1. Introduction
Ovarian cancer is one of the most common gynecological malignancies, with 313,959 new cases and 207,252 deaths worldwide in 2020 [
1]. The high fatality rate accounts for 2.5% of all malignancies among females, but 5% of cancer-related deaths. More than 70% of patients are diagnosed at stage III or IV because of a lack in exhibiting typical symptoms and effective screening. For all stages of ovarian cancer, the 5-year survival rate is approximately 46%. The 5-year survival rate is less than 20% in women diagnosed with advanced stage (Federation of Gynecology and Obstetrics (FIGO) stage III or IV) invasive epithelial ovarian cancer (EOC); however, it exceeds 90% for those at stage I [
2]. Therefore, increasing early ovarian cancer detection is the primary strategy to improve ovarian cancer patient survival.
Ovarian cancer is highly heterogeneous and demonstrates various molecular, pathological, and other features. EOC is the most common histomorphological type and accounts for more than 90% of ovarian cancer. Currently, transvaginal ultrasound scan (TVS), level of serum carbohydrate antigen 125 (CA125), and pelvic examination are the most commonly used EOC diagnostic methods [
3]. Unfortunately, pelvic examination lacks objective data standards and depends on the proficiency of the attending physician. Moreover, patients with early disease are likely to be overlooked because of the lack of typical signs. TVS can provide valuable information on an ovarian mass, including its size, location, composition, morphology, and blood flow, and these features are frequently used to assess malignancy. Although TVS has a sensitivity of more than 90% for the differential diagnosis of ovarian masses, its performance largely depends on the skill and experience of the ultrasound specialists [
4]. Furthermore, CA125 is elevated in only about 80% of EOC and is associated with other diseases, resulting in an unsatisfactory diagnostic performance when served as the sole diagnostic indicator [
5]. Alternative to the single current indicator, many studies have also proposed the comprehensive model of multiple indicators for diagnosing ovarian cancer, including the risk of malignancy index (RMI), the risk of ovarian cancer algorithm (ROMA), and the One Variable-at-a-Time (OVAT1) [
6,
7,
8]. However, the application of larger population data demonstrated that these combined models failed to produce a more stable and reliable diagnostic performance than a single index. Therefore, a new biomarker with superior sensitivity and specificity is still urgently needed for ovarian cancer.
Extracellular vesicle (EV) is a generic term for particles naturally released from cells that are delimited by a lipid bilayer and cannot replicate. Small EVs (sEVs), with a diameter of <200 nm, are present in various bodily fluids such as blood, urine, saliva, and ascitic fluid [
9]. By delivering nucleic acids, proteins, lipids, ions, and other specific components, sEVs participate in different pathophysiological processes to maintain cell homeostasis and regulate intercellular communication [
10]. The diverse effects mediated by tumor-derived sEVs have gradually been discovered, including epithelial–mesenchymal transition, metastasis, angiogenesis, immune regulation, cell metabolism, pre-metastasis niche formation, and therapeutic resistance [
11,
12]. In addition to their role as biocommunication mediators, the great potential of sEVs as circulating biomarkers for the diagnosis and prognosis of various cancers has attracted considerable attention [
13]. For example, in ovarian cancer, sEVs can be isolated from the ascitic fluid and blood of patients, making them potentially less invasive biomarkers for its diagnosis and prognosis. Moreover, microRNA (miR-200a, miR-200b, miR-200c, and miR-373) levels in sEVs derived from the serum of EOC patient showed diagnostic and prognostic values in a study conducted by Meng et al. [
14]. Furthermore, proteins from sEVs, including claudin-4, transforming growth factor-β1 (TGF-β1), and melanoma-associated antigen 3 and 6 (MAGE3/6), were considered as candidate biomarkers because of their enrichment in the serum of ovarian cancer patients compared to that in the patients with benign tumors or healthy controls [
15,
16]. However, these indicators have not yet been verified with a large sample size and developed for further clinical applications, owing to their unsatisfactory diagnostic performance or the difficulty and poor stability in detection methods.
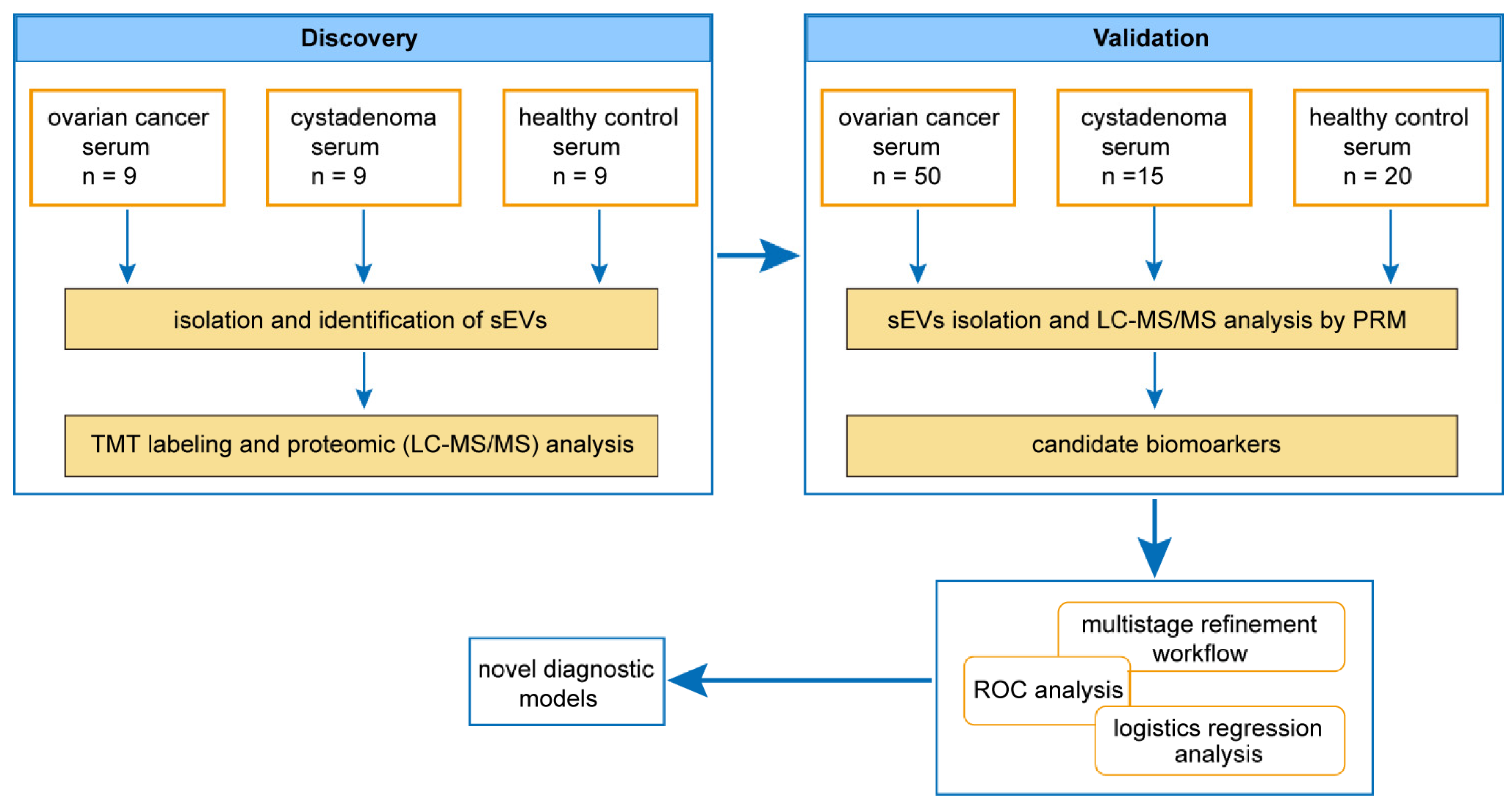
In this study, we performed tandem mass tag-based liquid chromatography/tandem mass spectrometry (TMT-LC-MS/MS) analysis to obtain the proteomic profiles of sEVs derived from patients with EOC. Our study aimed to obtain cancer-specific protein candidates in sEVs from the serum of ovarian cancer patients and develop a protein panel for ovarian cancer screening in the general population and differential diagnosis of ovarian masses.
2. Materials and Methods
2.1. Patients and Serum Samples
In total, 66 patients with ovarian masses (16 patients with cystadenoma and 50 patients with ovarian cancer) and 29 healthy subjects from the first affiliated hospital of the Sun Yat-sen University (Guangzhou, Guangdong Province, China) were enrolled in this study conducted between January 2018 and January 2020. All human blood samples were procured after obtaining the approval of the institutional review board of the first affiliated hospital of Sun Yat-sen University and informed consent from all participants. The detailed characteristics of all the participants are provided in
Table S1. Whole-blood samples were obtained in vacutainer serum blood collection tubes (BD Biosciences, Franklin Lakes, NJ, USA) from participants who were made to fast. After coagulation, the blood samples without hemolysis were centrifuged at 3000 rpm for 10 min at 4 °C to obtain serum samples. The supernatants were aliquoted and were stored at −80 °C for subsequent sEVs’ isolation.
2.2. Isolation of sEVs
The serum samples were removed from storage at −80 °C and centrifuged at 12,000×
g for 15 min at 4 °C. Next, the supernatants were transferred to a new centrifuge tube and filtered with a 0.22-μm microporous membrane. In HPLC/LC-MS/MS analysis and targeted proteomics, 1 mL aliquots of serum were subjected to the isolation of sEVs using commercially available qEVoriginal size-exclusion chromatography (SEC) columns (Izon Science, Christchurch, New Zealand), according to the manufacturer’s protocol [
17]. The purified sEVs were then used immediately or stored at −80 °C.
2.3. Transmission Electron Microscopy (TEM)
Isolated sEVs were resuspended in 50–100 μL of 2% paraformaldehyde solution, and 5 μL of this suspension was added to the Formvar–carbon copper grip and 100 μL PBS was added to the sealing film. The copper grid was placed on PBS drops and washed. The copper grid was placed on a 50 μL 1% glutaraldehyde droplet for 5 min and then washed with 100 μL double distilled water. Next, the copper grid was placed on 50 μL uranium oxalate drops for 75 min and then placed on 50 methylcellulose drops for 10 min (placed on ice). The excess liquid was absorbed on the filter paper and the copper grid was air-dried for 5–10 min. Finally, the copper grid was placed in the box, and electron microscopy photos were acquired at 80 kV.
2.4. Nanoparticle Tracking Analysis (NTA)
The particle size and concentration of sEVs were measured using NTA at Viva Cell Biosciences with ZetaView PMX 110 (Particle Metrix, Munich, Germany) and the ZetaView 8.04.02 software. The isolated sEVs were diluted 500 times with PBS before measurement. NTA measurements were recorded and analyzed at 11 positions. The ZetaView system was calibrated using 110 nm polystyrene particles. Temperature was maintained between 24.41 and 25.74 °C. Each process was repeated thrice.
2.5. Western Blotting Analysis
Isolated sEVs from 1 mL aliquots of serum were lysed in RIPA buffer with a protease inhibitor cocktail and quantified using a BCA protein assay reagent kit (Thermo Scientific, Waltham, MA, USA). The protein samples were denatured at 95 °C for 10 min in a 5× protein loading buffer. Equal amounts of the protein extracts were separated using 10% SDS-PAGE and transferred onto a PVDF membrane (Millipore, MA, USA). The membranes were blocked with 5% evaporated skimmed milk for 1 h in TBST at room temperature and then incubated overnight with primary antibodies against the following human proteins: TSG101 (#ab125011, 1:1000 dilution, Abcam, Cambridge, UK), CD63 (#ab134045, 1:5000 dilution, Abcam, Cambridge, UK), Albumin (#GTX102419, 1:1000 dilution, GeneTex, Irvine, CA, USA), FGG (#GTX108640, 1:5000 dilution, GeneTex, Irvine, CA, USA), MUC16 (#20077-1-AP, 1:500 dilution, Proteintech, Wuhan, China), and APOA4 (#17996-1-AP, 1:8000 dilution, Proteintech, Wuhan, China) at 4 °C. The membranes were then incubated with HRP-conjugated secondary antibody for 1 h at room temperature. The protein bands were visualized using enhanced chemiluminescence detection reagents (Thermo Scientific) following the manufacturer’s instructions.
2.6. Protein Extraction and Digestion
For protein lysis, an 8 M urea and 1% protease inhibitor cocktail was added to the sEVs, followed by sonication thrice on ice using a high-intensity ultrasonic processor. Subsequently, the protein concentration was determined using the BCA kit, following the manufacturer’s instructions. For digestion, the protein solution was reduced with 5 mM dithiothreitol for 30 min at 56 °C and alkylated with 11 mM iodoacetamide for 15 min at room temperature in the dark. The protein sample was then diluted by adding 100 mM TEAB to achieve a urea concentration less than 2 M. Finally, trypsin was added at a 1:50 trypsin-to-protein mass ratio for the first digestion overnight and a 1:100 trypsin-to-protein mass ratio for the second 4 h digestion.
2.7. Tandem Mass Tag (TMT) Labeling, HPLC Fractionation, and LC-MS/MS Analysis
After trypsin digestion, the peptide was desalted using Strata X C18 SPE column (Phenomenex Inc., Torrance, CA, USA) and vacuum dried. Based on the results of peptides’ quantification, the least number of peptides, 14 μg peptides, were sampled for labeling. The peptide was reconstituted in 0.5 M TEAB and processed following the manufacturer’s protocol for the TMT kit, and 2 μg peptides were sampled from each sample and mixed as MIX samples. After thorough mixing, 14 μg peptides were sampled from the MIX samples. Following the standardized quality inspection, the labeled samples of each group were mixed and subjected to HPLC fractionation. The tryptic peptides were fractionated using high pH reverse-phase HPLC using Agilent 300Extend C18 column. The peptides were subjected to the NSI source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo Scientific), coupled online to the UPLC. The resulting MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8) (Max-Planck Institute of Biochemistry, Munich, Germany). Tandem mass spectra were searched against the SwissProt human database concatenated with a reverse decoy database.
2.8. Targeted Proteomics
For the validation study, sEVs isolated from 1 mL aliquots of serum were lysed and digested, as described above. After digestion, the peptides were quantified. Then, 1.5 ug peptides from each sample were analyzed with LC-MS operated using the parallel reaction monitoring (PRM) acquisition scheme, a targeted proteomics technique based on high-resolution and high-precision mass spectrometry. Finally, the resulting MS data were processed using Skyline (v.3.6) (MacCoss Lab Software, University of Washington, Seattle, WA, USA).
2.9. Bioinformatic Analysis
The software used for the bioinformatics analysis is listed in
Table S2. Gene ontology (GO) annotation proteome was derived from the UniProt–GOA database. Proteins were classified into three GO annotation categories: biological process, cellular compartment, and molecular function. For each category, a two-tailed Fisher’s exact test was employed to test the enrichment of the differentially expressed proteins against all identified proteins. The GO term with an adjusted
p-value < 0.05 was considered significant [
18]. The reactome pathway database was used to identify enriched pathways using a two-tailed Fisher’s exact test for differentially expressed protein. The pathway with an adjusted
p-value < 0.05 was considered significant. Finally, the STRING database was used for protein–protein interaction (PPI) network analysis. The medium confidence was set as 0.4, and line thickness indicated the strength of data support.
2.10. Statistical Analyses
All statistical analyses were performed using the R version 4.1.3 (available online:
https://www.R-project.org/ (accessed on 10 March 2022)) and SPSS 25.0 (SPSS Inc., Chicago, IL, USA). Continuous variables are described in terms of medians (interquartile range (IQR)). Group differences in protein expression were analyzed using the two-tailed
t-test. ROC curve analyses were employed to assess the diagnostic efficacy. Univariate and multivariate logistic regression analyses were used to develop a diagnostic model. Estimates were presented as odds ratios (ORs) and 95% confidence intervals (CIs). All significance tests were two-tailed and conducted at a
p-value < 0.05.
4. Discussion
In the present study, we performed TMT-based LC-MS/MS analysis to obtain the proteomic profiles of serum sEVs for EOC. Potential biomarkers were identified and further validated via a targeted proteomic approach. A panel composed of three proteins (FGG, MUC16, and APOA4) in serum sEVs showed remarkable performance for ovarian cancer screening in populations. Moreover, a novel panel integrated with serum CA125, sEV-CD5L, and sEV-APOA4 showed potential value for the differential diagnosis between ovarian cancer and ovarian cystadenoma, even at their early stages.
Recently, great advances have been made in ovarian cancer screening and prevention, including the use of tailored prevention and screening methods that combine genetic and epidemiological factors to predict the individual risk of ovarian cancer [
19,
20]. No definitive mortality reduction was reported for screening compared with no screening until now, suggesting that there is a need to find screening strategies better than CA125 that are noninvasive, simple, and specific for detecting ovarian cancer, especially in its early stage. By far, a major focus of ovarian cancer biomarker discovery is tumor-specific biomarkers in plasma. Examination of various information on tumor cells in the blood represents the new diagnostic tool, namely, liquid biopsy, which quickly identifies biological behaviors of cancer cells, including cell death, clonal evolution, and drug resistance [
21,
22]. Although many potential benefits of circulating tumor cells (CTCs) have been demonstrated in ovarian cancer patients, the major challenge is processing and analyzing CTCs [
23]. In the study conducted by Shao, cell-free DNA carried genetic and epigenetic changes that mimic tumor cells and may be used as a tumor-specific biomarker with higher sensitivity and specificity than CA125 [
24]. Inconsistent results have been demonstrated for cfmiRNA in the diagnosis of cancers, which may be attributed to difficulties in sample collection, processing, and RNA stability and quantification, limiting the opportunity to use cfmiRNA as a promising cancer biomarker [
25]. EVs provide a protective environment for miRNAs against RNase destruction and are, therefore, considered promising targets for liquid biopsies [
26]. Moreover, typical characteristics of sEVs such as size, appearance, specific biomarkers, circulation stability, and the wide availability of sEV isolation kits make it a more suitable target for liquid biopsy than others.
In ovarian cancer, protein and miRNA are the main objects of sEV components considered as biomarkers for diagnosis and/or prognosis and are closely correlated to drug resistance, tumor microenvironment, and immune regulation [
27]. We established a three-protein model for liquid biopsy for ovarian cancer with minimal loss of diagnostic efficacy, which showed good diagnostic efficacy even in the early-stage disease. In addition to early diagnosis, differential diagnosis of ovarian masses is also an important reference for preoperative therapeutic options [
28]. Elevated CA125 level is associated with other conditions besides ovarian cancer, indicating that the differential diagnosis of benign and malignant ovarian tumors is still worth further optimization [
29]. For this purpose, we also established a three-protein panel whose performance is comparable to CA125 using tumor-specific sEVs proteomics. Once serumCA125 was included in the protein candidates, a new integrative model with improved sensitivity and accuracy was generated, with APOA4, CD5L, and serumCA125 replacing sEV-MUC16. This may be due to the poor encapsulation of MUC16 as a macromolecule in sEVs and the limitations of detection methods. Importantly, this new integrated model still has superior AUC, sensitivity, specificity, and accuracy in the differential diagnosis between early-stage ovarian cancer and cystadenoma.
As mediators of cellular communication, sEVs may play the same role in similar biological behaviors of different types of cancer. For example, ALCAM/CD166 mediates the docking and uptake of cancer cell derived EVs and promotes the peritoneal metastasis cascade in colorectal cancer and ovarian cancer [
30]. For diagnosis of cancer, biomarkers often contain the characteristic information of different cancer types. A recent study trying to diagnose multiple types of early-stage cancer isolated and purified EVs in plasma to determine the levels of potential protein markers by immunoassay. By utilizing artificial intelligence, 13 EV protein markers were analyzed to predict the likelihood of malignancy [
31]. CA19-9 is commonly used to monitor the therapeutic efficacy and recurrence of pancreatic cancer. The novel diagnostic method contains CA19-9 and has excellent diagnostic performance for early-stage pancreatic cancer (95.7% detection rate), while detection of early-stage ovarian cancer and bladder cancer were 74.4% and 43.8% [
31], suggesting the importance of characteristic biomarkers for diagnosing specific cancer type. The novel diagnostic protein panel in our study included CA125, a specific marker for ovarian cancer, and its performance in diagnosing ovarian cancer including their early-stage disease and discriminating between benign and malignant ovarian tumors in the population was more effective than CA125. Aiming at identification of sEV proteins as diagnostic biomarkers for ovarian cancer, early studies have shown that some proteins from EVs such as claudin-4, TGF-β1 and MAGE3/6 may have certain diagnostic value in ovarian cancer [
15,
16], but no further demonstration was made in a larger population sample. A recent study utilized mass spectrometry to detect the specific protein profiles in serum exosomes of ovarian cancer and revealed that the coagulation pathway was significantly enriched and that those cascade-related proteins present diagnostic and prognostic values [
32]. Our study also discovers relevant changes in the coagulation pathway, and our conclusions are more convincing because of the larger sample size, the stringent controls, an integrative multi-index model to optimize diagnostic performance, and the validation using targeted proteomics.
Circulating proteins within sEVs carry tumor-specific signals and communicate extensively with tissues throughout the body, thus mediating important events in tumor growth, cancer-associated microenvironment, and tumor evolution [
33]. The elucidation of the signaling network can provide new perspectives for the treatment of ovarian cancer. Among the biomarker candidates we screened, MUC16 has been proposed to exert roles in the innate defense of tracheal epithelium or the immune microenvironment of cancer [
34]. The FGG gene encodes the gamma chain of fibrinogen, a major component of blood clots, which promotes coagulation. Furthermore, fibrinogen has also been related to leukocyte migration, phagocytosis of microorganisms, tumor growth and metastasis, chemoresistance, and epithelial-mesenchymal transition [
35,
36,
37,
38]. APOA4 is not only a lipid-binding protein, but also has a role in the immune response [
39,
40]. Our model not only reflects the key role of CA125 for diagnosing ovarian cancer, but also suggests that the occurrence and development of ovarian cancer may be accompanied by the remodeling of cellular lipid metabolism and the complex interaction between the tumor and their hosts’ immune system [
41,
42,
43,
44]. These findings suggest that these proteins within sEVs found in our study can serve as diagnostic markers for ovarian cancer and provide new perspectives for therapeutic research on ovarian cancer. However, the specific molecular mechanism and the functional roles of sEVs still need to be further studied, complementing the limitations of the current study. The sample size of the study was limited, and more patients should be included to reduce bias and prove the universality of the model. Moreover, it is our next research plan to establish a standardized process for detection of multiple indicators through ELISA and perform integrative analyses of quantitative data. Until now, more research is still needed to collect sufficient serum samples and establish a more robust testing process.