The Clonal Diversity of Peripheral B Cell Receptor Immune Repertoire Impaired by Residual Malignant B Cells Predicts Treatment Efficacy in B Cell Lymphoma Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells Lines and Cell Culture
2.2. Lentiviral Transduction of Cell Lines
2.3. In Vitro Culture of Human GC B Cells with and without Tumor Cells
2.4. Flow Cytometry (FCM) Analysis
2.5. RNA-Seq Analysis
2.6. Patients’ Characteristics and Clinical Samples Collection Procedure
2.7. BCR IR Analysis
3. Results
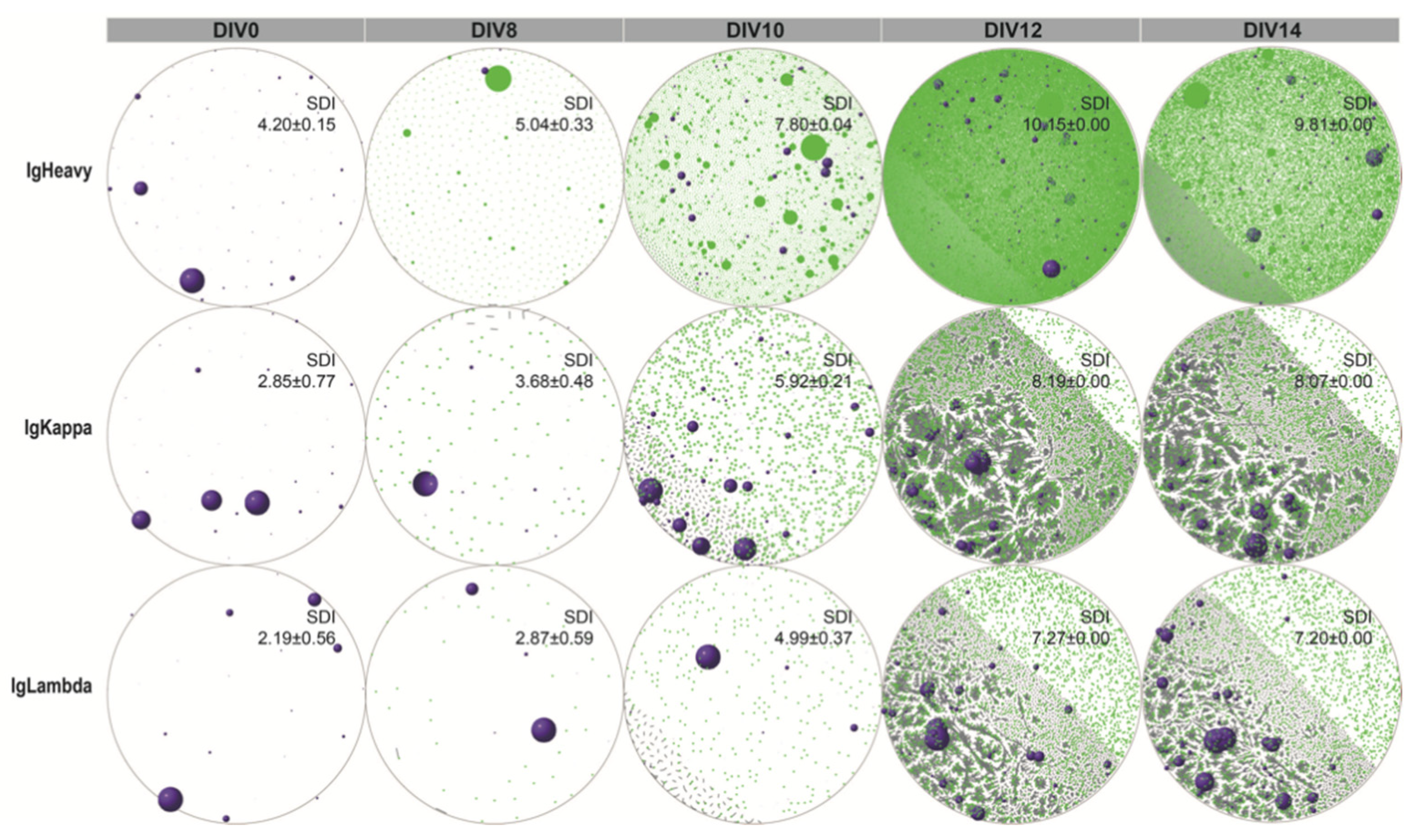
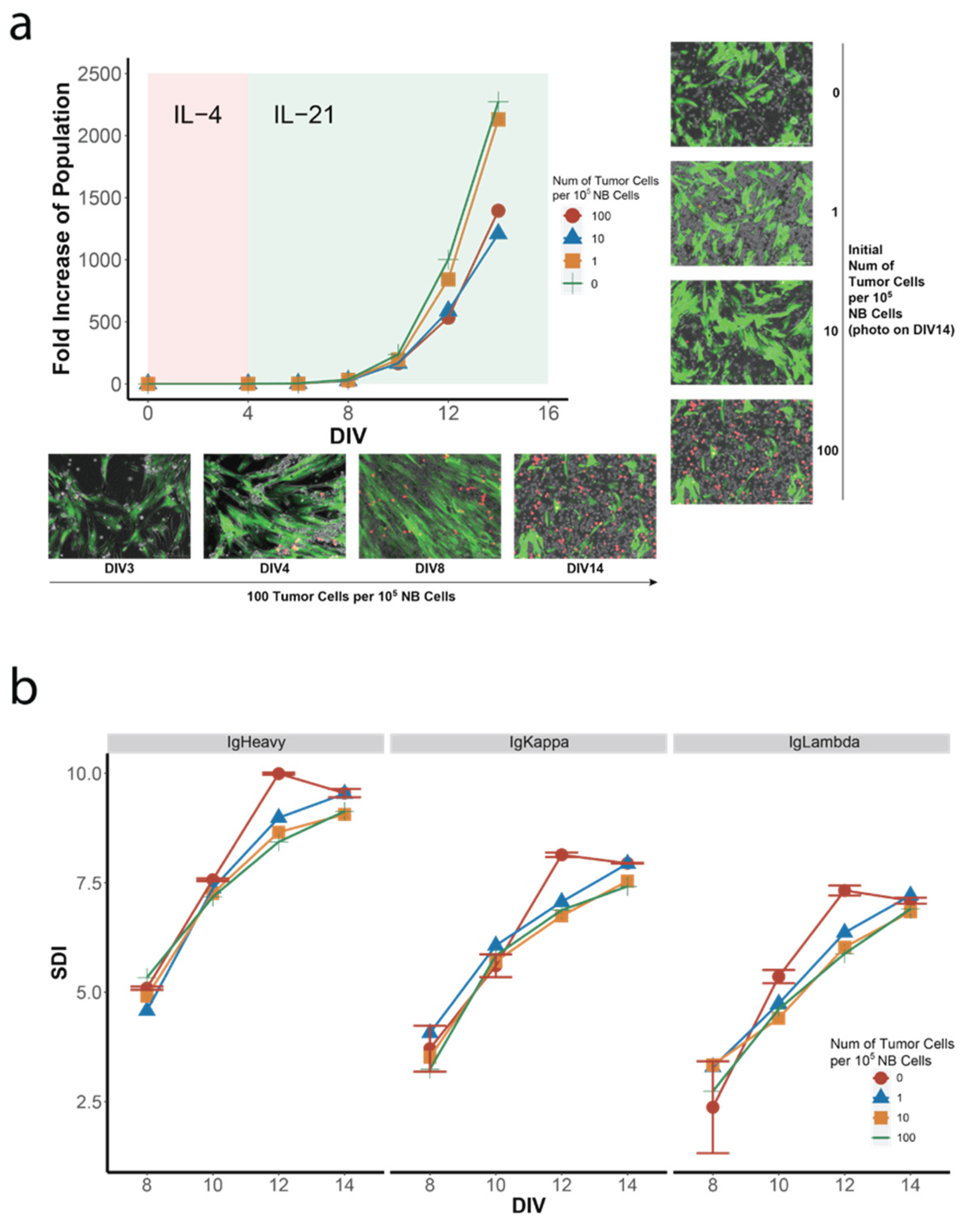
3.1. Establishment of a Human In Vitro hiGC Model
3.2. Co-Culture of NB Cells with GC B-Cell (GCB) Type DLBCL Cells in the hiGC
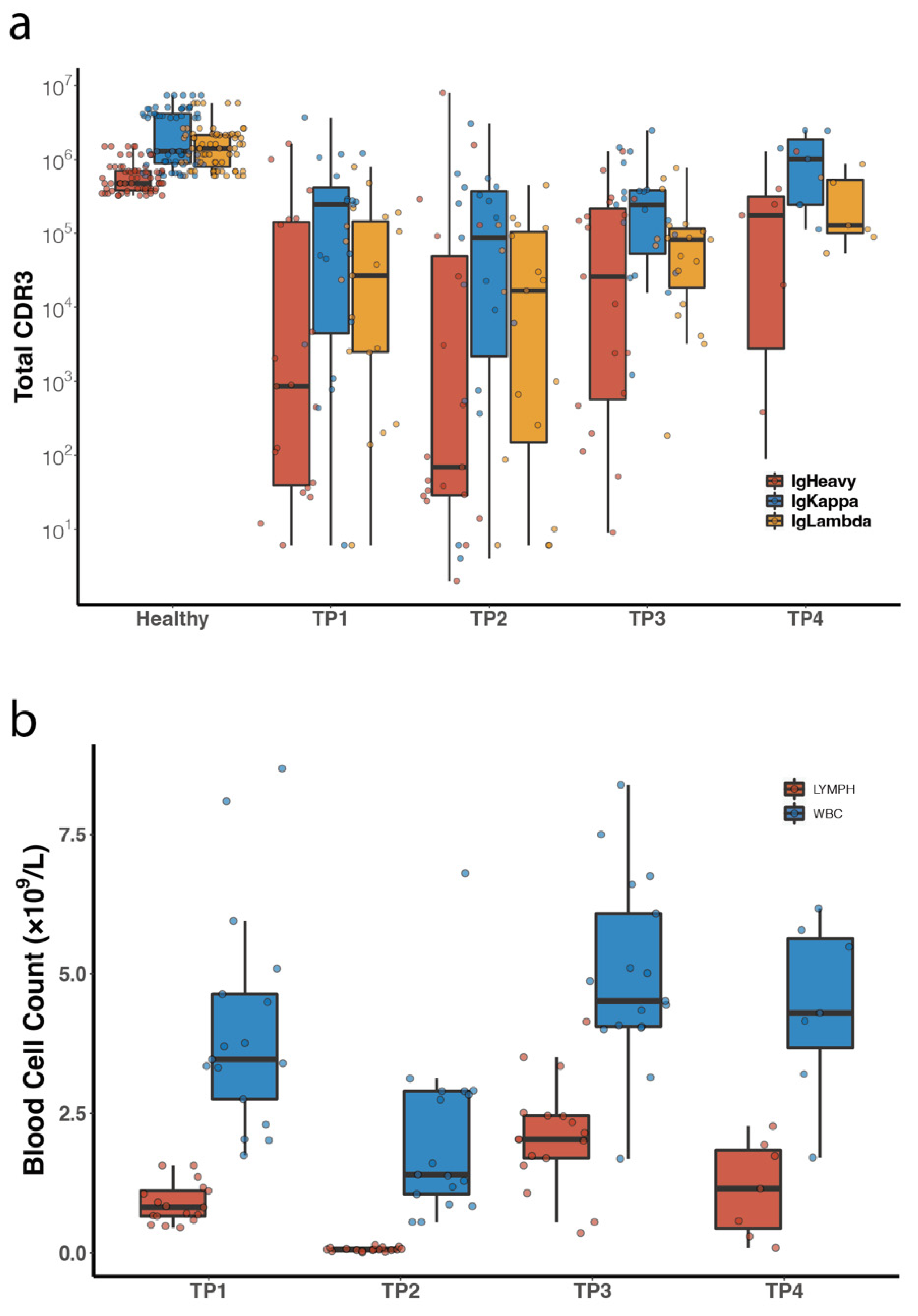
3.3. Comparison of Peripheral Blood BCR IR between DLBCL Patients and Healthy Volunteers
3.4. Characterization of Tumor BCR Clones in Peripheral BCR IR
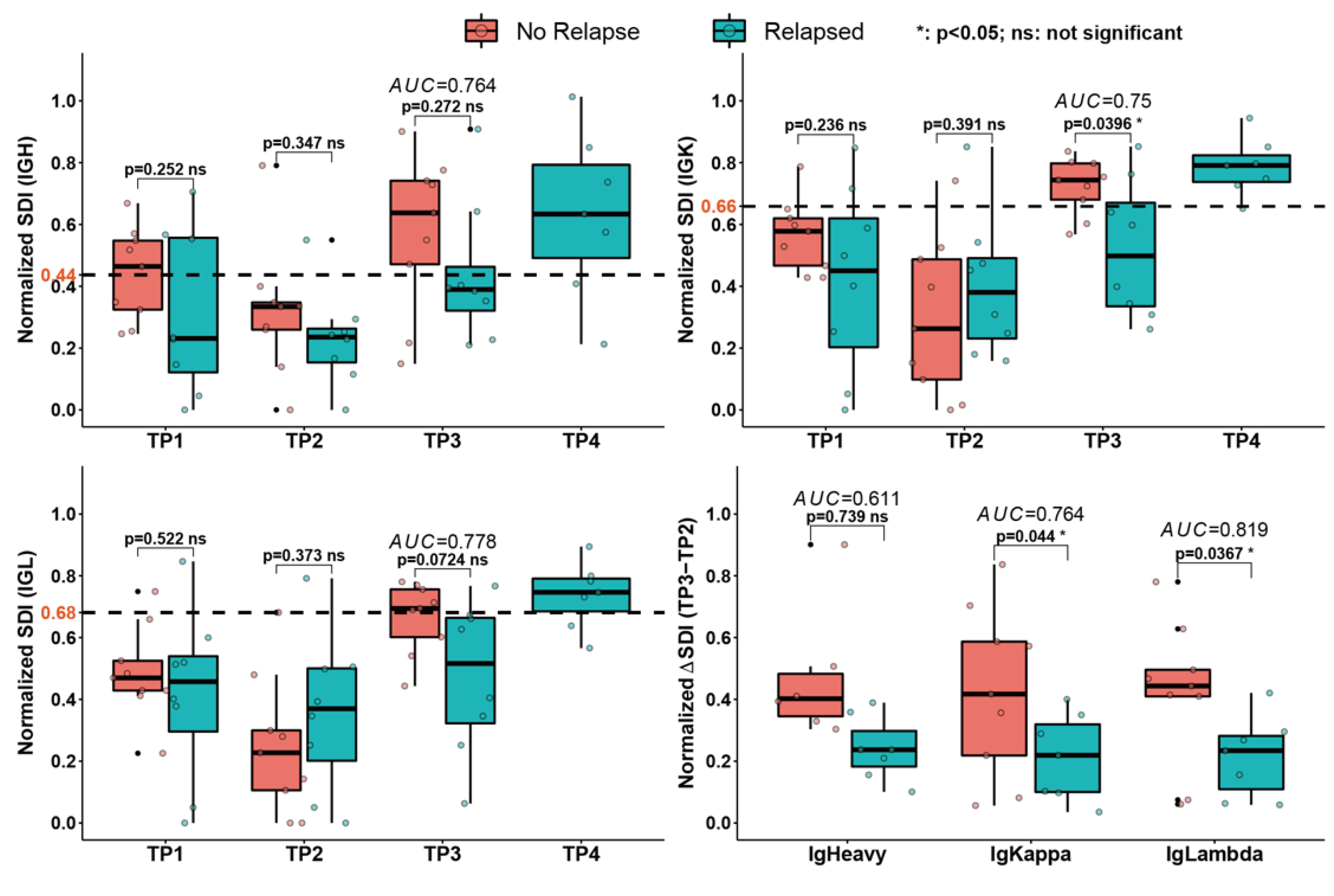
3.5. Diversity Analysis of All Peripheral Blood Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Silva, N.S.; Klein, U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 2015, 15, 137–148. [Google Scholar] [CrossRef]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef]
- Stebegg, M.; Kumar, S.D.; Silva-Cayetano, A.; Fonseca, V.R.; Linterman, M.A.; Graca, L. Regulation of the Germinal Center Response. Front. Immunol. 2018, 9, 2469. [Google Scholar] [CrossRef]
- Smith, K.G.; Light, A.; O’Reilly, L.A.; Ang, S.M.; Strasser, A.; Tarlinton, D. bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J. Exp. Med. 2000, 191, 475–484. [Google Scholar] [CrossRef]
- Klein, U.; Dalla-Favera, R. Germinal centres: Role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008, 8, 22–33. [Google Scholar] [CrossRef]
- Mlynarczyk, C.; Fontán, L.; Melnick, A. Germinal center-derived lymphomas: The darkest side of humoral immunity. Immunol. Rev. 2019, 288, 214–239. [Google Scholar] [CrossRef]
- Basso, K.; Dalla-Favera, R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015, 15, 172–184. [Google Scholar] [CrossRef]
- Hao, Z.; Duncan, G.S.; Seagal, J.; Su, Y.W.; Hong, C.; Haight, J.; Chen, N.J.; Elia, A.; Wakeham, A.; Li, W.Y.; et al. Fas receptor expression in germinal-center B cells is essential for T and B lymphocyte homeostasis. Immunity 2008, 29, 615–627. [Google Scholar] [CrossRef]
- Minton, K. Lymphocyte responses: Germinal-centre B cells take control. Nat. Rev. Immunol. 2008, 8, 826–827. [Google Scholar] [CrossRef]
- Koncz, G.; Hueber, A.O. The Fas/CD95 Receptor Regulates the Death of Autoreactive B Cells and the Selection of Antigen-Specific B Cells. Front. Immunol. 2012, 3, 207. [Google Scholar] [CrossRef]
- Lossos, I.S.; Levy, R.; Alizadeh, A.A. AID is expressed in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas and is not correlated with intraclonal heterogeneity. Leukemia 2004, 18, 1775–1779. [Google Scholar] [CrossRef]
- Greeve, J.; Philipsen, A.; Krause, K.; Klapper, W.; Heidorn, K.; Castle, B.E.; Janda, J.; Marcu, K.B.; Parwaresch, R. Expression of activation-induced cytidine deaminase in human B-cell non-Hodgkin lymphomas. Blood 2003, 101, 3574–3580. [Google Scholar] [CrossRef]
- Moffett, H.F.; Harms, C.K.; Fitzpatrick, K.S.; Tooley, M.R.; Boonyaratanakornkit, J.; Taylor, J.J. B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 2019, 4, eaax0644. [Google Scholar] [CrossRef]
- Purwada, A.; Jaiswal, M.K.; Ahn, H.; Nojima, T.; Kitamura, D.; Gaharwar, A.K.; Cerchietti, L.; Singh, A. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 2015, 63, 24–34. [Google Scholar] [CrossRef]
- Calado, D.P. Germinal Centers: Methods and Protocols; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017. [Google Scholar]
- Matsuda, Y.; Haneda, M.; Kadomatsu, K.; Kobayashi, T. A proliferation-inducing ligand sustains the proliferation of human naïve (CD27−) B cells and mediates their differentiation into long-lived plasma cells in vitro via transmembrane activator and calcium modulator and cyclophilin ligand interactor and B-cell mature antigen. Cell. Immunol. 2015, 295, 127–136. [Google Scholar] [CrossRef][Green Version]
- Haniuda, K.; Nojima, T.; Kitamura, D. In Vitro-Induced Germinal Center B Cell Culture System. Methods Mol. Biol. 2017, 1623, 125–133. [Google Scholar] [CrossRef]
- Nojima, T.; Haniuda, K.; Moutai, T.; Matsudaira, M.; Mizokawa, S.; Shiratori, I.; Azuma, T.; Kitamura, D. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat. Commun. 2011, 2, 465. [Google Scholar] [CrossRef]
- Haniuda, K.; Kitamura, D. Induced Germinal Center B Cell Culture System. Bio-Protocol 2019, 9, e3163. [Google Scholar] [CrossRef]
- Wagar, L.E.; Salahudeen, A.; Constantz, C.M.; Wendel, B.S.; Lyons, M.M.; Mallajosyula, V.; Jatt, L.P.; Adamska, J.Z.; Blum, L.K.; Gupta, N.; et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 2021, 27, 125–135. [Google Scholar] [CrossRef]
- Meyer, S.N.; Koul, S.; Pasqualucci, L. Mouse Models of Germinal Center Derived B-Cell Lymphomas. Front. Immunol. 2021, 12, 710711. [Google Scholar] [CrossRef]
- Galson, J.D.; Trück, J.; Fowler, A.; Clutterbuck, E.A.; Münz, M.; Cerundolo, V.; Reinhard, C.; van der Most, R.; Pollard, A.J.; Lunter, G.; et al. Analysis of B Cell Repertoire Dynamics Following Hepatitis B Vaccination in Humans, and Enrichment of Vaccine-specific Antibody Sequences. EBioMedicine 2015, 2, 2070–2079. [Google Scholar] [CrossRef]
- Bashford-Rogers, R.J.M.; Bergamaschi, L.; McKinney, E.F.; Pombal, D.C.; Mescia, F.; Lee, J.C.; Thomas, D.C.; Flint, S.M.; Kellam, P.; Jayne, D.R.W.; et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature 2019, 574, 122–126. [Google Scholar] [CrossRef]
- Ma, K.Y.; He, C.; Wendel, B.S.; Williams, C.M.; Xiao, J.; Yang, H.; Jiang, N. Immune Repertoire Sequencing Using Molecular Identifiers Enables Accurate Clonality Discovery and Clone Size Quantification. Front. Immunol. 2018, 9, 33. [Google Scholar] [CrossRef]
- Miho, E.; Roškar, R.; Greiff, V.; Reddy, S.T. Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat. Commun. 2019, 10, 1321. [Google Scholar] [CrossRef]
- Simon, J.S.; Botero, S.; Simon, S.M. Sequencing the peripheral blood B and T cell repertoire—Quantifying robustness and limitations. J. Immunol. Methods 2018, 463, 137–147. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Cheson, B.D.; Fisher, R.I.; Barrington, S.F.; Cavalli, F.; Schwartz, L.H.; Zucca, E.; Lister, T.A. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: The Lugano classification. J. Clin. Oncol. 2014, 32, 3059–3068. [Google Scholar] [CrossRef]
- Song, L.; Cohen, D.; Ouyang, Z.; Cao, Y.; Hu, X.; Liu, X.S. TRUST4: Immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 2021, 18, 627–630. [Google Scholar] [CrossRef]
- Bashford-Rogers, R.J.; Palser, A.L.; Huntly, B.J.; Rance, R.; Vassiliou, G.S.; Follows, G.A.; Kellam, P. Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Res. 2013, 23, 1874–1884. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University Of Illinois Press: Champaign, IL, USA, 1949; Volume 60. [Google Scholar]
- Caso, C.; Angeles Gil, M. The gini-simpson index of diversity: Estimation in the stratified sampling. Commun. Stat. Theory Methods 1988, 17, 2981–2995. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. Pipe-Friendly Framework for Basic Statistical Tests, version 0.6.0; R package rstatix; R Foundation for Statistical computing: Vienna, Austria, 2021. [Google Scholar]
- Glass, D.R.; Tsai, A.G.; Oliveria, J.P.; Hartmann, F.J.; Kimmey, S.C.; Calderon, A.A.; Borges, L.; Glass, M.C.; Wagar, L.E.; Davis, M.M.; et al. An Integrated Multi-omic Single-Cell Atlas of Human B Cell Identity. Immunity 2020, 53, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.B.; Corinaldesi, C.; Shen, Q.; Kumar, R.; Compagno, N.; Wang, Z.; Nitzan, M.; Grunstein, E.; Pasqualucci, L.; Dalla-Favera, R.; et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J. Exp. Med. 2020, 217, e20200483. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; Tu, Y.; Stolovitzky, G.A.; Keller, J.L.; Haddad, J., Jr.; Miljkovic, V.; Cattoretti, G.; Califano, A.; Dalla-Favera, R. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA 2003, 100, 2639–2644. [Google Scholar] [CrossRef] [PubMed]
- Sprangers, M.; Feldhahn, N.; Liedtke, S.; Jumaa, H.; Siebert, R.; Müschen, M. SLP65 deficiency results in perpetual V(D)J recombinase activity in pre-B-lymphoblastic leukemia and B-cell lymphoma cells. Oncogene 2006, 25, 5180–5186. [Google Scholar] [CrossRef][Green Version]
- Plumas, J.; Jacob, M.C.; Chaperot, L.; Molens, J.P.; Sotto, J.J.; Bensa, J.C. Tumor B cells from non-Hodgkin’s lymphoma are resistant to CD95 (Fas/Apo-1)-mediated apoptosis. Blood 1998, 91, 2875–2885. [Google Scholar] [CrossRef]
- Scherer, F.; Kurtz, D.M.; Newman, A.M.; Stehr, H.; Craig, A.F.; Esfahani, M.S.; Lovejoy, A.F.; Chabon, J.J.; Klass, D.M.; Liu, C.L.; et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016, 8, 364ra155. [Google Scholar] [CrossRef]
- Kurtz, D.M.; Green, M.R.; Bratman, S.V.; Scherer, F.; Liu, C.L.; Kunder, C.A.; Takahashi, K.; Glover, C.; Keane, C.; Kihira, S.; et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015, 125, 3679–3687. [Google Scholar] [CrossRef]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef]


| No. | Age | Sex | Subtypes | Status | Conditioning Regimen | Complication during ASCR | Progression | PFS |
|---|---|---|---|---|---|---|---|---|
| 598 | 36 | M | GCB | Salvage | CBV | Pneumonia | No | 35.5 |
| 592 | 51 | M | non-GCB | Salvage | BEAC | - | Yes | 9.3 |
| 602 | 29 | M | non-GCB | Salvage | CBV | - | No | 34.0 |
| 535 | 23 | F | GCB | Consolidation | CBV | - | Yes | 4.9 |
| 457 | 45 | M | non-GCB | Salvage | CBV | - | Yes | 12.6 |
| 473 | 50 | F | non-GCB | Salvage | CBV | - | Yes | 7.1 |
| 512 | 45 | M | non-GCB | Salvage | CBV | - | No | 46.6 |
| 519 | 44 | M | GCB | Consolidation | CBV | - | No | 42.5 |
| 383 | 40 | F | non-GCB | Consolidation | R-CBV | - | Yes | 23.5 |
| 419 | 62 | M | non-GCB | Salvage | BEAM | Pneumonia & AF | Yes | 18.8 |
| 486 | 49 | M | non-GCB | Salvage | CBV | Diarrhea | Yes | 6.7 |
| 615 | 55 | F | non-GCB | Salvage | CBV | - | Yes | 6.0 |
| 548 | 65 | M | non-GCB | Salvage | CBV | - | No | 32.5 |
| 559 | 32 | M | non-GCB | Salvage | CBV | - | No | 38.3 |
| 601 | 20 | M | GCB | Salvage | CBV | - | No | 30.2 |
| 617 | 52 | F | non-GCB | Salvage | CBV | - | No | 21.8 |
| 620 | 47 | M | non-GCB | Salvage | CBV | - | No | 17.5 |
| Patient No. | Primary Tumor CDR3 | TP | LD | Ratio |
|---|---|---|---|---|
| 419 | TGTCAGCAAAGTTACAGTATTCCTCGGACGTTC | 1 | 0 | 86.2% |
| 2 | 0 | |||
| 3 | 0 | |||
| TGTCAGCAAAGTTACAGTATTCCTCGGACCTTC | 1 | 1 | 7.9% | |
| 2 | 0 | |||
| 3 | 1 | |||
| 457 | CAGCAGTATGGTAGCTCACCGGCGACG | 1 | 1 | 92.5% |
| 2 | 1 | |||
| 3 | 0 | |||
| CAGCAGTATGGTAGCTCACCGGCGACC | 1 | 1 | 6.4% | |
| 2 | 1 | |||
| 3 | 1 | |||
| 548 | TGTCAGCAGATTGATACCTGGCCTCGAACCTTC | 1 | N/A | 97.3% |
| 2 | ||||
| 3 | ||||
| TGTCTGCAGCATAATAGATACCCGCTCACTTTC | 1 | 0.6% | ||
| 2 | ||||
| 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Zhang, J.; Wang, Y.; Mi, L.; Wang, X.; Liu, W.; Fu, J.; Song, H.; Song, Y.; Zhu, J. The Clonal Diversity of Peripheral B Cell Receptor Immune Repertoire Impaired by Residual Malignant B Cells Predicts Treatment Efficacy in B Cell Lymphoma Patients. Cancers 2022, 14, 4628. https://doi.org/10.3390/cancers14194628
Wu M, Zhang J, Wang Y, Mi L, Wang X, Liu W, Fu J, Song H, Song Y, Zhu J. The Clonal Diversity of Peripheral B Cell Receptor Immune Repertoire Impaired by Residual Malignant B Cells Predicts Treatment Efficacy in B Cell Lymphoma Patients. Cancers. 2022; 14(19):4628. https://doi.org/10.3390/cancers14194628
Chicago/Turabian StyleWu, Meng, Jing Zhang, Yi Wang, Lan Mi, Xiaopei Wang, Weiping Liu, Jie Fu, Haifeng Song, Yuqin Song, and Jun Zhu. 2022. "The Clonal Diversity of Peripheral B Cell Receptor Immune Repertoire Impaired by Residual Malignant B Cells Predicts Treatment Efficacy in B Cell Lymphoma Patients" Cancers 14, no. 19: 4628. https://doi.org/10.3390/cancers14194628
APA StyleWu, M., Zhang, J., Wang, Y., Mi, L., Wang, X., Liu, W., Fu, J., Song, H., Song, Y., & Zhu, J. (2022). The Clonal Diversity of Peripheral B Cell Receptor Immune Repertoire Impaired by Residual Malignant B Cells Predicts Treatment Efficacy in B Cell Lymphoma Patients. Cancers, 14(19), 4628. https://doi.org/10.3390/cancers14194628





