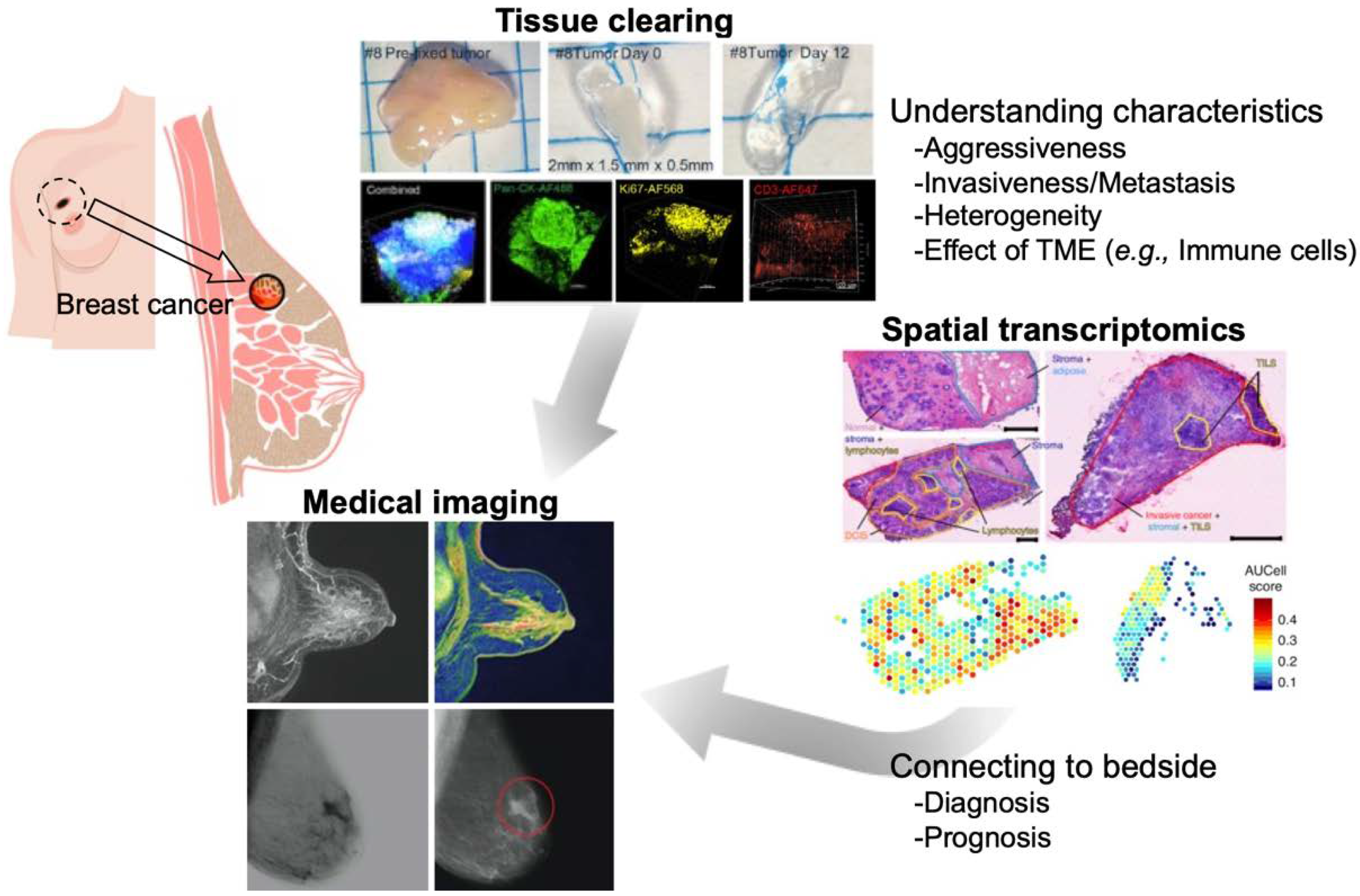
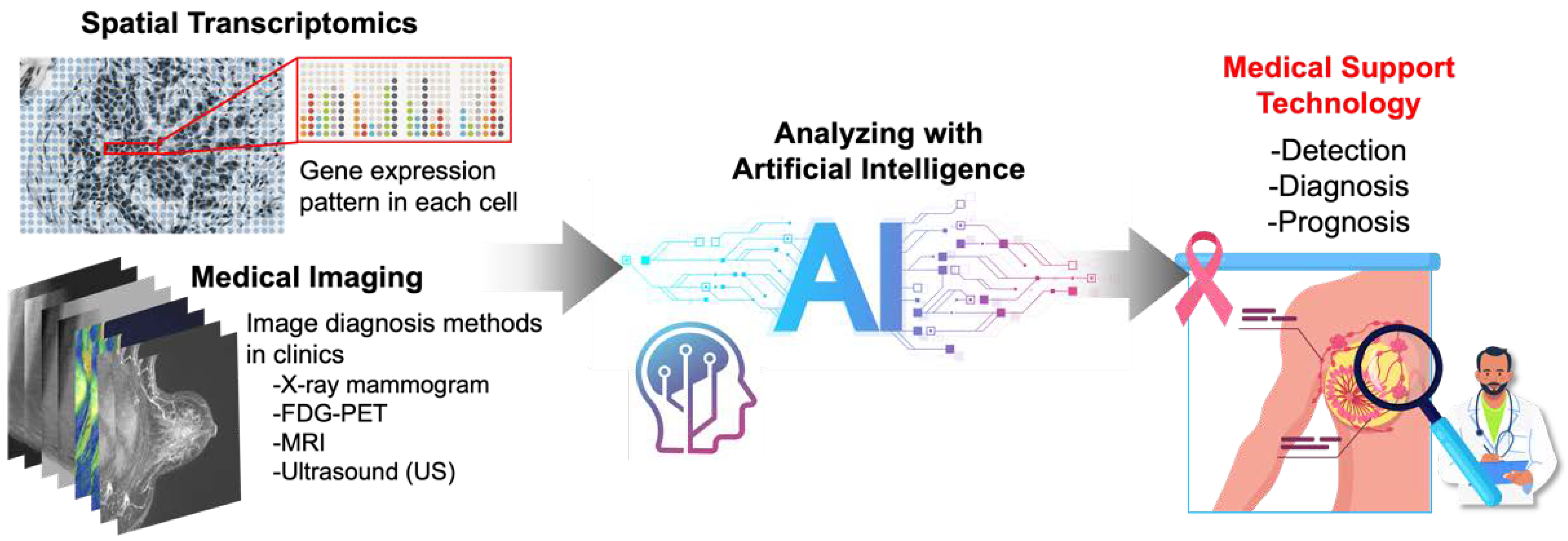
Understanding Breast Cancers through Spatial and High-Resolution Visualization Using Imaging Technologies
Abstract
Simple Summary
Abstract
1. Introduction
2. Tissue Clearing and Imaging
2.1. Principals and Methods of Tissue Clearing
2.2. Application to Diseased Tissue Specimens
2.3. Application to Breast Cancers
2.4. Optical Imaging
3. Spatial Transcriptomics
3.1. Transcriptomics Analysis in Single-Cell Resolution
3.2. Spatial Gene Expression Analysis of Cancer Tissue
4. Medical Imaging
4.1. Recent Advances of Medical Imaging for Breast Cancer
4.2. Photoacoustic Imaging
5. AI-Based Analysis of Spatial Transcriptomics and Medical Images
5.1. Spatial Transcriptomics and Artificial Intelligence (AI)
5.2. Medical Imaging and Artificial Intelligence (AI)
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies-An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef]
- Lüönd, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal 2020, 18, 59. [Google Scholar] [CrossRef]
- Lim, B.; Woodward, W.A.; Wang, X.; Reuben, J.M.; Ueno, N.T. Inflammatory breast cancer biology: The tumour microenvironment is key. Nat. Rev. Cancer 2018, 18, 485–499. [Google Scholar] [CrossRef]
- Barriga, V.; Kuol, N.; Nurgali, K.; Apostolopoulos, V. The Complex Interaction between the Tumor Micro-Environment and Immune Checkpoints in Breast Cancer. Cancers 2019, 11, 1205. [Google Scholar] [CrossRef]
- Pepperkok, R.; Ellenberg, J. High-throughput fluorescence microscopy for systems biology. Nat. Rev. Mol. Cell Biol. 2006, 7, 690–696. [Google Scholar] [CrossRef]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell. Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef]
- Richardson, D.S.; Lichtman, J.W. Clarifying Tissue Clearing. Cell 2015, 162, 246–257. [Google Scholar] [CrossRef]
- Tian, T.; Yang, Z.; Li, X. Tissue clearing technique: Recent progress and biomedical applications. J. Anat. 2021, 238, 489–507. [Google Scholar] [CrossRef]
- Almagro, J.; Messal, H.A.; Zaw Thin, M.; van Rheenen, J.; Behrens, A. Tissue clearing to examine tumour complexity in three dimensions. Nat. Rev. Cancer 2021, 21, 718–730. [Google Scholar] [CrossRef]
- Waylen, L.N.; Nim, H.T.; Martelotto, L.G.; Ramialison, M. From whole-mount to single-cell spatial assessment of gene expression in 3D. Commun. Biol. 2020, 3, 602. [Google Scholar] [CrossRef]
- Rao, A.; Barkley, D.; França, G.S.; Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 2021, 596, 211–220. [Google Scholar] [CrossRef]
- Longo, S.K.; Guo, M.G.; Ji, A.L.; Khavari, P.A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 2021, 22, 627–644. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, Q.; White, S.L.; Gokmen-Polar, Y.; Badve, S.; Goodman, L.J. Three-dimensional imaging and quantitative analysis in CLARITY processed breast cancer tissues. Sci. Rep. 2019, 9, 5624. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Molbay, M.; Kolabas, Z.I.; Todorov, M.I.; Ohn, T.L.; Ertürk, A. A guidebook for DISCO tissue clearing. Mol. Syst. Biol. 2021, 17, e9807. [Google Scholar] [CrossRef]
- Nojima, S.; Susaki, E.A.; Yoshida, K.; Takemoto, H.; Tsujimura, N.; Iijima, S.; Takachi, K.; Nakahara, Y.; Tahara, S.; Ohshima, K.; et al. CUBIC pathology: Three-dimensional imaging for pathological diagnosis. Sci. Rep. 2017, 7, 9269. [Google Scholar] [CrossRef]
- Rakha, E.A.; Putti, T.C.; Abd El-Rehim, D.M.; Paish, C.; Green, A.R.; Powe, D.G.; Lee, A.H.; Robertson, J.F.; Ellis, I.O. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J. Pathol. 2006, 208, 495–506. [Google Scholar] [CrossRef]
- Bombonati, A.; Sgroi, D.C. The molecular pathology of breast cancer progression. J. Pathol. 2011, 223, 307–317. [Google Scholar] [CrossRef]
- Lloyd-Lewis, B.; Davis, F.M.; Harris, O.B.; Hitchcock, J.R.; Lourenco, F.C.; Pasche, M.; Watson, C.J. Imaging the mammary gland and mammary tumours in 3D: Optical tissue clearing and immunofluorescence methods. Breast Cancer Res. 2016, 18, 127. [Google Scholar] [CrossRef]
- Rios, A.C.; Capaldo, B.D.; Vaillant, F.; Pal, B.; van Ineveld, R.; Dawson, C.A.; Chen, Y.; Nolan, E.; Fu, N.Y.; Jackling, F.C.; et al. Intraclonal Plasticity in Mammary Tumors Revealed through Large-Scale Single-Cell Resolution 3D Imaging. Cancer Cell 2019, 35, 618–632.e616. [Google Scholar] [CrossRef]
- Sabdyusheva Litschauer, I.; Becker, K.; Saghafi, S.; Ballke, S.; Bollwein, C.; Foroughipour, M.; Gaugeler, J.; Schavelová, V.; László, V.; Döme, B.; et al. 3D histopathology of human tumours by fast clearing and ultramicroscopy. Sci. Rep. 2020, 10, 17619. [Google Scholar] [CrossRef]
- Chung, K.; Wallace, J.; Kim, S.Y.; Kalyanasundaram, S.; Andalman, A.S.; Davidson, T.J.; Mirzabekov, J.J.; Zalocusky, K.A.; Mattis, J.; Denisin, A.K.; et al. Structural and molecular interrogation of intact biological systems. Nature 2013, 497, 332–337. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Alieva, M.; Wellens, L.M.; Ariese, H.C.R.; Jamieson, P.R.; Vonk, A.M.; Amatngalim, G.D.; Hu, H.; Oost, K.C.; Snippert, H.J.G.; et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019, 14, 1756–1771. [Google Scholar] [CrossRef]
- Grist, S.M.; Nasseri, S.S.; Poon, T.; Roskelley, C.; Cheung, K.C. On-chip clearing of arrays of 3-D cell cultures and micro-tissues. Biomicrofluidics 2016, 10, 044107. [Google Scholar] [CrossRef]
- Jonkman, J.; Brown, C.M.; Wright, G.D.; Anderson, K.I.; North, A.J. Tutorial: Guidance for quantitative confocal microscopy. Nat. Protoc. 2020, 15, 1585–1611. [Google Scholar] [CrossRef]
- Haddad, T.S.; Friedl, P.; Farahani, N.; Treanor, D.; Zlobec, I.; Nagtegaal, I. Tutorial: Methods for three-dimensional visualization of archival tissue material. Nat. Protoc. 2021, 16, 4945–4962. [Google Scholar] [CrossRef]
- Piston, D.W. When two is better than one: Elements of intravital microscopy. PLoS Biol. 2005, 3, e207. [Google Scholar] [CrossRef]
- Franke, T.; Rhode, S. Two-photon microscopy for deep tissue imaging of living specimens. Microsc. Today 2012, 20, 12–16. [Google Scholar] [CrossRef]
- Wan, Y.; McDole, K.; Keller, P.J. Light-Sheet Microscopy and Its Potential for Understanding Developmental Processes. Annu. Rev. Cell Dev. Biol. 2019, 35, 655–681. [Google Scholar] [CrossRef]
- Poola, P.K.; Afzal, M.I.; Yoo, Y.; Kim, K.H.; Chung, E. Light sheet microscopy for histopathology applications. Biomed. Eng. Lett. 2019, 9, 279–291. [Google Scholar] [CrossRef]
- Kristensen, V.N.; Lingjærde, O.C.; Russnes, H.G.; Vollan, H.K.; Frigessi, A.; Børresen-Dale, A.L. Principles and methods of integrative genomic analyses in cancer. Nat. Rev. Cancer 2014, 14, 299–313. [Google Scholar] [CrossRef]
- Kamps, R.; Brandão, R.D.; Bosch, B.J.; Paulussen, A.D.; Xanthoulea, S.; Blok, M.J.; Romano, A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int. J. Mol. Sci. 2017, 18, 308. [Google Scholar] [CrossRef]
- Gupta, R.; Kala, N.; Pai, A.; Malviya, R. Bioinformatics Approach for Data Capturing: The Case of Breast Cancer. Curr. Cancer Ther. Rev. 2021, 17, 261–266. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 2020, 14, 1–24. [Google Scholar] [CrossRef]
- Kagohara, L.T.; Stein-O’Brien, G.L.; Kelley, D.; Flam, E.; Wick, H.C.; Danilova, L.V.; Easwaran, H.; Favorov, A.V.; Qian, J.; Gaykalova, D.A.; et al. Epigenetic regulation of gene expression in cancer: Techniques, resources and analysis. Brief Funct Genom. 2018, 17, 49–63. [Google Scholar] [CrossRef]
- Akbani, R.; Ng, P.K.; Werner, H.M.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef]
- Hackl, H.; Charoentong, P.; Finotello, F.; Trajanoski, Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nat. Rev. Genet. 2016, 17, 441–458. [Google Scholar] [CrossRef]
- Meng, C.; Zeleznik, O.A.; Thallinger, G.G.; Kuster, B.; Gholami, A.M.; Culhane, A.C. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief Bioinform. 2016, 17, 628–641. [Google Scholar] [CrossRef]
- Kim, D.; Li, R.; Dudek, S.M.; Ritchie, M.D. Predicting censored survival data based on the interactions between meta-dimensional omics data in breast cancer. J. Biomed. Inform. 2015, 56, 220–228. [Google Scholar] [CrossRef]
- Guffanti, A.; Iacono, M.; Pelucchi, P.; Kim, N.; Soldà, G.; Croft, L.J.; Taft, R.J.; Rizzi, E.; Askarian-Amiri, M.; Bonnal, R.J.; et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genom. 2009, 10, 163. [Google Scholar] [CrossRef]
- Heng, Y.J.; Lester, S.C.; Tse, G.M.; Factor, R.E.; Allison, K.H.; Collins, L.C.; Chen, Y.Y.; Jensen, K.C.; Johnson, N.B.; Jeong, J.C.; et al. The molecular basis of breast cancer pathological phenotypes. J. Pathol. 2017, 241, 375–391. [Google Scholar] [CrossRef]
- Peri, S.; de Cicco, R.L.; Santucci-Pereira, J.; Slifker, M.; Ross, E.A.; Russo, I.H.; Russo, P.A.; Arslan, A.A.; Belitskaya-Lévy, I.; Zeleniuch-Jacquotte, A.; et al. Defining the genomic signature of the parous breast. BMC Med. Genom. 2012, 5, 46. [Google Scholar] [CrossRef]
- Suo, C.; Hrydziuszko, O.; Lee, D.; Pramana, S.; Saputra, D.; Joshi, H.; Calza, S.; Pawitan, Y. Integration of somatic mutation, expression and functional data reveals potential driver genes predictive of breast cancer survival. Bioinformatics 2015, 31, 2607–2613. [Google Scholar] [CrossRef]
- Niida, A.; Smith, A.D.; Imoto, S.; Aburatani, H.; Zhang, M.Q.; Akiyama, T. Gene set-based module discovery in the breast cancer transcriptome. BMC Bioinform. 2009, 10, 71. [Google Scholar] [CrossRef]
- Martínez-Canales, S.; Cifuentes, F.; López De Rodas Gregorio, M.; Serrano-Oviedo, L.; Galán-Moya, E.M.; Amir, E.; Pandiella, A.; Győrffy, B.; Ocaña, A. Transcriptomic immunologic signature associated with favorable clinical outcome in basal-like breast tumors. PLoS ONE 2017, 12, e0175128. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, L.; Shi, L.; Yun, F.; Liu, X.; Chen, Y.; Chen, C.; Ren, Y.; Jia, Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol. Biol. Lett. 2019, 24, 38. [Google Scholar] [CrossRef]
- Liu, S.; Liu, X.; Wu, J.; Zhou, W.; Ni, M.; Meng, Z.; Jia, S.; Zhang, J.; Guo, S.; Lu, S.; et al. Identification of candidate biomarkers correlated with the pathogenesis and prognosis of breast cancer via integrated bioinformatics analysis. Medicine (Baltimore) 2020, 99, e23153. [Google Scholar] [CrossRef]
- Albiges, L.; Goubar, A.; Scott, V.; Vicier, C.; Lefèbvre, C.; Alsafadi, S.; Commo, F.; Saghatchian, M.; Lazar, V.; Dessen, P.; et al. Chk1 as a new therapeutic target in triple-negative breast cancer. Breast 2014, 23, 250–258. [Google Scholar] [CrossRef]
- Aswad, L.; Yenamandra, S.P.; Ow, G.S.; Grinchuk, O.; Ivshina, A.V.; Kuznetsov, V.A. Genome and transcriptome delineation of two major oncogenic pathways governing invasive ductal breast cancer development. Oncotarget 2015, 6, 36652–36674. [Google Scholar] [CrossRef]
- Rosati, D.; Giordano, A. Single-cell RNA sequencing and bioinformatics as tools to decipher cancer heterogenicity and mechanisms of drug resistance. Biochem. Pharmacol. 2022, 195, 114811. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Zhu, D.; Zhao, Z.; Cui, G.; Chang, S.; Hu, L.; See, Y.X.; Lim, M.G.L.; Guo, D.; Chen, X.; Poudel, B.; et al. Single-Cell Transcriptome Analysis Reveals Estrogen Signaling Coordinately Augments One-Carbon, Polyamine, and Purine Synthesis in Breast Cancer. Cell Rep. 2018, 25, 2285–2298.e2284. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef]
- Duffy, M.J.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef]
- Toss, A.; Cristofanilli, M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015, 17, 60. [Google Scholar] [CrossRef]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer-expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef]
- Danenberg, E.; Bardwell, H.; Zanotelli, V.R.T.; Provenzano, E.; Chin, S.F.; Rueda, O.M.; Green, A.; Rakha, E.; Aparicio, S.; Ellis, I.O.; et al. Breast tumor microenvironment structures are associated with genomic features and clinical outcome. Nat. Genet. 2022, 54, 660–669. [Google Scholar] [CrossRef]
- Hutchinson, K.E.; Yost, S.E.; Chang, C.W.; Johnson, R.M.; Carr, A.R.; McAdam, P.R.; Halligan, D.L.; Chang, C.C.; Schmolze, D.; Liang, J.; et al. Comprehensive Profiling of Poor-Risk Paired Primary and Recurrent Triple-Negative Breast Cancers Reveals Immune Phenotype Shifts. Clin. Cancer Res. 2020, 26, 657–668. [Google Scholar] [CrossRef]
- Wang, Q.; Guldner, I.H.; Golomb, S.M.; Sun, L.; Harris, J.A.; Lu, X.; Zhang, S. Single-cell profiling guided combinatorial immunotherapy for fast-evolving CDK4/6 inhibitor-resistant HER2-positive breast cancer. Nat. Commun. 2019, 10, 3817. [Google Scholar] [CrossRef]
- Han, Y.; Wang, D.; Peng, L.; Huang, T.; He, X.; Wang, J.; Ou, C. Single-cell sequencing: A promising approach for uncovering the mechanisms of tumor metastasis. J. Hematol. Oncol. 2022, 15, 59. [Google Scholar] [CrossRef]
- Ward, S.; Scope, A.; Rafia, R.; Pandor, A.; Harnan, S.; Evans, P.; Wyld, L. Gene expression profiling and expanded immunohistochemistry tests to guide the use of adjuvant chemotherapy in breast cancer management: A systematic review and cost-effectiveness analysis. Health Technol. Assess 2013, 17, 1–302. [Google Scholar] [CrossRef]
- Harbeck, N.; Sotlar, K.; Wuerstlein, R.; Doisneau-Sixou, S. Molecular and protein markers for clinical decision making in breast cancer: Today and tomorrow. Cancer Treat. Rev. 2014, 40, 434–444. [Google Scholar] [CrossRef]
- Sun, L.; Wu, A.; Bean, G.R.; Hagemann, I.S.; Lin, C.Y. Molecular Testing in Breast Cancer: Current Status and Future Directions. J. Mol. Diagn. 2021, 23, 1422–1432. [Google Scholar] [CrossRef]
- Oliveira, L.J.C.; Amorim, L.C.; Megid, T.B.C.; de Resende, C.A.A.; Mano, M.S. Gene expression signatures in early breast cancer: Better together with clinicopathological features. Crit. Rev. Oncol. Hematol. 2022, 175, 103708. [Google Scholar] [CrossRef]
- Varga, Z.; Sinn, P.; Seidman, A.D. Summary of head-to-head comparisons of patient risk classifications by the 21-gene Recurrence Score® (RS) assay and other genomic assays for early breast cancer. Int. J. Cancer 2019, 145, 882–893. [Google Scholar] [CrossRef]
- Matikas, A.; Foukakis, T.; Swain, S.; Bergh, J. Avoiding over- and undertreatment in patients with resected node-positive breast cancer with the use of gene expression signatures: Are we there yet? Ann. Oncol. 2019, 30, 1044–1050. [Google Scholar] [CrossRef]
- Kim, H.K.; Park, K.H.; Kim, Y.; Park, S.E.; Lee, H.S.; Lim, S.W.; Cho, J.H.; Kim, J.Y.; Lee, J.E.; Ahn, J.S.; et al. Discordance of the PAM50 Intrinsic Subtypes Compared with Immunohistochemistry-Based Surrogate in Breast Cancer Patients: Potential Implication of Genomic Alterations of Discordance. Cancer Res. Treat. 2019, 51, 737–747. [Google Scholar] [CrossRef]
- Giesen, C.; Wang, H.A.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schüffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Asp, M.; Bergenstråhle, J.; Lundeberg, J. Spatially Resolved Transcriptomes-Next Generation Tools for Tissue Exploration. Bioessays 2020, 42, e1900221. [Google Scholar] [CrossRef]
- Jackson, H.W.; Fischer, J.R.; Zanotelli, V.R.T.; Ali, H.R.; Mechera, R.; Soysal, S.D.; Moch, H.; Muenst, S.; Varga, Z.; Weber, W.P.; et al. The single-cell pathology landscape of breast cancer. Nature 2020, 578, 615–620. [Google Scholar] [CrossRef]
- Crosetto, N.; Bienko, M.; van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2015, 16, 57–66. [Google Scholar] [CrossRef]
- Wellings, E.; Vassiliades, L.; Abdalla, R. Breast Cancer Screening for High-Risk Patients of Different Ages and Risk-Which Modality Is Most Effective? Cureus 2016, 8, e945. [Google Scholar] [CrossRef]
- Pinsky, R.W.; Helvie, M.A. Mammographic breast density: Effect on imaging and breast cancer risk. J. Natl. Compr. Canc. Netw. 2010, 8, 1157–1164. [Google Scholar] [CrossRef]
- McCormack, V.A.; dos Santos Silva, I. Breast density and parenchymal patterns as markers of breast cancer risk: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1159–1169. [Google Scholar] [CrossRef]
- Sardanelli, F.; Boetes, C.; Borisch, B.; Decker, T.; Federico, M.; Gilbert, F.J.; Helbich, T.; Heywang-Köbrunner, S.H.; Kaiser, W.A.; Kerin, M.J.; et al. Magnetic resonance imaging of the breast: Recommendations from the EUSOMA working group. Eur. J. Cancer 2010, 46, 1296–1316. [Google Scholar] [CrossRef]
- Mann, R.M.; Kuhl, C.K.; Kinkel, K.; Boetes, C. Breast MRI: Guidelines from the European Society of Breast Imaging. Eur. Radiol. 2008, 18, 1307–1318. [Google Scholar] [CrossRef]
- Mango, V.L.; Morris, E.A.; David Dershaw, D.; Abramson, A.; Fry, C.; Moskowitz, C.S.; Hughes, M.; Kaplan, J.; Jochelson, M.S. Abbreviated protocol for breast MRI: Are multiple sequences needed for cancer detection? Eur. J. Radiol. 2015, 84, 65–70. [Google Scholar] [CrossRef]
- Heacock, L.; Melsaether, A.N.; Heller, S.L.; Gao, Y.; Pysarenko, K.M.; Babb, J.S.; Kim, S.G.; Moy, L. Evaluation of a known breast cancer using an abbreviated breast MRI protocol: Correlation of imaging characteristics and pathology with lesion detection and conspicuity. Eur. J. Radiol. 2016, 85, 815–823. [Google Scholar] [CrossRef]
- Ozmen, N.; Dapp, R.; Zapf, M.; Gemmeke, H.; Ruiter, N.V.; van Dongen, K.W. Comparing different ultrasound imaging methods for breast cancer detection. IEEE Trans. Ultrason. Ferroelectr Freq. Control. 2015, 62, 637–646. [Google Scholar] [CrossRef]
- Berg, W.A.; Zhang, Z.; Lehrer, D.; Jong, R.A.; Pisano, E.D.; Barr, R.G.; Böhm-Vélez, M.; Mahoney, M.C.; Evans, W.P.; Larsen, L.H.; et al. Detection of breast cancer with addition of annual screening ultrasound or a single screening MRI to mammography in women with elevated breast cancer risk. JAMA 2012, 307, 1394–1404. [Google Scholar] [CrossRef]
- Kuhl, C.; Weigel, S.; Schrading, S.; Arand, B.; Bieling, H.; König, R.; Tombach, B.; Leutner, C.; Rieber-Brambs, A.; Nordhoff, D.; et al. Prospective multicenter cohort study to refine management recommendations for women at elevated familial risk of breast cancer: The EVA trial. J. Clin. Oncol. 2010, 28, 1450–1457. [Google Scholar] [CrossRef]
- Basu, S.; Mavi, A.; Cermik, T.; Houseni, M.; Alavi, A. Implications of standardized uptake value measurements of the primary lesions in proven cases of breast carcinoma with different degree of disease burden at diagnosis: Does 2-deoxy-2-[F-18]fluoro-D-glucose-positron emission tomography predict tumor biology? Mol. Imaging Biol. 2008, 10, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Schwarzbach, M.H.; Hinz, U.; Dimitrakopoulou-Strauss, A.; Willeke, F.; Cardona, S.; Mechtersheimer, G.; Lehnert, T.; Strauss, L.G.; Herfarth, C.; Büchler, M.W. Prognostic significance of preoperative [18-F] fluorodeoxyglucose (FDG) positron emission tomography (PET) imaging in patients with resectable soft tissue sarcomas. Ann. Surg. 2005, 241, 286–294. [Google Scholar] [CrossRef] [PubMed]
- van Baardwijk, A.; Dooms, C.; van Suylen, R.J.; Verbeken, E.; Hochstenbag, M.; Dehing-Oberije, C.; Rupa, D.; Pastorekova, S.; Stroobants, S.; Buell, U.; et al. The maximum uptake of (18) F-deoxyglucose on positron emission tomography scan correlates with survival, hypoxia inducible factor-1alpha and GLUT-1 in non-small cell lung cancer. Eur. J. Cancer 2007, 43, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Pawaskar, A.; Basu, S.; Rajan, M.G.; Asopa, R.V.; Arora, B.; Nair, N.; Banavali, S. Potential role of FDG PET imaging in predicting metastatic potential and assessment of therapeutic response to neoadjuvant chemotherapy in Ewing sarcoma family of tumors. Clin. Nucl. Med. 2011, 36, 973–977. [Google Scholar] [CrossRef]
- Zimny, M.; Gagel, B.; DiMartino, E.; Hamacher, K.; Coenen, H.H.; Westhofen, M.; Eble, M.; Buell, U.; Reinartz, P. FDG—A marker of tumour hypoxia? A comparison with [18F] fluoromisonidazole and pO2-polarography in metastatic head and neck cancer. Eur. J. Nucl. Med. Mol. Imaging 2006, 33, 1426–1431. [Google Scholar] [CrossRef]
- Zhuang, H.; Pourdehnad, M.; Lambright, E.S.; Yamamoto, A.J.; Lanuti, M.; Li, P.; Mozley, P.D.; Rossman, M.D.; Albelda, S.M.; Alavi, A. Dual time point 18F-FDG PET imaging for differentiating malignant from inflammatory processes. J. Nucl. Med. 2001, 42, 1412–1417. [Google Scholar]
- Chenevert, T.L.; McKeever, P.E.; Ross, B.D. Monitoring early response of experimental brain tumors to therapy using diffusion magnetic resonance imaging. Clin. Cancer Res. 1997, 3, 1457–1466. [Google Scholar]
- Chenevert, T.L.; Stegman, L.D.; Taylor, J.M.; Robertson, P.L.; Greenberg, H.S.; Rehemtulla, A.; Ross, B.D. Diffusion magnetic resonance imaging: An early surrogate marker of therapeutic efficacy in brain tumors. J. Natl. Cancer Inst. 2000, 92, 2029–2036. [Google Scholar] [CrossRef]
- Winnard, P.T.; Pathak, A.P.; Dhara, S.; Cho, S.Y.; Raman, V.; Pomper, M.G. Molecular imaging of metastatic potential. J. Nucl. Med. 2008, 49 (Suppl. S2), 96S–112S. [Google Scholar] [CrossRef]
- Monteil, J.; Maubon, A.; Leobon, S.; Roux, S.; Marin, B.; Renaudie, J.; Genet, D.; Fermeaux, V.; Aubard, Y.; Tubiana-Mathieu, N. Lymph node assessment with (18)F-FDG-PET and MRI in uterine cervical cancer. Anticancer Res. 2011, 31, 3865–3871. [Google Scholar]
- Song, B.I.; Lee, S.W.; Jeong, S.Y.; Chae, Y.S.; Lee, W.K.; Ahn, B.C.; Lee, J. 18F-FDG uptake by metastatic axillary lymph nodes on pretreatment PET/CT as a prognostic factor for recurrence in patients with invasive ductal breast cancer. J. Nucl. Med. 2012, 53, 1337–1344. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klerkx, W.M.; Bax, L.; Veldhuis, W.B.; Heintz, A.P.; Mali, W.P.; Peeters, P.H.; Moons, K.G. Detection of lymph node metastases by gadolinium-enhanced magnetic resonance imaging: Systematic review and meta-analysis. J. Natl. Cancer Inst. 2010, 102, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Moy, L.; Noz, M.E.; Maguire, G.Q.; Melsaether, A.; Deans, A.E.; Murphy-Walcott, A.D.; Ponzo, F. Role of fusion of prone FDG-PET and magnetic resonance imaging of the breasts in the evaluation of breast cancer. Breast J. 2010, 16, 369–376. [Google Scholar] [CrossRef]
- Margolis, N.E.; Moy, L.; Sigmund, E.E.; Freed, M.; McKellop, J.; Melsaether, A.N.; Kim, S.G. Assessment of Aggressiveness of Breast Cancer Using Simultaneous 18F-FDG-PET and DCE-MRI: Preliminary Observation. Clin. Nucl. Med. 2016, 41, e355–e361. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, N.; Suzuki, A.; Sobue, T.; Kawai, M.; Yamamoto, S.; Zheng, Y.F.; Shiono, Y.N.; Saito, H.; Kuriyama, S.; Tohno, E.; et al. Sensitivity and specificity of mammography and adjunctive ultrasonography to screen for breast cancer in the Japan Strategic Anti-cancer Randomized Trial (J-START): A randomised controlled trial. Lancet 2016, 387, 341–348. [Google Scholar] [CrossRef]
- Xia, J.; Yao, J.; Wang, L.V. Photoacoustic tomography: Principles and advances. Electromagn. Waves (Camb.) 2014, 147, 1–22. [Google Scholar] [CrossRef]
- Beard, P. Biomedical photoacoustic imaging. Interface Focus 2011, 1, 602–631. [Google Scholar] [CrossRef]
- Tromberg, B.J.; Pogue, B.W.; Paulsen, K.D.; Yodh, A.G.; Boas, D.A.; Cerussi, A.E. Assessing the future of diffuse optical imaging technologies for breast cancer management. Med. Phys. 2008, 35, 2443–2451. [Google Scholar] [CrossRef]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef]
- Hoelen, C.G.; de Mul, F.F.; Pongers, R.; Dekker, A. Three-dimensional photoacoustic imaging of blood vessels in tissue. Opt. Lett. 1998, 23, 648–650. [Google Scholar] [CrossRef]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- McDougall, S.R.; Anderson, A.R.; Chaplain, M.A. Mathematical modelling of dynamic adaptive tumour-induced angiogenesis: Clinical implications and therapeutic targeting strategies. J. Theor. Biol. 2006, 241, 564–589. [Google Scholar] [CrossRef]
- Ensenyat-Mendez, M.; Llinàs-Arias, P.; Orozco, J.I.J.; Íñiguez-Muñoz, S.; Salomon, M.P.; Sesé, B.; DiNome, M.L.; Marzese, D.M. Current Triple-Negative Breast Cancer Subtypes: Dissecting the Most Aggressive Form of Breast Cancer. Front. Oncol. 2021, 11, 681476. [Google Scholar] [CrossRef] [PubMed]
- Menezes, G.L.G.; Mann, R.M.; Meeuwis, C.; Bisschops, B.; Veltman, J.; Lavin, P.T.; van de Vijver, M.J.; Pijnappel, R.M. Optoacoustic imaging of the breast: Correlation with histopathology and histopathologic biomarkers. Eur. Radiol. 2019, 29, 6728–6740. [Google Scholar] [CrossRef] [PubMed]
- Neuschler, E.I.; Butler, R.; Young, C.A.; Barke, L.D.; Bertrand, M.L.; Böhm-Vélez, M.; Destounis, S.; Donlan, P.; Grobmyer, S.R.; Katzen, J.; et al. A Pivotal Study of Optoacoustic Imaging to Diagnose Benign and Malignant Breast Masses: A New Evaluation Tool for Radiologists. Radiology 2018, 287, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Huang, C.; Maslov, K.; Anastasio, M.A.; Wang, L.V. Enhancement of photoacoustic tomography by ultrasonic computed tomography based on optical excitation of elements of a full-ring transducer array. Opt. Lett. 2013, 38, 3140–3143. [Google Scholar] [CrossRef] [PubMed]
- Merčep, E.; Herraiz, J.L.; Deán-Ben, X.L.; Razansky, D. Transmission-reflection optoacoustic ultrasound (TROPUS) computed tomography of small animals. Light Sci. Appl. 2019, 8, 18. [Google Scholar] [CrossRef]
- Nyayapathi, N.; Xia, J. Photoacoustic imaging of breast cancer: A mini review of system design and image features. J. Biomed. Opt. 2019, 24, 1–13. [Google Scholar] [CrossRef]
- Manohar, S.; Dantuma, M. Current and future trends in photoacoustic breast imaging. Photoacoustics 2019, 16, 100134. [Google Scholar] [CrossRef]
- Levy-Jurgenson, A.; Tekpli, X.; Kristensen, V.N.; Yakhini, Z. Spatial transcriptomics inferred from pathology whole-slide images links tumor heterogeneity to survival in breast and lung cancer. Sci. Rep. 2020, 10, 18802. [Google Scholar] [CrossRef]
- He, B.; Bergenstråhle, L.; Stenbeck, L.; Abid, A.; Andersson, A.; Borg, Å.; Maaskola, J.; Lundeberg, J.; Zou, J. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat. Biomed. Eng. 2020, 4, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Monjo, T.; Koido, M.; Nagasawa, S.; Suzuki, Y.; Kamatani, Y. Efficient prediction of a spatial transcriptomics profile better characterizes breast cancer tissue sections without costly experimentation. Sci. Rep. 2022, 12, 4133. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wei, Z.; Yu, W.; Yin, R.; Yuan, Y.; Li, B.; Tang, Z.; Lu, Y.; Yang, Y. Spatial transcriptomics prediction from histology jointly through Transformer and graph neural networks. Brief Bioinform. 2022, bbac297. [Google Scholar] [CrossRef] [PubMed]
- Yoosuf, N.; Navarro, J.F.; Salmén, F.; Ståhl, P.L.; Daub, C.O. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res. 2020, 22, 6. [Google Scholar] [CrossRef]
- Mitchell, T. Machine Learning; McGraw Hill: New York, NY, USA, 1997. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Salama, W.M.; Aly, M.H. Deep learning in mammography images segmentation and classification: Automated CNN approach. Alex. Eng. J. 2021, 60, 4701–4709. [Google Scholar] [CrossRef]
- Jiang, Y.; Inciardi, M.F.; Edwards, A.V.; Papaioannou, J. Interpretation Time Using a Concurrent-Read Computer-Aided Detection System for Automated Breast Ultrasound in Breast Cancer Screening of Women with Dense Breast Tissue. AJR Am. J. Roentgenol. 2018, 211, 452–461. [Google Scholar] [CrossRef]
- Fan, M.; Li, Y.; Zheng, S.; Peng, W.; Tang, W.; Li, L. Computer-aided detection of mass in digital breast tomosynthesis using a faster region-based convolutional neural network. Methods 2019, 166, 103–111. [Google Scholar] [CrossRef]
- Redondo, A.; Comas, M.; Macià, F.; Ferrer, F.; Murta-Nascimento, C.; Maristany, M.T.; Molins, E.; Sala, M.; Castells, X. Inter- and intraradiologist variability in the BI-RADS assessment and breast density categories for screening mammograms. Br. J. Radiol. 2012, 85, 1465–1470. [Google Scholar] [CrossRef]
- Barinov, L.; Jairaj, A.; Paster, L.; Hulbert, W.; Mammone, R.; Podilchuk, C. Decision quality support in diagnostic breast ultrasound through Artificial Intelligence. In Proceedings of the 2016 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 3 December 2016; pp. 1–4. [Google Scholar]
- Lehman, C.D.; Wellman, R.D.; Buist, D.S.; Kerlikowske, K.; Tosteson, A.N.; Miglioretti, D.L.; Consortium, B.C.S. Diagnostic Accuracy of Digital Screening Mammography with and Without Computer-Aided Detection. JAMA Intern. Med. 2015, 175, 1828–1837. [Google Scholar] [CrossRef]
- Al-Antari, M.A.; Al-Masni, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int. J. Med. Inform. 2018, 117, 44–54. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, H.; Kawahara, D.; Kikuchi, Y. Understanding Breast Cancers through Spatial and High-Resolution Visualization Using Imaging Technologies. Cancers 2022, 14, 4080. https://doi.org/10.3390/cancers14174080
Takahashi H, Kawahara D, Kikuchi Y. Understanding Breast Cancers through Spatial and High-Resolution Visualization Using Imaging Technologies. Cancers. 2022; 14(17):4080. https://doi.org/10.3390/cancers14174080
Chicago/Turabian StyleTakahashi, Haruko, Daisuke Kawahara, and Yutaka Kikuchi. 2022. "Understanding Breast Cancers through Spatial and High-Resolution Visualization Using Imaging Technologies" Cancers 14, no. 17: 4080. https://doi.org/10.3390/cancers14174080
APA StyleTakahashi, H., Kawahara, D., & Kikuchi, Y. (2022). Understanding Breast Cancers through Spatial and High-Resolution Visualization Using Imaging Technologies. Cancers, 14(17), 4080. https://doi.org/10.3390/cancers14174080




