Early Dynamics of Quantitative SEPT9 and SHOX2 Methylation in Circulating Cell-Free Plasma DNA during Prostate Biopsy for Prostate Cancer Diagnosis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. TCGA (Tissue) Cohort
2.2. University Hospital Bonn (Plasma) Cohort
2.3. Investigation of ccfDNA Levels during Prostate Biopsy
2.4. Statistics
3. Results
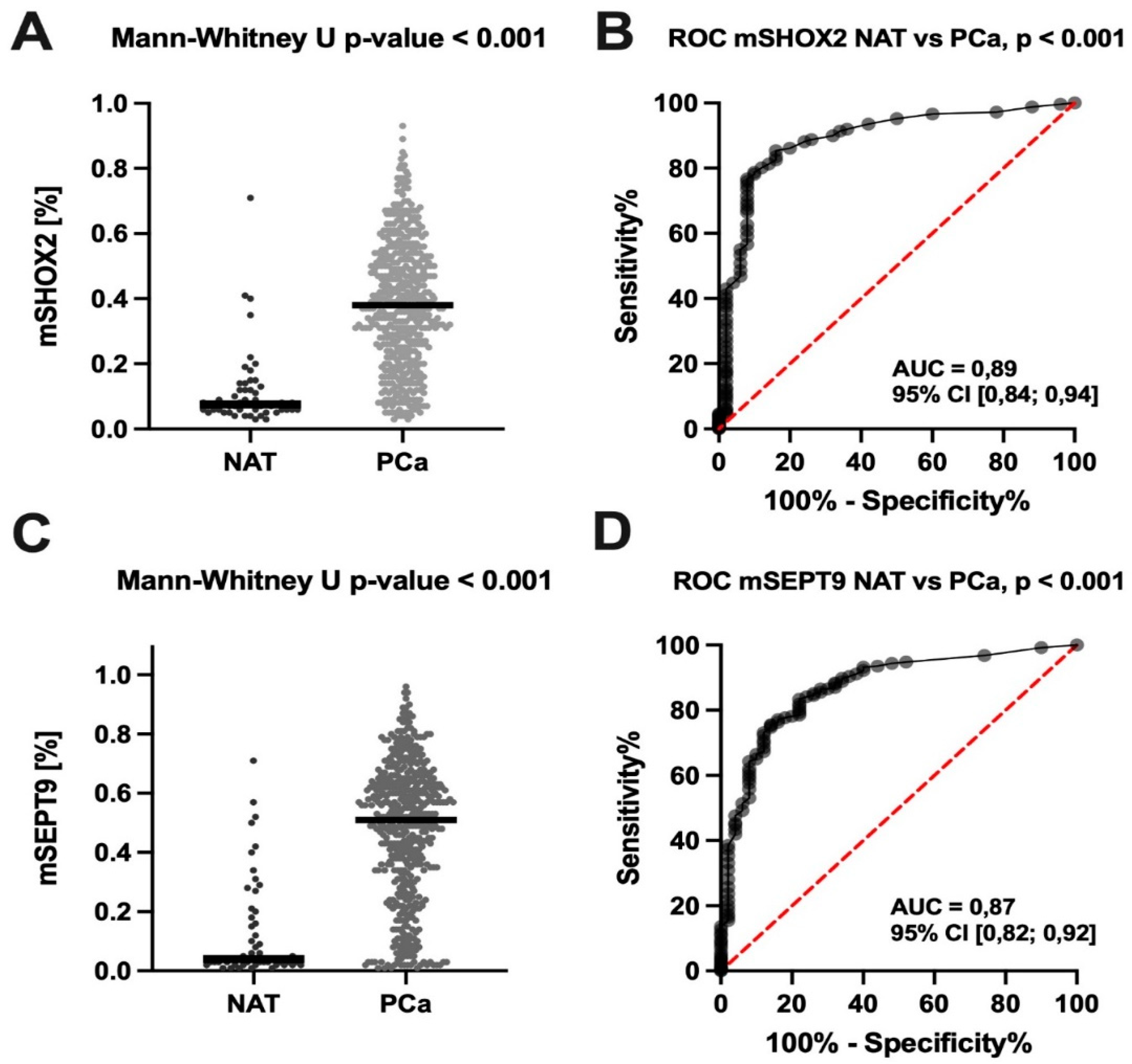
3.1. TCGA (Tissue) Cohort
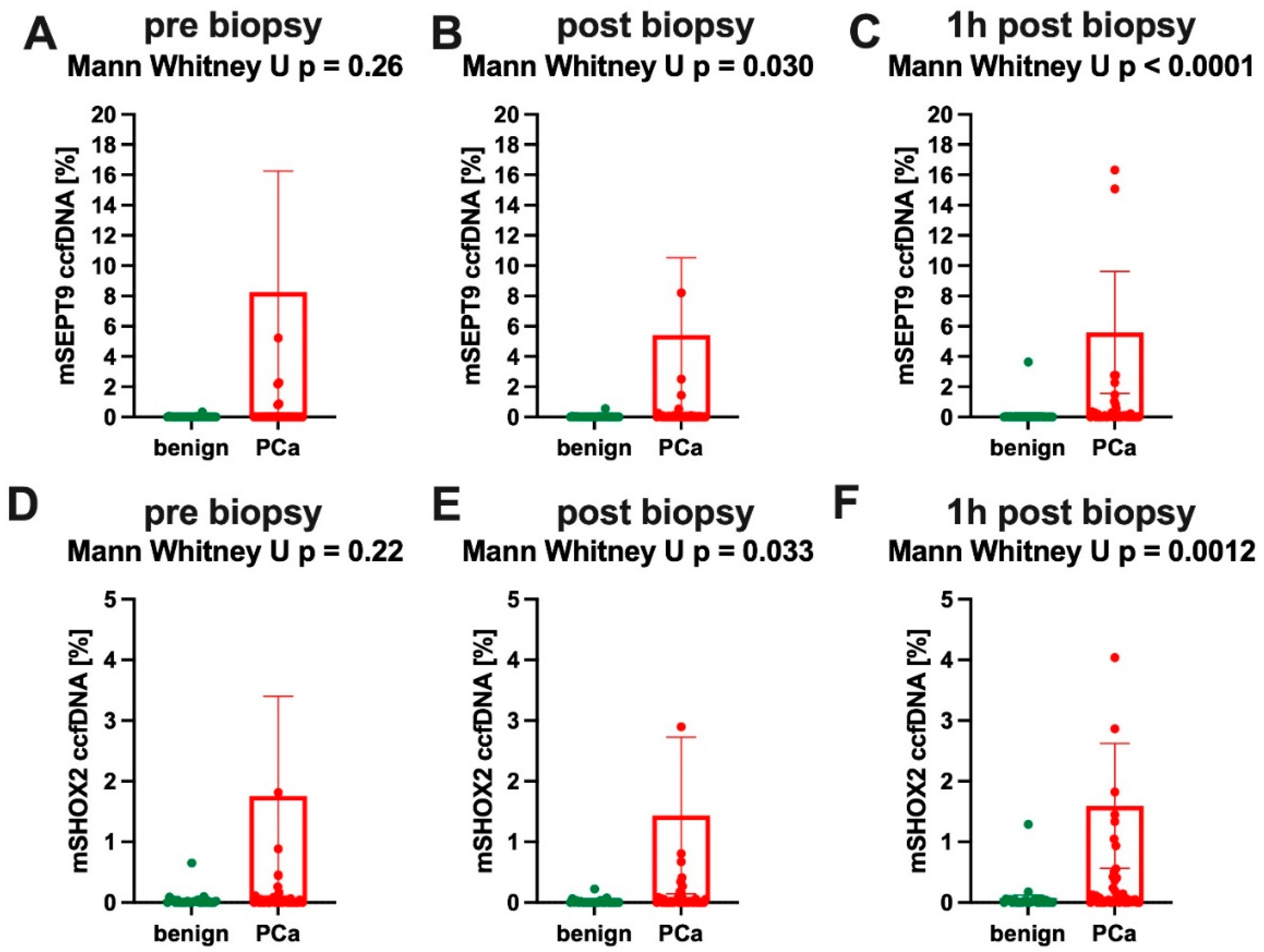
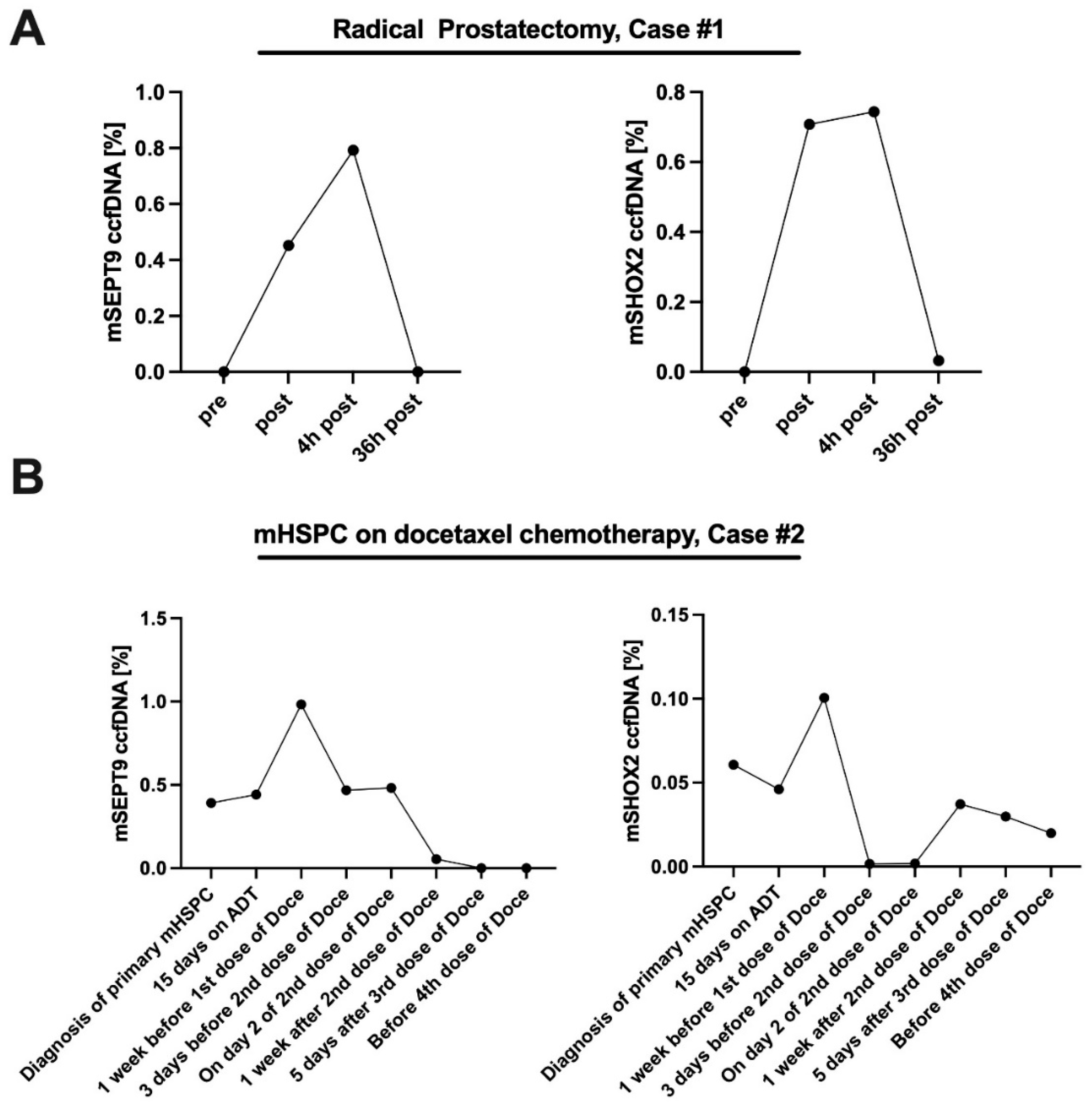
3.2. University Hospital Bonn (Plasma) Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, A.J.; Weinberger, M.I.; Nelson, C.J. Prostate Cancer: Psychosocial Implications and Management. Future Oncol. 2008, 4, 561–568. [Google Scholar] [CrossRef]
- Hövels, A.; Heesakkers, R.; Adang, E.; Jager, G.; Strum, S.; Hoogeveen, Y.; Severens, J.; Barentsz, J. The Diagnostic Accuracy of CT and MRI in the Staging of Pelvic Lymph Nodes in Patients with Prostate Cancer: A Meta-Analysis. Clin. Radiol. 2008, 63, 387–395. [Google Scholar] [CrossRef]
- Ahdoot, M.; Wilbur, A.R.; Reese, S.E.; Lebastchi, A.H.; Mehralivand, S.; Gomella, P.T.; Bloom, J.; Gurram, S.; Siddiqui, M.; Pinsky, P.; et al. Mri-Targeted, Systematic, And Combined Biopsy for Prostate Cancer Diagnosis. N. Engl. J. Med. 2020, 382, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Krausewitz, P.; Schmeller, H.; Luetkens, J.; Dabir, D.; Ellinger, J.; Ritter, M.; Conrad, R. Prospective Analysis of Pain Expectancy and Experience during Mr-Fusion Prostate Biopsy: Does Reality Match Patients’ Expectancy? World J. Urol. 2022, 40, 2239–2244. [Google Scholar] [CrossRef]
- Joosse, S.A.; Beyer, B.; Gasch, C.; Nastaly, P.; Kuske, A.; Isbarn, H.; Horst, L.J.; Hille, C.; Gorges, T.M.; Cayrefourcq, L.; et al. Tumor-Associated Release of Prostatic Cells into the Blood after Transrectal Ultrasound-Guided Biopsy in Patients with Histologically Confirmed Prostate Cancer. Clin. Chem. 2020, 66, 161–168. [Google Scholar] [CrossRef]
- Kuske, A.; Gorges, T.M.; Tennstedt, P.; Tiebel, A.-K.; Pompe, R.S.; Preißer, F.; Prues, S.; Mazel, M.; Markou, A.; Lianidou, E.; et al. Improved Detection of Circulating Tumor Cells in Non-Metastatic High-Risk Prostate Cancer Patients. Sci. Rep. 2016, 6, 39736. [Google Scholar] [CrossRef] [PubMed]
- Litwin, M.S.; Tan, H.-J. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA 2017, 317, 2532–2542. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, H.; Long, Y.; Li, P.; Gu, Y. The Main Sources of Circulating Cell-Free DNA: Apoptosis, Necrosis and Active Secretion. Crit. Rev. Oncol. Hematol. 2021, 157, 103166. [Google Scholar] [CrossRef]
- Galardi, F.; De Luca, F.; Romagnoli, D.; Biagioni, C.; Moretti, E.; Biganzoli, L.; Di Leo, A.; Migliaccio, I.; Malorni, L.; Benelli, M. Cell-Free DNA-Methylation-Based Methods and Applications in Oncology. Biomolecules 2020, 10, 1677. [Google Scholar] [CrossRef]
- Palanca-Ballester, C.; Rodriguez-Casanova, A.; Torres, S.; Calabuig-Fariñas, S.; Exposito, F.; Serrano, D.; Redin, E.; Valencia, K.; Jantus-Lewintre, E.; Diaz-Lagares, A.; et al. Cancer Epigenetic Biomarkers in Liquid Biopsy for High Incidence Malignancies. Cancers 2021, 13, 3016. [Google Scholar] [CrossRef] [PubMed]
- Vde Vos, L.; Jung, M.; Koerber, R.M.; Bawden, E.G.; Holderried, T.A.; Dietrich, J.; Bootz, F.; Brossart, P.; Kristiansen, G.; Dietrich, D. Treatment Response Monitoring in Patients with Advanced Malignancies Using Cell-Free Shox2 and Sept9 Dna Methylation in Blood: An Observational Prospective Study. J. Mol. Diagn. Jmd 2020, 22, 920–933. [Google Scholar] [CrossRef]
- Weiss, G.; Schlegel, A.; Kottwitz, D.; König, T.; Tetzner, R. Validation Of The Shox2/Ptger4 Dna Methylation Marker Panel for Plasma-Based Discrimination Between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2017, 12, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castaños-Vélez, E.; Blumenstein, B.A.; Rösch, T.; Osborn, N.; et al. Prospective Evaluation of Methylated Sept9 in Plasma for Detection of Asymptomatic Colorectal Cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Angulo, J.C.; Andrés, G.; Ashour, N.; Sánchez-Chapado, M.; López, J.I.; Ropero, S. Development of Castration Resistant Prostate Cancer Can Be Predicted by a Dna Hypermethylation Profile. J. Urol. 2016, 195, 619–626. [Google Scholar] [CrossRef]
- Angulo, J.C.; Andres, G.; Ashour, N.; Sánchez-Chapado, M.; Lopez, J.I.; Ropero, S. Free-Circulating Methylated DNA in Blood for Diagnosis, Staging, Prognosis, and Monitoring of Head and Neck Squamous Cell Carcinoma Patients: An Observational Prospective Cohort Study. Clin. Chem. 2017, 63, 1288–1296. [Google Scholar]
- Jung, M.; Kristiansen, G.; Dietrich, D. DNA Methylation Analysis of Free-Circulating DNA in Body Fluids. Methods Mol. Biol. 2018, 1708, 621–641. [Google Scholar] [PubMed]
- Dietrich, D.; Kneip, C.; Raji, O.; Liloglou, T.; Seegebarth, A.; Schlegel, T.; Flemming, N.; Rausch, S.; Distler, J.; Fleischhacker, M.; et al. Performance Evaluation of the DNA Methylation Biomarker Shox2 for the Aid in Diagnosis of Lung Cancer Based on the Analysis of Bronchial Aspirates. Int. J. Oncol. 2012, 40, 825–832. [Google Scholar] [CrossRef]
- de Vos, L.; Gevensleben, H.; Schröck, A.; Franzen, A.; Kristiansen, G.; Bootz, F.; Dietrich, D. Comparison of Quantification Algorithms for Circulating Cell-Free DNA Methylation Biomarkers in Blood Plasma from Cancer Patients. Clin. Epigenetics 2017, 9, 125. [Google Scholar] [CrossRef]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. Eau-Eanm-Estro-Esur-Siog Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences, 2nd ed.; Routledge Academic: New York, NY, USA; Lawrence Erlbaum Associates: New York, NY, USA, 1988. [Google Scholar]
- Oderda, M.; Rosazza, M.; Agnello, M.; Barale, M.; Calleris, G.; Daniele, L.; Delsedime, L.; Falcone, M.; Faletti, R.; Filippini, C.; et al. Natural History of Widespread High Grade Prostatic Intraepithelial Neoplasia and Atypical Small Acinar Proliferation: Should We Rebiopsy Them All? Scand. J. Urol. 2021, 55, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Uhr, A.; Glick, L.; Gomella, L.G. An Overview of Biomarkers in the Diagnosis and Management of Prostate Cancer. Can. J. Urol. 2020, 27 (Suppl. S3), 24–27. [Google Scholar] [PubMed]
- Li, N.; Zeng, Y.; Tai, M.; Lin, B.; Zhu, D.; Luo, Y.; Ren, X.; Zhu, X.; Li, L.; Wu, H.; et al. Analysis of the Prognostic Value and Gene Expression Mechanism of Shox2 in Lung Adenocarcinoma. Front. Mol. Biosci. 2021, 8, 688274. [Google Scholar] [CrossRef] [PubMed]
- Birtle, A.J.; Freeman, A.; Masters, J.R.W.; Payne, H.A.; Harland, S.J. Clinical Features of Patients Who Present with Metastatic Prostate Carcinoma and Serum Prostate-Specific Antigen (Psa) Levels < 10 Ng/Ml: The “Psa Negative” Patients. Cancer 2003, 98, 2362–2367. [Google Scholar] [PubMed]
- Haese, A.; Trooskens, G.; Steyaert, S.; Hessels, D.; Brawer, M.; Vlaeminck-Guillem, V.; Ruffion, A.; Tilki, D.; Schalken, J.; Groskopf, J.; et al. Multicenter Optimization and Validation of a 2-Gene Mrna Urine Test for Detection of Clinically Significant Prostate Cancer before Initial Prostate Biopsy. J. Urol. 2019, 202, 256–263. [Google Scholar] [CrossRef]


| All Men (n = 72) | |
|---|---|
| Age (years) | 70.5 ± 7.6 |
| PSA (ng/mL) | 10.6 ± 15.7 |
| Prostate volume (mL) | 51.7 ± 27.6 |
| Abnormal DRE (%) | 43.0 |
| Abnormal US (%) | 33.3 |
| PI-RADS < 3 (%) | 16.7 |
| PI-RADS 3 (%) | 19.4 |
| PI-RADS 4 (%) | 33.3 |
| PI-RADS 5 (%) | 30.6 |
| Re-Biopsy (%) | 29.2 |
| Total number of tumor bearing cores/patient | 4.6 ± 4.9 |
| PCa detection (%) | 65.3 |
| csPCa detection (%) | 62.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krausewitz, P.; Kluemper, N.; Richter, A.-P.; Büttner, T.; Kristiansen, G.; Ritter, M.; Ellinger, J. Early Dynamics of Quantitative SEPT9 and SHOX2 Methylation in Circulating Cell-Free Plasma DNA during Prostate Biopsy for Prostate Cancer Diagnosis. Cancers 2022, 14, 4355. https://doi.org/10.3390/cancers14184355
Krausewitz P, Kluemper N, Richter A-P, Büttner T, Kristiansen G, Ritter M, Ellinger J. Early Dynamics of Quantitative SEPT9 and SHOX2 Methylation in Circulating Cell-Free Plasma DNA during Prostate Biopsy for Prostate Cancer Diagnosis. Cancers. 2022; 14(18):4355. https://doi.org/10.3390/cancers14184355
Chicago/Turabian StyleKrausewitz, Philipp, Niklas Kluemper, Ayk-Peter Richter, Thomas Büttner, Glen Kristiansen, Manuel Ritter, and Jörg Ellinger. 2022. "Early Dynamics of Quantitative SEPT9 and SHOX2 Methylation in Circulating Cell-Free Plasma DNA during Prostate Biopsy for Prostate Cancer Diagnosis" Cancers 14, no. 18: 4355. https://doi.org/10.3390/cancers14184355
APA StyleKrausewitz, P., Kluemper, N., Richter, A.-P., Büttner, T., Kristiansen, G., Ritter, M., & Ellinger, J. (2022). Early Dynamics of Quantitative SEPT9 and SHOX2 Methylation in Circulating Cell-Free Plasma DNA during Prostate Biopsy for Prostate Cancer Diagnosis. Cancers, 14(18), 4355. https://doi.org/10.3390/cancers14184355







