Single Shot vs. Cocktail: A Comparison of Mono- and Combinative Application of miRNA-Targeted Mesyl Oligonucleotides for Efficient Antitumor Therapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotide Synthesis
2.2. Transfection of Tumor Cells with Oligonucleotides
2.3. xCelligence Real-Time Analysis of Cell Proliferation and Viability
2.4. Scratch Assay
2.5. Mice
2.6. Antitumor Studies Ex Vivo
2.7. Preparation of the Complexes of Cationic Liposomes and Oligonucleotides
2.8. Antitumor Studies In Vivo
2.9. Histology, Immunohistochemistry and Morphometry
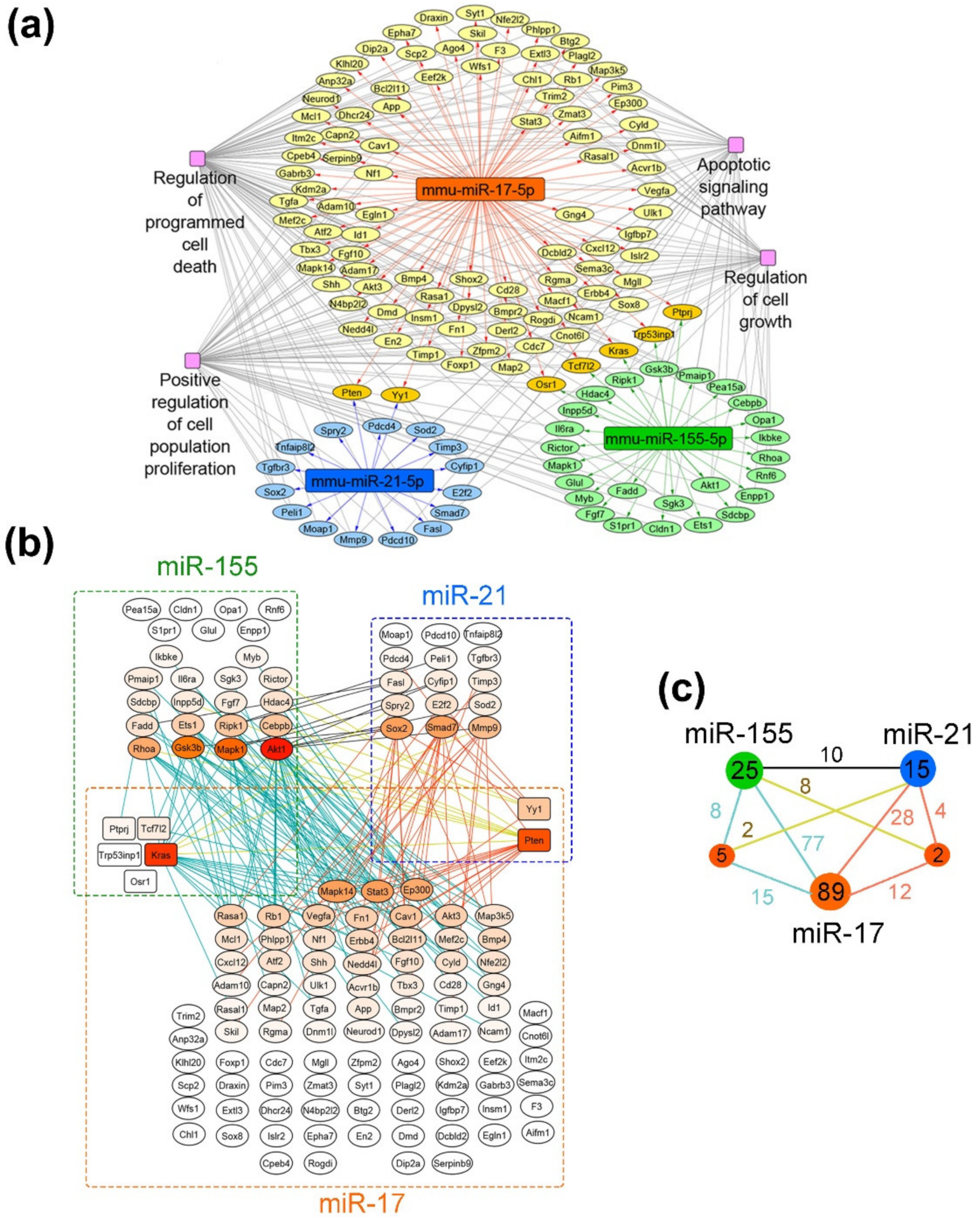
2.10. Reconstruction of miRNA-Target Networks and Their Functional Annotation
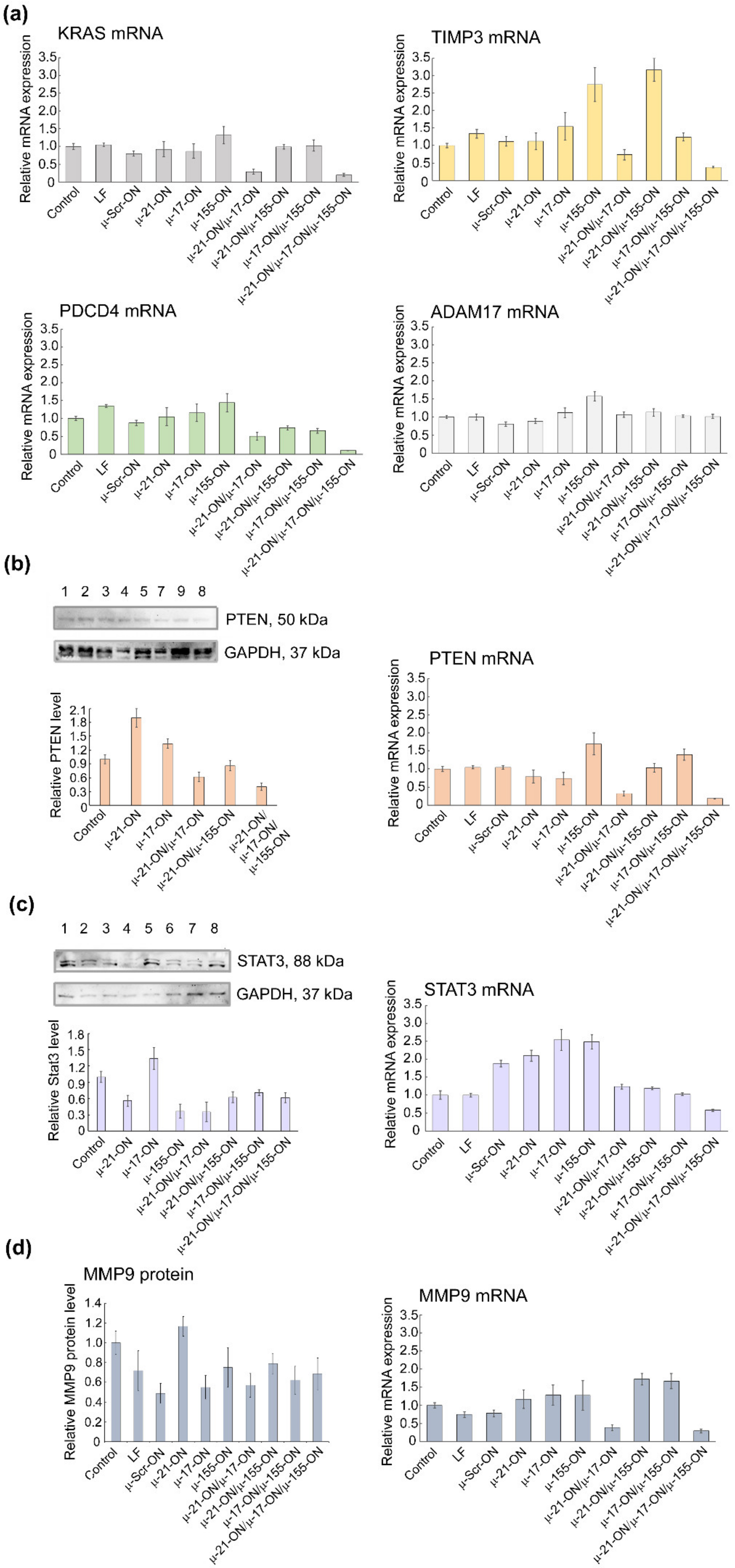
2.11. qPCR
2.12. Western Blot
2.13. Protein Level Measurement Using Flow Cytometry
2.14. Statistics
3. Results
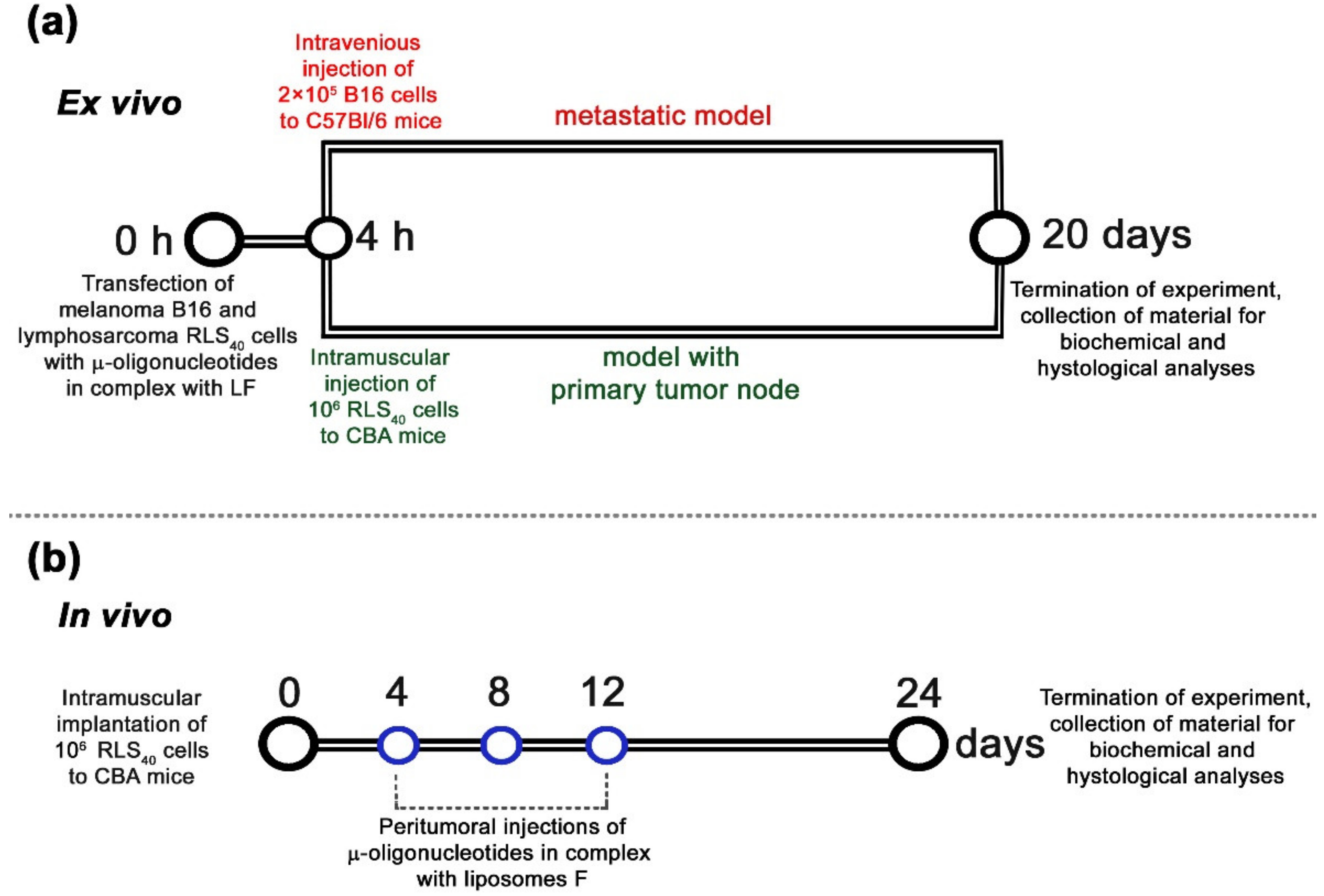
3.1. Conceptual Design of the Study
3.2. Antiproliferative Effect of Mono- and Combination Therapy with µ-Oligonucleotides Targeted to miR-21, miR-17 and miR-155
3.3. Antimigrative Effect of Mono- and Combination Therapy with µ-Oligonucleotides Targeted to miR-21, miR-17 and miR-155
3.4. Antitumor and Antimetastatic Activity of µ-Oligonucleotides in Murine Models
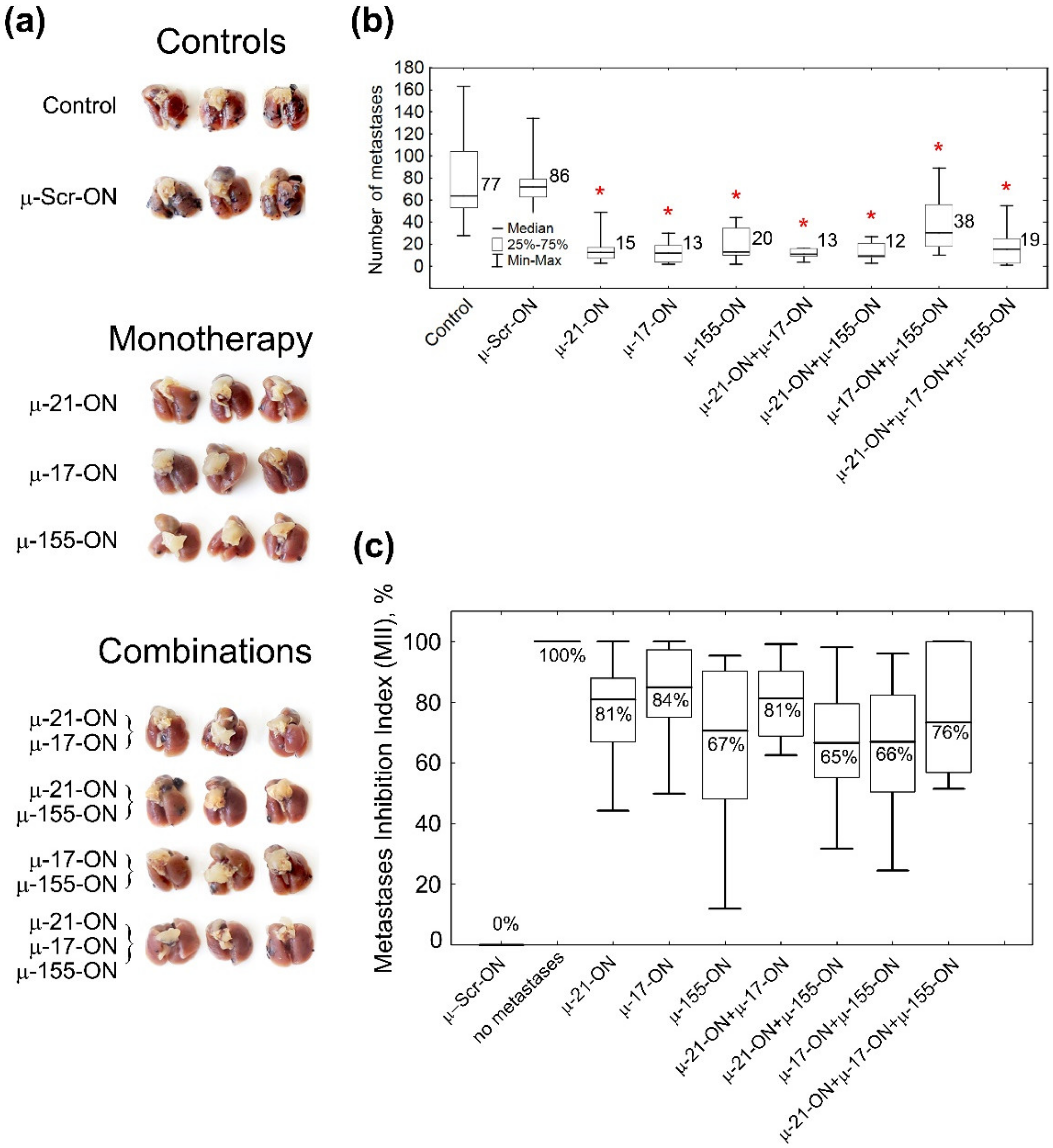
3.5. The Effect of Anti-miRNA Mono- and Combination Therapy with µ-Oligonucleotides on Melanoma B16 Metastases Development in Mice (Ex Vivo Experiment)
3.6. The Effect of Anti-miRNA Mono- and Combination Therapy with µ-Oligonucleotides on Lymphosarcoma RLS40 Tumor Growth (Ex Vivo Experiment)
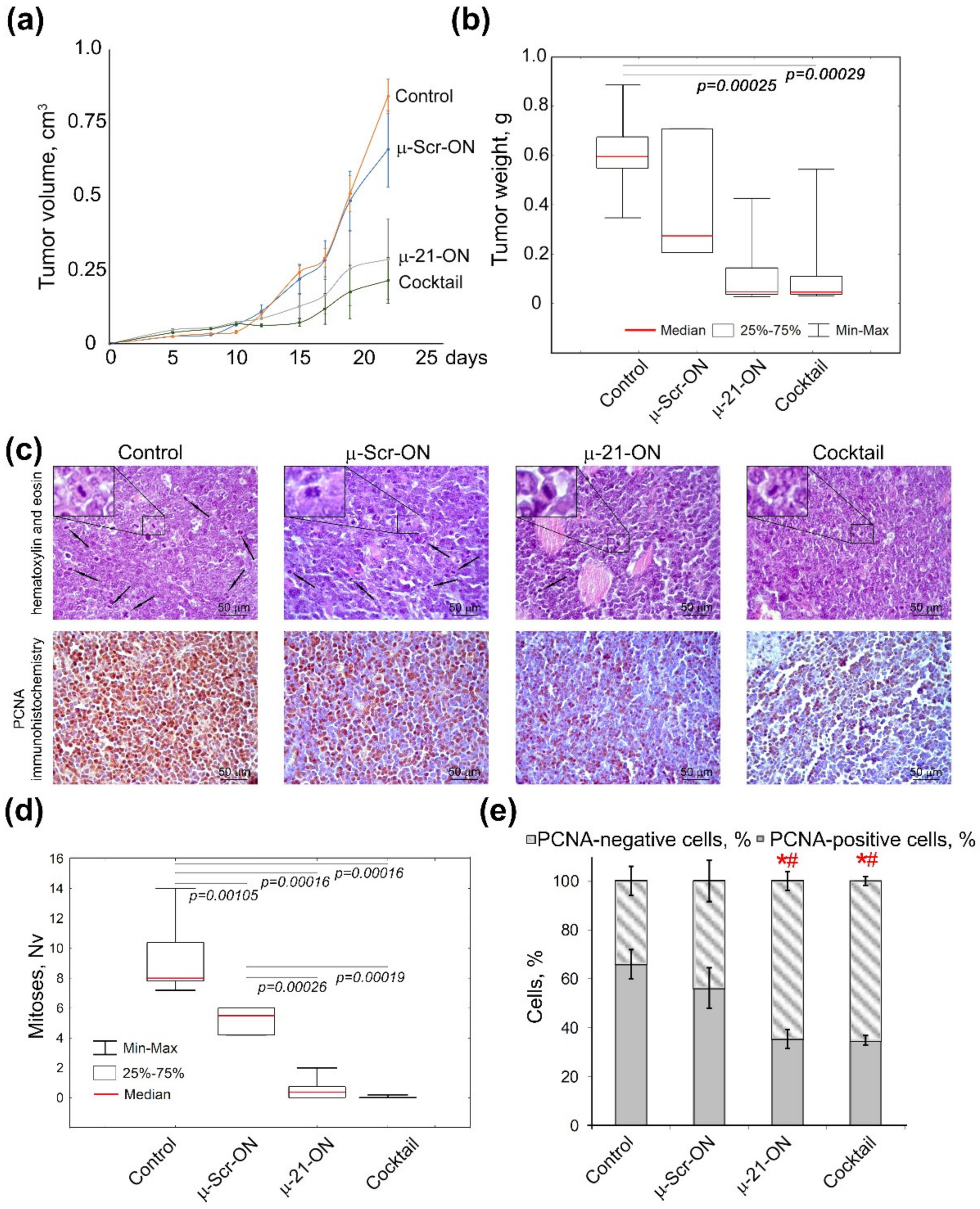
3.7. Efficiency of Anti-miRNA Therapy with µ-Oligonucleotides on Lymphosarcoma RLS40 Tumor Growth In Vivo
3.8. Toxicity of Anti-miRNA Combinative Treatment in Mice
3.9. Bioinformatic Analysis of Possible Molecular Interactions Underlying the Effects of Mono- and Combination Therapy with µ-Oligonucleotides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vaghari-Tabari, M.; Ferns, G.A.; Qujeq, D.; Andevari, A.N.; Sabahi, Z.; Moein, S. Signaling, Metabolism, and Cancer: An Important Relationship for Therapeutic Intervention. J. Cell. Physiol. 2021, 236, 5512–5532. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, N.; Coppola, V.; Bonci, D.; Miele, F.; Costantini, A.; Guadagnoli, M.; Bonanno, E.; Muto, G.; Frajese, G.V.; De Maria, R.; et al. The Inhibition of the Highly Expressed Mir-221 and Mir-222 Impairs the Growth of Prostate Carcinoma Xenografts in Mice. PLoS ONE 2008, 3, e4029. [Google Scholar] [CrossRef] [PubMed]
- Boutla, A.; Delidakis, C.; Tabler, M. Developmental Defects by Antisense-Mediated Inactivation of Micro-RNAs 2 and 13 in Drosophila and the Identification of Putative Target Genes. Nucleic Acids Res. 2003, 31, 4973–4980. [Google Scholar] [CrossRef]
- Raue, R.; Frank, A.C.; Syed, S.N.; Brüne, B. Therapeutic Targeting of Micrornas in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 2210. [Google Scholar] [CrossRef]
- Veliceasa, D.; Biyashev, D.; Qin, G.; Misener, S.; Mackie, A.R.; Kishore, R.; Volpert, O.V. Therapeutic Manipulation of Angiogenesis with MiR-27b. Vasc. Cell 2015, 7, 6. [Google Scholar] [CrossRef]
- Liu, Y.S.; Lin, H.Y.; Lai, S.W.; Huang, C.Y.; Huang, B.R.; Chen, P.Y.; Wei, K.C.; Lu, D.Y. MIR-181b Modulates EGFR-Dependent VCAM-1 Expression and Monocyte Adhesion in Glioblastoma. Oncogene 2017, 36, 5006–5022. [Google Scholar] [CrossRef]
- Mignacca, L.; Saint-Germain, E.; Benoit, A.; Bourdeau, V.; Moro, A.; Ferbeyre, G. Sponges against MiR-19 and MiR-155 Reactivate the P53-Socs1 Axis in Hematopoietic Cancers. Cytokine 2016, 82, 80–86. [Google Scholar] [CrossRef]
- Li, S.; Chen, T.; Zhong, Z.; Wang, Y.; Li, Y.; Zhao, X. MicroRNA-155 Silencing Inhibits Proliferation and Migration and Induces Apoptosis by Upregulating BACH1 in Renal Cancer Cells. Mol. Med. Rep. 2012, 5, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Buscaglia, L.E.B.; Li, Y. Apoptosis and the Target Genes of MicroRNA-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Li, D.; Sha, J.; Sun, P.; Huang, Y. MicroRNA-21 Directly Targets MARCKS and Promotes Apoptosis Resistance and Invasion in Prostate Cancer Cells. Biochem. Biophys. Res. Commun. 2009, 383, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Miroshnichenko, S.; Patutina, O. Enhanced Inhibition of Tumorigenesis Using Combinations of MiRNA-Targeted Therapeutics. Front. Pharmacol. 2019, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.K.; Mondal, G.; Kumar, V.; Kattel, K.; Mahato, R.I. Chemosensitization and Inhibition of Pancreatic Cancer Stem Cell Proliferation by Overexpression of MicroRNA-205. Cancer Lett. 2017, 402, 1–8. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Li, J.; Zhang, Z.; Huang, C.; Lian, G.; Yang, K.; Chen, S.; Lin, Y.; Wang, L.; et al. Co-Delivery of MicroRNA-21 Antisense Oligonucleotides and Gemcitabine Using Nanomedicine for Pancreatic Cancer Therapy. Cancer Sci. 2017, 108, 1493–1503. [Google Scholar] [CrossRef]
- Kasinski, A.L.; Kelnar, K.; Stahlhut, C.; Orellana, E.; Zhao, J.; Shimer, E.; Dysart, S.; Chen, X.; Bader, A.G.; Slack, F.J. A Combinatorial MicroRNA Therapeutics Approach to Suppressing Non-Small Cell Lung Cancer. Oncogene 2015, 34, 3547–3555. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, Y.; Yang, Q.; Liu, B.; Wu, J.; Zhang, Y.; Yang, C.; Jiang, Y. MiR-497 and MiR-34a Retard Lung Cancer Growth by Co-Inhibiting Cyclin E1 (CCNE1). Oncotarget 2015, 6, 13149–13163. [Google Scholar] [CrossRef]
- Zhang, R.; Li, F.; Wang, W.; Wang, X.; Li, S.; Liu, J. The Effect of Antisense Inhibitor of MiRNA 106b∼25 on the Proliferation, Invasion, Migration, and Apoptosis of Gastric Cancer Cell. Tumor Biol. 2016, 37, 10507–10515. [Google Scholar] [CrossRef]
- Dong, C.G.; Wu, W.K.K.; Feng, S.Y.; Wang, X.J.; Shao, J.F.; Qiao, J. Co-Inhibition of MicroRNA-10b and MicroRNA-21 Exerts Synergistic Inhibition on the Proliferation and Invasion of Human Glioma Cells. Int. J. Oncol. 2012, 41, 1005–1012. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Fan, W.; Wang, Y.; Huang, L.; Jiang, Y.; Shi, L.; Mckinley, D.A.; Tan, W.; Tan, C. Combined Delivery of Let-7b MicroRNA and Paclitaxel via Biodegradable Nanoassemblies for the Treatment of KRAS Mutant Cancer. Mol. Pharm. 2016, 13, 520–533. [Google Scholar] [CrossRef]
- Fan, L.; Yang, Q.; Tan, J.; Qiao, Y.; Wang, Q.; He, J.; Wu, H.; Zhang, Y. Dual Loading MiR-218 Mimics and Temozolomide Using AuCOOH@FA-CS Drug Delivery System: Promising Targeted Anti-Tumor Drug Delivery System with Sequential Release Functions. J. Exp. Clin. Cancer Res. 2015, 34, 106. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Ren, W.; Huang, B.; Yi, L.; Zhu, H. MicroRNA-183/182/96 Cooperatively Regulates the Proliferation of Colon Cancer Cells. Mol. Med. Rep. 2015, 12, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Kang, C.; You, Y.; Pu, P.; Yang, W.; Zhao, P.; Wang, G.; Wang, A.; Jia, Z.; Han, L.; et al. Co-Suppression of MiR-221/222 Cluster Suppresses Human Glioma Cell Growth by Targeting P27kip1 in Vitro and in Vivo. Int. J. Oncol. 2009, 34, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Brognara, E.; Fabbri, E.; Montagner, G.; Gasparello, J.; Manicardi, A.; Corradini, R.; Bianchi, N.; Finotti, A.; Breveglieri, G.; Borgatti, M.; et al. High Levels of Apoptosis Are Induced in Human Glioma Cell Lines by Co-Administration of Peptide Nucleic Acids Targeting MiR-221 and MiR-222. Int. J. Oncol. 2016, 48, 1029–1038. [Google Scholar] [CrossRef]
- Miroshnichenko, S.K.; Patutina, O.A.; Burakova, E.A.; Chelobanov, B.P.; Fokina, A.A.; Vlassov, V.V.; Altman, S.; Zenkova, M.A.; Stetsenko, D.A. Mesyl Phosphoramidate Antisense Oligonucleotides as an Alternative to Phosphorothioates with Improved Biochemical and Biological Properties. Proc. Natl. Acad. Sci. USA 2019, 116, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Patutina, O.A.; Gaponova, S.K.; Sen’kova, A.V.; Savin, I.A.; Gladkikh, D.V.; Burakova, E.A.; Fokina, A.A.; Maslov, M.A.; Shmendel’, E.V.; Wood, M.J.A.; et al. Mesyl Phosphoramidate Backbone Modified Antisense Oligonucleotides Targeting MiR-21 with Enhanced in Vivo Therapeutic Potency. Proc. Natl. Acad. Sci. USA 2020, 117, 32370–32379. [Google Scholar] [CrossRef]
- Hammond, S.M.; Sergeeva, O.V.; Melnikov, P.A.; Goli, L.; Stoodley, J.; Zatsepin, T.S.; Stetsenko, D.A.; Wood, M.J.A. Mesyl Phosphoramidate Oligonucleotides as Potential Splice-Switching Agents: Impact of Backbone Structure on Activity and Intracellular Localization. Nucleic Acid Ther. 2021, 31, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.A.; Freestone, G.C.; Low, A.; De-Hoyos, C.L.; Iii, W.J.D.; Østergaard, M.E.; Migawa, M.T.; Fazio, M.; Wan, W.B.; Berdeja, A.; et al. Towards next Generation Antisense Oligonucleotides: Mesylphosphoramidate Modification Improves Therapeutic Index and Duration of Effect of Gapmer Antisense Oligonucleotides. Nucleic Acids Res. 2021, 49, 9026–9041. [Google Scholar] [CrossRef]
- Mironova, N.; Shklyaeva, O.; Andreeva, E.; Popova, N.; Kaledin, V.; Nikolin, V.; Vlassov, V.; Zenkova, M. Animal Model of Drug-Resistant Tumor Progression. Ann. N. Y. Acad. Sci. 2006, 1091, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. SynergyFinder 2.0: Visual Analytics of Multi-Drug Combination Synergies. Nucleic Acids Res. 2020, 48, W488–W493. [Google Scholar] [CrossRef]
- Patutina, O.A.; Bichenkova, E.V.; Miroshnichenko, S.K.; Mironova, N.L.; Trivoluzzi, L.T.; Burusco, K.K.; Bryce, R.A.; Vlassov, V.V.; Zenkova, M.A. MiRNases: Novel Peptide-Oligonucleotide Bioconjugates That Silence MiR-21 in Lymphosarcoma Cells. Biomaterials 2017, 122, 163–178. [Google Scholar] [CrossRef] [Green Version]
- Kabilova, T.O.; Shmendel, E.V.; Gladkikh, D.V.; Chernolovskaya, E.L.; Markov, O.V.; Morozova, N.G.; Maslov, M.A.; Zenkova, M.A. Targeted Delivery of Nucleic Acids into Xenograft Tumors Mediated by Novel Folate-Equipped Liposomes. Eur. J. Pharm. Biopharm. 2018, 123, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.D.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. MiRTarBase 2020: Updates to the Experimentally Validated MicroRNA-Target Interaction Database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed]
- Kutmon, M.; Kelder, T.; Mandaviya, P.; Evelo, C.T.A.; Coort, S.L. CyTargetLinker: A Cytoscape App to Integrate Regulatory Interactions in Network Analysis. PLoS ONE 2013, 8, e82160. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.E.S.E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Hellens, R.P. Quantitative Stem-Loop RT-PCR for Detection of MicroRNAs. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2011; Volume 744, pp. 145–157. [Google Scholar]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem-Loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Patutina, O.A.; Bazhenov, M.A.; Miroshnichenko, S.K.; Mironova, N.L.; Pyshnyi, D.V.; Vlassov, V.V.; Zenkova, M.A. Peptide-Oligonucleotide Conjugates Exhibiting Pyrimidine-X Cleavage Specificity Efficiently Silence MiRNA Target Acting Synergistically with RNase H. Sci. Rep. 2018, 8, 14990. [Google Scholar] [CrossRef]
- Zou, M.; Zhang, Q. MiR-17-5p Accelerates Cervical Cancer Cells Migration and Invasion via the TIMP2/MMPs Signaling Cascade. Cytotechnology 2021, 73, 619–627. [Google Scholar] [CrossRef]
- Minami, Y.; Kohsaka, S.; Tsuda, M.; Yachi, K.; Hatori, N.; Tanino, M.; Kimura, T.; Nishihara, H.; Minami, A.; Iwasaki, N.; et al. SS18-SSX-Regulated MiR-17 Promotes Tumor Growth of Synovial Sarcoma by Inhibiting P21WAF1/CIP1. Cancer Sci. 2014, 105, 1152–1159. [Google Scholar] [CrossRef]
- Huang, Y. The Novel Regulatory Role of LncRNA-MiRNA-MRNA Axis in Cardiovascular Diseases. J. Cell. Mol. Med. 2018, 22, 5768–5775. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.P.; Liu, G.L.; Li, S.; Liu, X.L. MiR-17-5p Knockdown Inhibits Proliferation, Autophagy and Promotes Apoptosis in Thyroid Cancer via Targeting PTEN. Neoplasma 2020, 67, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Arghiani, N.; Matin, M.M. MiR-21: A Key Small Molecule with Great Effects in Combination Cancer Therapy. Nucleic Acid Ther. 2021, 31, 271–283. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K. MiR-155 in Cancer Drug Resistance and as Target for MiRNA-Based Therapeutics. Cancer Metastasis Rev. 2018, 37, 33–44. [Google Scholar] [CrossRef]
- González-Magaña, A.; Blanco, F.J. Human PCNA Structure, Function, and Interactions. Biomolecules 2020, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Gladkikh, D.V.; Sen′ Kova, A.V.; Chernikov, I.V.; Kabilova, T.O.; Popova, N.A.; Nikolin, V.P.; Shmendel, E.V.; Maslov, M.A.; Vlassov, V.V.; Zenkova, M.A.; et al. Folate-Equipped Cationic Liposomes Deliver Anti-Mdr1-Sirna to the Tumor and Increase the Efficiency of Chemotherapy. Pharmaceutics 2021, 13, 1252. [Google Scholar] [CrossRef]
- Lawrie, C.H. MicroRNA Expression in Lymphoma. Expert Opin. Biol. Ther. 2007, 7, 1363–1374. [Google Scholar] [CrossRef]
- Mihailovich, M.; Bremang, M.; Spadotto, V.; Musiani, D.; Vitale, E.; Varano, G.; Zambelli, F.; Mancuso, F.M.; Cairns, D.A.; Pavesi, G.; et al. MiR-17-92 Fine-Tunes MYC Expression and Function to Ensure Optimal B Cell Lymphoma Growth. Nat. Commun. 2015, 6, 8725. [Google Scholar] [CrossRef]
- Haidar, M.; Jacquemin, P. Past and Future Strategies to Inhibit Membrane Localization of the KRAS Oncogene. Int. J. Mol. Sci. 2021, 22, 13193. [Google Scholar] [CrossRef]
- Bononi, A.; Pinton, P. Study of PTEN Subcellular Localization. Methods 2015, 77–78, 92–103. [Google Scholar] [CrossRef]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The Functions and Regulation of the PTEN Tumour Suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jeong, A.J.; Ye, S.K. Highlighted STAT3 as a Potential Drug Target for Cancer Therapy. BMB Rep. 2019, 52, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E. Stat3 as an Oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef]
- Lu, J.; Xu, S.; Huo, Y.; Sun, D.; Hu, Y.; Wang, J.; Zhang, X.; Wang, P.; Li, Z.; Liang, M.; et al. Sorting Nexin 3 Induces Heart Failure via Promoting Retromer-Dependent Nuclear Trafficking of STAT3. Cell Death Differ. 2021, 28, 2871–2887. [Google Scholar] [CrossRef] [PubMed]
- Tamahashi, U.; Kumagai, J.; Takizawa, T.; Sekine, M.; Eishi, Y. Expression and Intracellular Localization of Matrix Metalloproteinases in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Virchows Arch. 2008, 453, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Adhikari, N.; Banerjee, S.; Amin, S.A.; Jha, T. Matrix Metalloproteinase-9 (MMP-9) and Its Inhibitors in Cancer: A Minireview. Eur. J. Med. Chem. 2020, 194, 112260. [Google Scholar] [CrossRef]
- Rosewell, K.L.; Li, F.; Puttabyatappa, M.; Akin, J.W.; Brännström, M.; Curry, T.E. Ovarian Expression, Localization, and Function of Tissue Inhibitor of Metalloproteinase 3 (TIMP3) During the Periovulatory Period of the Human Menstrual Cycle. Biol. Reprod. 2013, 89, 121. [Google Scholar] [CrossRef]
- Tran, T.T.; Rane, C.K.; Zito, C.R.; Weiss, S.A.; Jessel, S.; Lucca, L.; Lu, B.Y.; Oria, V.O.; Adeniran, A.; Chiang, V.L.; et al. Clinical Significance of PDCD4 in Melanoma by Subcellular Expression and in Tumor-Associated Immune Cells. Cancers 2021, 13, 1049. [Google Scholar] [CrossRef]
- Pavlenko, E.; Cabron, A.S.; Arnold, P.; Dobert, J.P.; Rose-John, S.; Zunke, F. Functional Characterization of Colon Cancer-Associated Mutations in ADAM17: Modifications in the Pro-Domain Interfere with Trafficking and Maturation. Int. J. Mol. Sci. 2019, 20, 2198. [Google Scholar] [CrossRef]
- Düsterhöft, S.; Lokau, J.; Garbers, C. The Metalloprotease ADAM17 in Inflammation and Cancer. Pathol. Res. Pract. 2019, 215, 152410. [Google Scholar] [CrossRef]
- Saad, M.I.; Rose-John, S.; Jenkins, B.J. ADAM17: An Emerging Therapeutic Target for Lung Cancer. Cancers 2019, 11, 1218. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.Y.; Wu, S.Q.; Xu, Z.Z.; Lin, J.; Zhan, R. Anti-Leukemia Mechanism of MiR-17 and MiR-20a Silencing Mediated by MiRNA Sponge. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2014, 22, 932–937. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Xue, J.; Yang, S.; Chen, Y.; Wang, Y.; Zhu, Y.; Wang, X.; Kuang, D.; Ruan, Q.; Duan, Y.; et al. Co-Targeting of IGF1R/MTOR Pathway by MiR-497 and MiR-99a Impairs Hepatocellular Carcinoma Development. Oncotarget 2017, 8, 47984–47997. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhao, Y.; Wang, P. MiR-204 Inhibits Angiogenesis and Promotes Sensitivity to Cetuximab in Head and Neck Squamous Cell Carcinoma Cells by Blocking JAK2-STAT3 Signaling. Biomed. Pharmacother. 2018, 99, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xiao, J.; Lin, H.; Bai, Y.; Luo, X.; Wang, Z.; Yang, B. A Single Anti-MicroRNA Antisense Oligodeoxyribonucleotide (AMO) Targeting Multiple MicroRNAs Offers an Improved Approach for MicroRNA Interference. Nucleic Acids Res. 2009, 37, e24. [Google Scholar] [CrossRef]
- Matsubara, H.; Takeuchi, T.; Nishikawa, E.; Yanagisawa, K.; Hayashita, Y.; Ebi, H.; Yamada, H.; Suzuki, M.; Nagino, M.; Nimura, Y.; et al. Apoptosis Induction by Antisense Oligonucleotides against MiR-17-5p and MiR-20a in Lung Cancers Overexpressing MiR-17-92. Oncogene 2007, 26, 6099–6105. [Google Scholar] [CrossRef]
- Shishodia, G.; Shukla, S.; Srivastava, Y.; Masaldan, S.; Mehta, S.; Bhambhani, S.; Sharma, S.; Mehrotra, R.; Das, B.C.; Bharti, A.C. Alterations in MicroRNAs MiR-21 and Let-7a Correlate with Aberrant STAT3 Signaling and Downstream Effects during Cervical Carcinogenesis. Mol. Cancer 2015, 14, 116. [Google Scholar] [CrossRef]
- Mao, X.H.; Chen, M.; Wang, Y.; Cui, P.G.; Liu, S.B.; Xu, Z.Y. MicroRNA-21 Regulates the ERK/NF-ΚB Signaling Pathway to Affect the Proliferation, Migration, and Apoptosis of Human Melanoma A375 Cells by Targeting SPRY1, PDCD4, and PTEN. Mol. Carcinog. 2017, 56, 886–894. [Google Scholar] [CrossRef]
- Ince, F.A.; Shariev, A.; Dixon, K. PTEN as a Target in Melanoma. J. Clin. Pathol. 2022, 75, 581–584. [Google Scholar] [CrossRef]
- Feng, L.; Li, J.; Bu, X.; Zuo, Y.; Shen, L.; Qu, X. BRAF V600E Dictates Cell Survival via C-Myc-Dependent Induction of Skp2 in Human Melanoma. Biochem. Biophys. Res. Commun. 2020, 524, 28–35. [Google Scholar] [CrossRef]
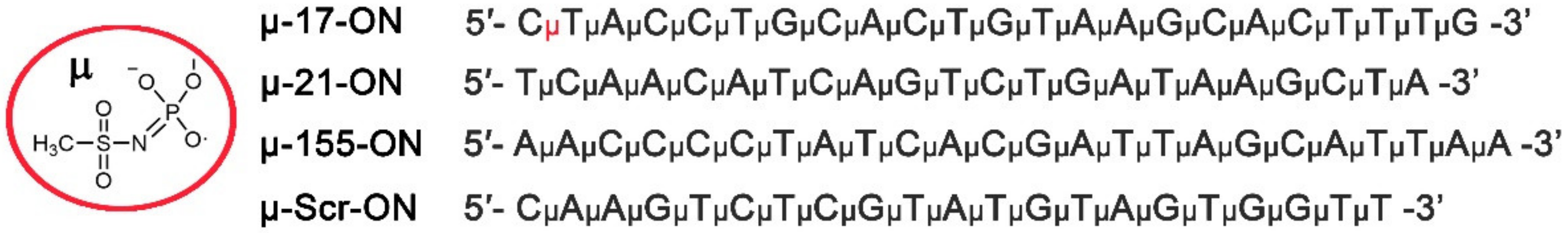




| Healthy | Control | µ-Scr-ON | µ-21-ON | Cocktail | |
|---|---|---|---|---|---|
| Normal hepatocytes, Vv, % | 72.2 ± 1.6 | 30.5 ± 3.3 † | 31.1 ± 3.3 † | 35.9 ± 3.3 † | 59 ± 3.5 †*# |
| Dystrophy, Vv, % | 7.2 ± 1 | 29.8 ± 2.9 † | 28.1 ± 1.9 † | 31.3 ± 2.6† | 13.7 ± 1.3 †*# |
| Necrosis, Vv, % | 7.8 ± 1.3 | 27.8 ± 1.5 † | 29.3 ± 3.3 † | 21.1 ± 1.4 †*# | 14.3 ± 1.6 †*# |
| Total destructive changes, Vv, % | 15 ± 2.1 | 57.6 ± 3.3 † | 57.3 ± 3.8 † | 52.4 ± 3.6† | 28 ± 2.5 †*# |
| Stroma, Vv, % | 12 ± 1.7 | 11.9 ± 1.1 | 11.3 ± 1.2 | 11.5 ± 1.3 | 10.1 ± 1.2 |
| Binuclear hepatocytes, Nv | 4.4 ± 1.1 | 3.4 ± 0.7 | 4.7 ± 0.9 | 7.1 ± 1.1 †* | 4.9 ± 0.7 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaponova, S.; Patutina, O.; Sen’kova, A.; Burakova, E.; Savin, I.; Markov, A.; Shmendel, E.; Maslov, M.; Stetsenko, D.; Vlassov, V.; et al. Single Shot vs. Cocktail: A Comparison of Mono- and Combinative Application of miRNA-Targeted Mesyl Oligonucleotides for Efficient Antitumor Therapy. Cancers 2022, 14, 4396. https://doi.org/10.3390/cancers14184396
Gaponova S, Patutina O, Sen’kova A, Burakova E, Savin I, Markov A, Shmendel E, Maslov M, Stetsenko D, Vlassov V, et al. Single Shot vs. Cocktail: A Comparison of Mono- and Combinative Application of miRNA-Targeted Mesyl Oligonucleotides for Efficient Antitumor Therapy. Cancers. 2022; 14(18):4396. https://doi.org/10.3390/cancers14184396
Chicago/Turabian StyleGaponova, Svetlana, Olga Patutina, Aleksandra Sen’kova, Ekaterina Burakova, Innokenty Savin, Andrey Markov, Elena Shmendel, Mikhail Maslov, Dmitry Stetsenko, Valentin Vlassov, and et al. 2022. "Single Shot vs. Cocktail: A Comparison of Mono- and Combinative Application of miRNA-Targeted Mesyl Oligonucleotides for Efficient Antitumor Therapy" Cancers 14, no. 18: 4396. https://doi.org/10.3390/cancers14184396
APA StyleGaponova, S., Patutina, O., Sen’kova, A., Burakova, E., Savin, I., Markov, A., Shmendel, E., Maslov, M., Stetsenko, D., Vlassov, V., & Zenkova, M. (2022). Single Shot vs. Cocktail: A Comparison of Mono- and Combinative Application of miRNA-Targeted Mesyl Oligonucleotides for Efficient Antitumor Therapy. Cancers, 14(18), 4396. https://doi.org/10.3390/cancers14184396






