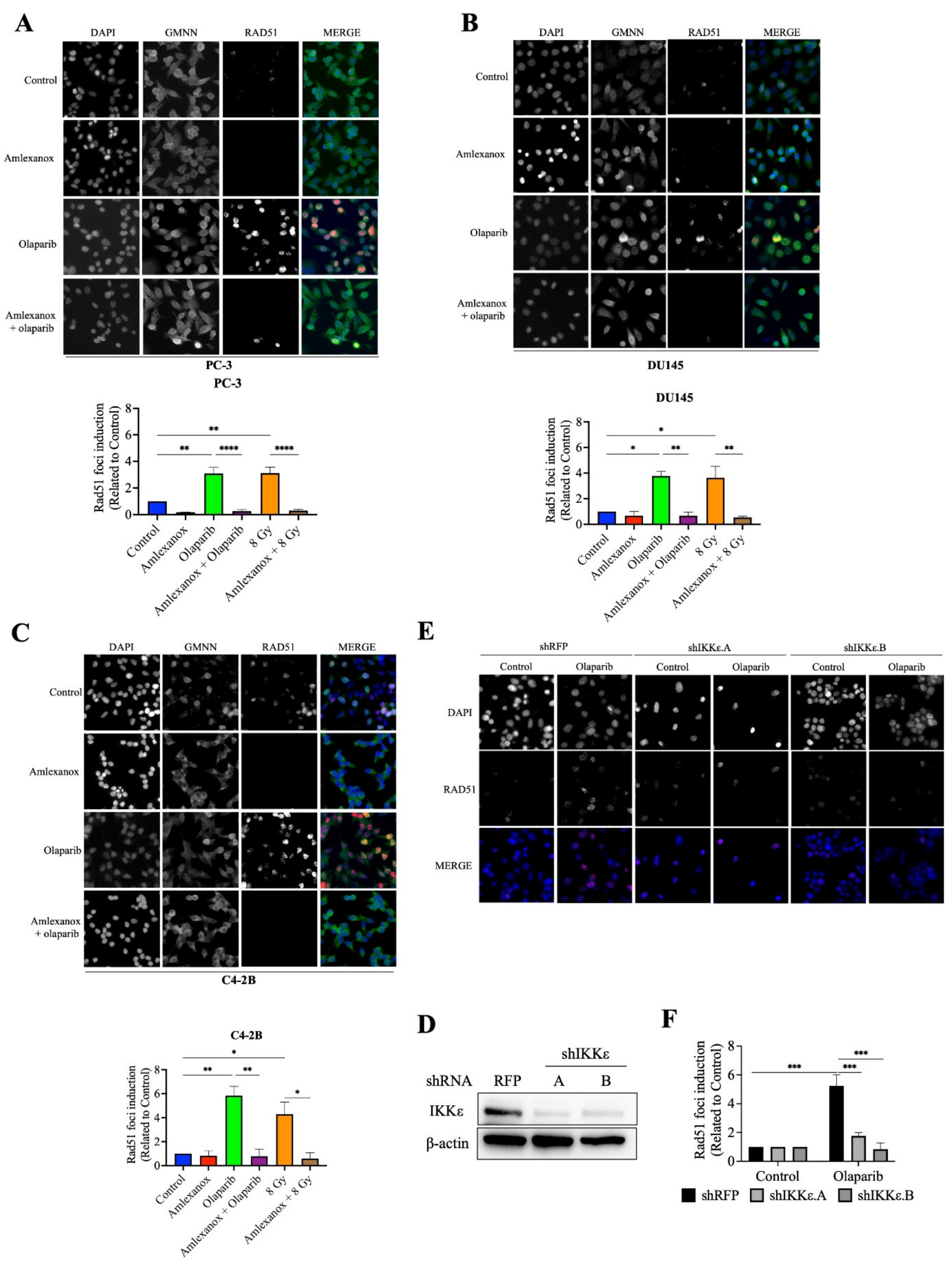
3.3. Amlexanox Inhibits Olaparib-Induced Rad51 Recruitment
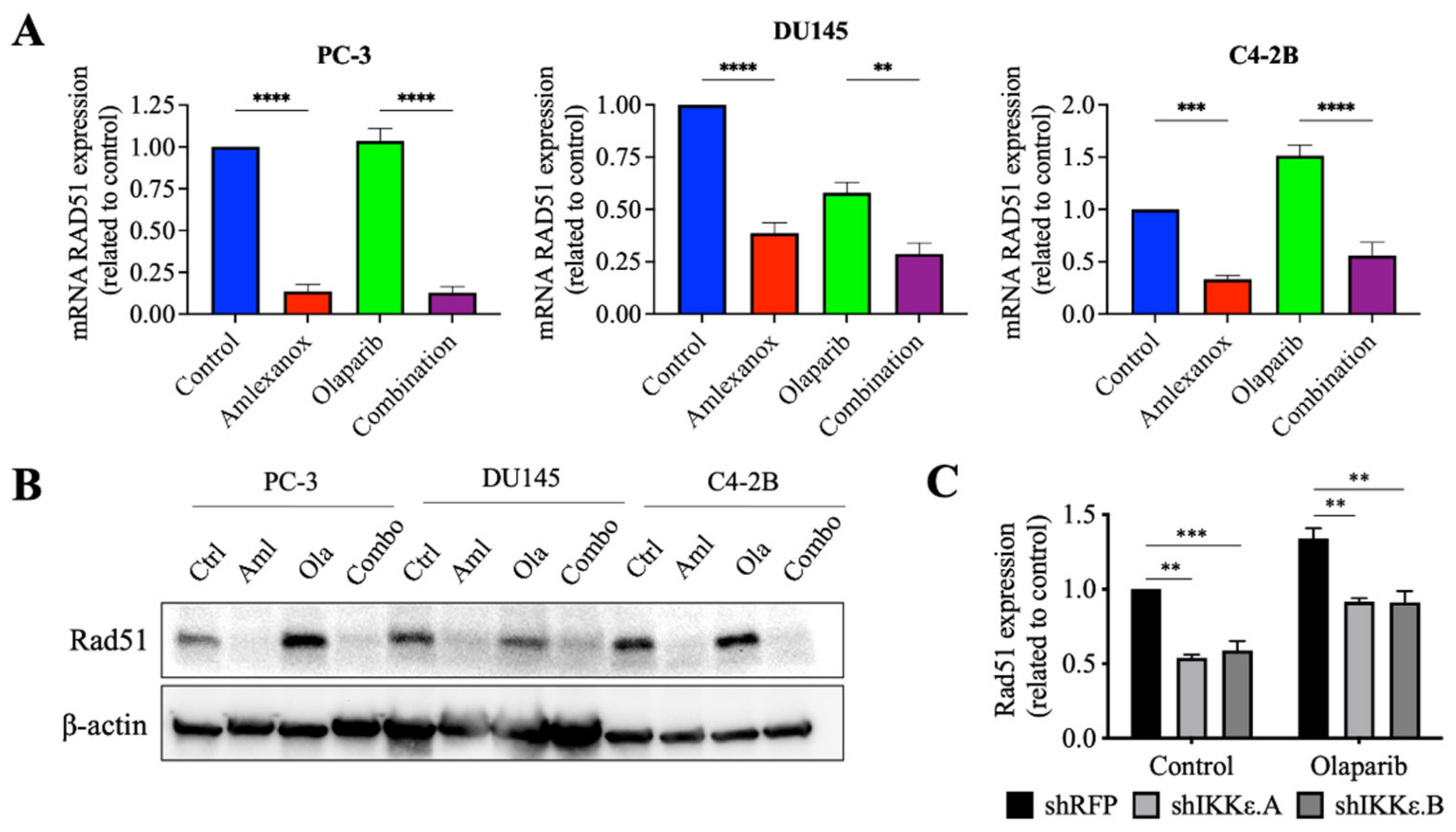
3.4. Effect of Amlexanox on the Rad51 Expression
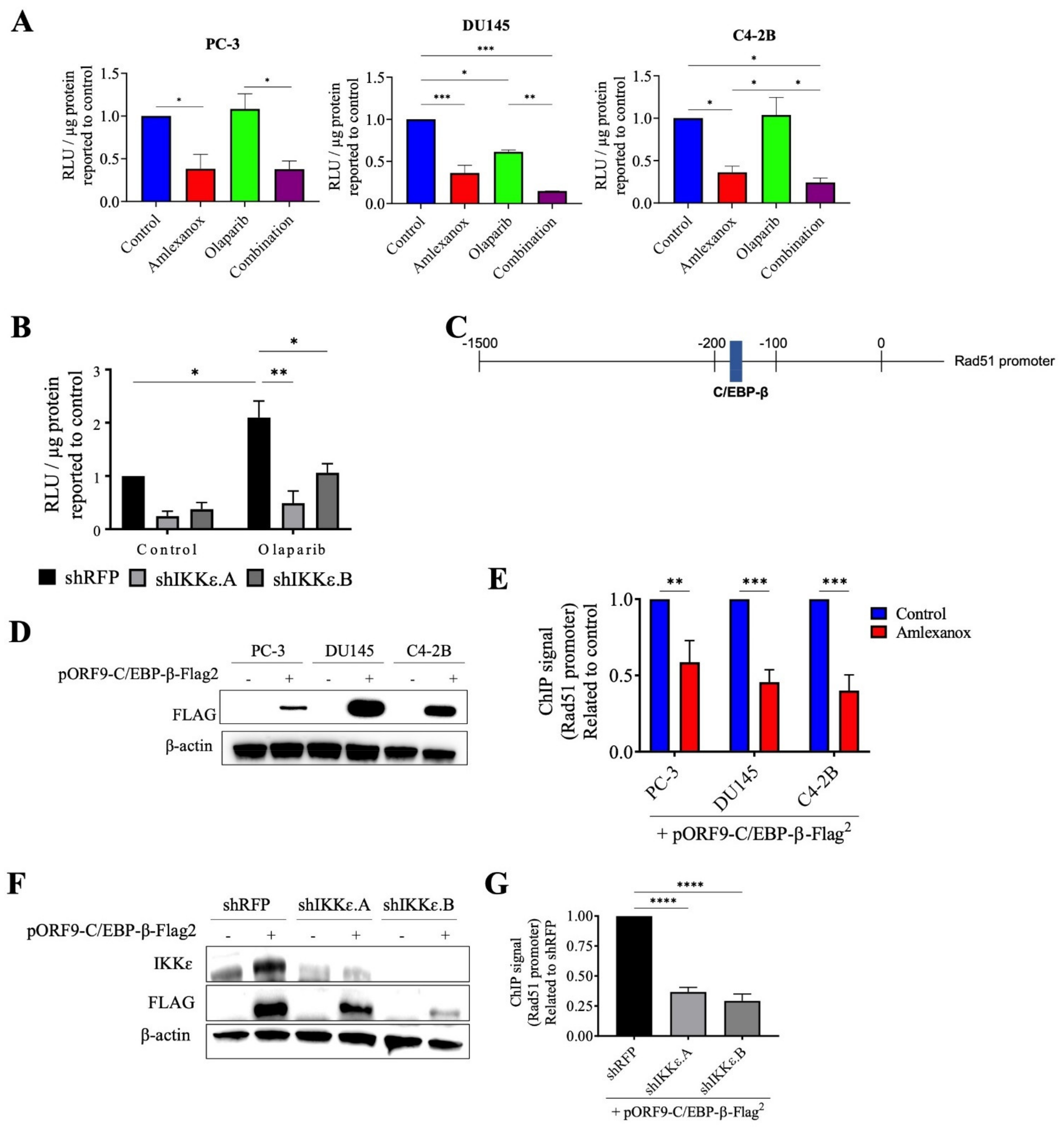
3.5. Effect of Amlexanox on the Regulation of Rad51 Mediated by C/EBP-β
3.6. Amlexanox-Olaparib Combination Impairs the Growth of CRPC Xenograft
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodriguez, M.I.; Majuelos-Melguizo, J.; Marti Martin-Consuegra, J.M.; Ruiz de Almodovar, M.; Lopez-Rivas, A.; Javier Oliver, F. Deciphering the insights of poly(ADP-ribosylation) in tumor progression. Med. Res. Rev. 2015, 35, 678–697. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Wang, P.Y.; Wang, Y.T.; Yang, G.F.; Zhang, A.; Miao, Z.H. An Update on Poly(ADP-ribose)polymerase-1 (PARP-1) Inhibitors: Opportunities and Challenges in Cancer Therapy. J. Med. Chem. 2016, 59, 9575–9598. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [Green Version]
- de Bono, J.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 382, 2091–2102. [Google Scholar] [CrossRef]
- Hussain, M.; Mateo, J.; Fizazi, K.; Saad, F.; Shore, N.; Sandhu, S.; Chi, K.N.; Sartor, O.; Agarwal, N.; Olmos, D.; et al. Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020, 383, 2345–2357. [Google Scholar] [CrossRef]
- Clarke, N.; Wiechno, P.; Alekseev, B.; Sala, N.; Jones, R.; Kocak, I.; Chiuri, V.E.; Jassem, J.; Flechon, A.; Redfern, C.; et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2018, 19, 975–986. [Google Scholar] [CrossRef]
- Sharma, S.; TenOever, B.R.; Grandvaux, N.; Zhou, G.P.; Lin, R.; Hiscott, J. Triggering the interferon antiviral response through an IKK-related pathway. Science 2003, 300, 1148–1151. [Google Scholar] [CrossRef] [PubMed]
- Hutti, J.E.; Shen, R.R.; Abbott, D.W.; Zhou, A.Y.; Sprott, K.M.; Asara, J.M.; Hahn, W.C.; Cantley, L.C. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell 2009, 34, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.P.; Tian, W.; Shu, S.; Xin, Y.; Shou, C.; Cheng, J.Q. IKBKE phosphorylation and inhibition of FOXO3a: A mechanism of IKBKE oncogenic function. PLoS ONE 2013, 8, e63636. [Google Scholar] [CrossRef] [Green Version]
- Hsu, S.; Kim, M.; Hernandez, L.; Grajales, V.; Noonan, A.; Anver, M.; Davidson, B.; Annunziata, C.M. IKK-epsilon coordinates invasion and metastasis of ovarian cancer. Cancer Res. 2012, 72, 5494–5504. [Google Scholar] [CrossRef] [Green Version]
- Peant, B.; Diallo, J.S.; Lessard, L.; Delvoye, N.; Le Page, C.; Saad, F.; Mes-Masson, A.M. Regulation of IkappaB kinase epsilon expression by the androgen receptor and the nuclear factor-kappaB transcription factor in prostate cancer. Mol. Cancer Res. 2007, 5, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peant, B.; Forest, V.; Trudeau, V.; Latour, M.; Mes-Masson, A.M.; Saad, F. IkappaB-Kinase-epsilon (IKKepsilon/IKKi/IkappaBKepsilon) expression and localization in prostate cancer tissues. Prostate 2011, 71, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Peant, B.; Gilbert, S.; Le Page, C.; Poisson, A.; L’Ecuyer, E.; Boudhraa, Z.; Bienz, M.N.; Delvoye, N.; Saad, F.; Mes-Masson, A.M. IkappaB-Kinase-epsilon (IKKepsilon) over-expression promotes the growth of prostate cancer through the C/EBP-beta dependent activation of IL-6 gene expression. Oncotarget 2017, 8, 14487–14501. [Google Scholar] [CrossRef] [Green Version]
- Burton, D.G.; Giribaldi, M.G.; Munoz, A.; Halvorsen, K.; Patel, A.; Jorda, M.; Perez-Stable, C.; Rai, P. Androgen deprivation-induced senescence promotes outgrowth of androgen-refractory prostate cancer cells. PLoS ONE 2013, 8, e68003. [Google Scholar] [CrossRef]
- Renner, F.; Moreno, R.; Schmitz, M.L. SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol. Cell 2010, 37, 503–515. [Google Scholar] [CrossRef]
- Peant, B.; Diallo, J.S.; Dufour, F.; Le Page, C.; Delvoye, N.; Saad, F.; Mes-Masson, A.M. Over-expression of IkappaB-kinase-epsilon (IKKepsilon/IKKi) induces secretion of inflammatory cytokines in prostate cancer cell lines. Prostate 2009, 69, 706–718. [Google Scholar] [CrossRef]
- Gilbert, S.; Peant, B.; Malaquin, N.; Tu, V.; Fleury, H.; Leclerc-Desaulniers, K.; Rodier, F.; Mes-Masson, A.M.; Saad, F. Targeting IKKepsilon in Androgen-Independent Prostate Cancer Causes Phenotypic Senescence and Genomic Instability. Mol. Cancer Ther. 2022, 21, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Fleury, H.; Malaquin, N.; Tu, V.; Gilbert, S.; Martinez, A.; Olivier, M.A.; Sauriol, A.; Communal, L.; Leclerc-Desaulniers, K.; Carmona, E.; et al. Exploiting interconnected synthetic lethal interactions between PARP inhibition and cancer cell reversible senescence. Nat. Commun. 2019, 10, 2556. [Google Scholar] [CrossRef] [Green Version]
- Valerie, K.; Povirk, L.F. Regulation and mechanisms of mammalian double-strand break repair. Oncogene 2003, 22, 5792–5812. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef] [Green Version]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef] [Green Version]
- Raderschall, E.; Golub, E.I.; Haaf, T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 1999, 96, 1921–1926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haaf, T.; Golub, E.I.; Reddy, G.; Radding, C.M.; Ward, D.C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 1995, 92, 2298–2302. [Google Scholar] [CrossRef] [Green Version]
- Zahnow, C.A. CCAAT/enhancer-binding protein beta: Its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev. Mol. Med. 2009, 11, e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.; Zheng, X.; Li, M.; Ye, F.; Song, C.; Xu, C.; Zhang, X.; Li, W.; Wang, Y.; Zeng, S.; et al. C/EBPbeta promotes poly(ADP-ribose) polymerase inhibitor resistance by enhancing homologous recombination repair in high-grade serous ovarian cancer. Oncogene 2021, 40, 3845–3858. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.F.; Guo, S.; Demicco, E.G.; Romieu-Mourez, R.; Landesman-Bollag, E.; Seldin, D.C.; Sonenshein, G.E. Inducible IkappaB kinase/IkappaB kinase epsilon expression is induced by CK2 and promotes aberrant nuclear factor-kappaB activation in breast cancer cells. Cancer Res. 2005, 65, 11375–11383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehm, J.S.; Zhao, J.J.; Yao, J.; Kim, S.Y.; Firestein, R.; Dunn, I.F.; Sjostrom, S.K.; Garraway, L.A.; Weremowicz, S.; Richardson, A.L.; et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 2007, 129, 1065–1079. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.P.; Shu, S.K.; He, L.; Lee, Y.C.; Kruk, P.A.; Grenman, S.; Nicosia, S.V.; Mor, G.; Schell, M.J.; Coppola, D.; et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am. J. Pathol. 2009, 175, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Chen, Y.; Zhang, J.; Hong, L.; Yuan, N.; Wang, X.; Lv, H. IKBKE Upregulation is Positively Associated with Squamous Cell Carcinoma of the Lung In Vivo and Malignant Transformation of Human Bronchial Epithelial Cells In Vitro. Med. Sci. Monit. 2015, 21, 1577–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, M.R.; Kim, M.S.; Kim, S.S.; Ahn, C.H.; Yoo, N.J.; Lee, S.H. NF-kappaB signalling proteins p50/p105, p52/p100, RelA, and IKKepsilon are over-expressed in oesophageal squamous cell carcinomas. Pathology 2009, 41, 622–625. [Google Scholar] [CrossRef]
- Geng, B.; Zhang, C.; Wang, C.; Che, Y.; Mu, X.; Pan, J.; Xu, C.; Hu, S.; Yang, J.; Zhao, T.; et al. IkappaB-kinase-epsilon in the tumor microenvironment is essential for the progression of gastric cancer. Oncotarget 2017, 8, 75298–75307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.E.; Hong, M.; Cho, J.; Lee, J.; Kim, K.M. IKKepsilon and TBK1 expression in gastric cancer. Oncotarget 2017, 8, 16233–16242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, B.; Cheng, K. Silencing of the IKKepsilon gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast Cancer Res. 2010, 12, R74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhao, J.; Chen, X.; Ding, C.; Lee, K.; Jia, Z.; Zhang, Y.; Zhou, Y.; Wei, C.; He, J.; et al. Hyper activation of beta-catenin signalling induced by IKKepsilon inhibition thwarts colorectal cancer cell proliferation. Cell Prolif. 2017, 50, e12350. [Google Scholar] [CrossRef] [PubMed]
- Barbie, T.U.; Alexe, G.; Aref, A.R.; Li, S.; Zhu, Z.; Zhang, X.; Imamura, Y.; Thai, T.C.; Huang, Y.; Bowden, M.; et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J. Clin. Investig. 2014, 124, 5411–5423. [Google Scholar] [CrossRef] [PubMed]
- Moller, M.; Wasel, J.; Schmetzer, J.; Weiss, U.; Meissner, M.; Schiffmann, S.; Weigert, A.; Moser, C.V.; Niederberger, E. The Specific IKKepsilon/TBK1 Inhibitor Amlexanox Suppresses Human Melanoma by the Inhibition of Autophagy, NF-kappaB and MAP Kinase Pathways. Int. J. Mol. Sci. 2020, 21, 4721. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, J.; Zhang, Z.; Zhu, L.; Dong, S.; Guo, G.; Li, R.; Nan, Y.; Yu, K.; Zhong, Y.; et al. Amlexanox, a selective inhibitor of IKBKE, generates anti-tumoral effects by disrupting the Hippo pathway in human glioblastoma cell lines. Cell Death Dis. 2017, 8, e3022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Ji, Z.; Sheng, Y.; Wang, J.; Sun, Y.; Zhao, H.; Li, X.; Wang, X.; He, Y.; Yao, J.; et al. Aphthous ulcer drug inhibits prostate tumor metastasis by targeting IKKvarepsilon/TBK1/NF-kappaB signaling. Theranostics 2018, 8, 4633–4648. [Google Scholar] [CrossRef] [PubMed]
- Bishop, R.T.; Marino, S.; de Ridder, D.; Allen, R.J.; Lefley, D.V.; Sims, A.H.; Wang, N.; Ottewell, P.D.; Idris, A.I. Pharmacological inhibition of the IKKepsilon/TBK-1 axis potentiates the anti-tumour and anti-metastatic effects of Docetaxel in mouse models of breast cancer. Cancer Lett. 2019, 450, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Challa, S.; Guo, J.P.; Ding, X.; Xu, C.X.; Li, Y.; Kim, D.; Smith, M.A.; Cress, D.W.; Coppola, D.; Haura, E.B.; et al. IKBKE Is a Substrate of EGFR and a Therapeutic Target in Non-Small Cell Lung Cancer with Activating Mutations of EGFR. Cancer Res. 2016, 76, 4418–4429. [Google Scholar] [CrossRef] [Green Version]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilbert, S.; Péant, B.; Mes-Masson, A.-M.; Saad, F. IKKε Inhibitor Amlexanox Promotes Olaparib Sensitivity through the C/EBP-β-Mediated Transcription of Rad51 in Castrate-Resistant Prostate Cancer. Cancers 2022, 14, 3684. https://doi.org/10.3390/cancers14153684
Gilbert S, Péant B, Mes-Masson A-M, Saad F. IKKε Inhibitor Amlexanox Promotes Olaparib Sensitivity through the C/EBP-β-Mediated Transcription of Rad51 in Castrate-Resistant Prostate Cancer. Cancers. 2022; 14(15):3684. https://doi.org/10.3390/cancers14153684
Chicago/Turabian StyleGilbert, Sophie, Benjamin Péant, Anne-Marie Mes-Masson, and Fred Saad. 2022. "IKKε Inhibitor Amlexanox Promotes Olaparib Sensitivity through the C/EBP-β-Mediated Transcription of Rad51 in Castrate-Resistant Prostate Cancer" Cancers 14, no. 15: 3684. https://doi.org/10.3390/cancers14153684






