Simple Summary
The clinical application of PARPis in patients with ovarian cancer has unresolved issues, and whether PARPis can have a similar first-line efficacy to that of platinum-based chemotherapy is still undefined. This study used the PDX model to explore the above problems. We demonstrated that the PDX model can reflect PARPi efficacy more accurately than BRCA mutation, homologous recombination deficiency positivity, and platinum sensitivity. Moreover, the novel clinical and molecular biomarkers suggested that KRAS overexpression was associated with PARPi sensitivity. Additionally, ATK1 enrichment could lead to PARPi resistance, and CA125 less than 10 U/mL during chemotherapy can be a potential indicator for the therapeutic use of PARPi. Above all, PARPis cannot replace platinum-based chemotherapy as first-line treatment in our preclinical trial, indicating that chemotherapy-free tests in the unselected population are not recommended.
Abstract
(1) The accuracy of patient-derived xenografts (PDXs) in predicting ADP-ribose polymerase inhibitor (PARPi) efficacy in ovarian cancer was tested, novel biomarkers were investigated, and whether PARPis could replace platinum-based chemotherapy as a first-line therapy was explored. (2) PDXs were reconstructed for 40 patients with ovarian cancer, and niraparib, olaparib and paclitaxel, and carboplatin (TC) sensitivity tests were conducted. Whole exon sequencing and homologous recombination deficiency (HRD) scores were performed, and patient clinical information was collected. The molecular biomarkers were identified by reverse-transcription quantitative PCR and immunoblotting. (3) Niraparib and olaparib sensitivity were tested in 26 patients and showed high consistency. Approximately half of BRCA wild-type, HRD-negative, and platinum-resistant patients may benefit from PARPis. AKT1 enrichment indicated PARPi resistance; high KRAS expression indicated PARPi sensitivity. CA125 below 10 U/mL during chemotherapy has a sensitivity and specificity similar to platinum sensitivity in predicting PARPi efficacy. Niraparib and TC sensitivity tests were performed on 23 patients, and TC showed a better response in this preclinical trial. (4) PDX can indicate individualized PARPi efficacy. Decreased CA125 levels and KRAS and ATK1 expression levels may be novel biomarkers. The preclinical evidence does not support the implementation of PARPis as the first-line treatment in an unselected population.
1. Introduction
Poly ADP-ribose polymerase inhibitors (PARPis) have inspired a new era in epithelial ovarian cancer treatment, with PARPis serving as targeted drugs that competitively inhibit the PARP family, kill tumor cells with homologous recombination deficiency (HRD), and benefit survival [1]. Niraparib and olaparib have been approved by the American Food and Drug Administration (FDA) as advanced post-first-line treatments and maintenance therapies for ovarian cancer, given the significant improvement in patient survival [2,3,4,5,6,7]. China has also recently decided to cover these two medicines with medical insurance for ovarian cancer patients.
Many questions have surfaced with the wide use of PARPis clinically. Most notably, gynecologic oncologists lack evaluation criteria of the clinical efficacy for PARPi maintenance therapy, and the current molecular and clinical indicators for PARPis, such as BRCA1/2 mutations (BRCA1/2muts), homologous repair deficiency (HRD) status, and platinum drug response, lack accuracy [8]. PARPis only benefited two-thirds of relapsed patients with BRCA1/2 mutations [9,10]. The HRD score is a sign of gene scarring. This score can accumulate over time and cannot reflect the real-time appearance of drug resistance, such as that of BRCA recovery mutations [11]. Platinum-sensitive patients have genomic instability due to interstrand crosslink repair deficiency, not HRD [12]. Moreover, wild-type BRCA1/2 (BRCA1/2wt), HRD-negative (HRD–), and platinum-resistant patients can still obtain a benefit from PARPis [13,14,15,16,17,18]. Interestingly, the OReO study demonstrated that BRCA1/2mut and HRD status lost their close relationship with therapeutic effect as PARPi treatment lines increased [19,20].
As a result, some patients have to endure the economic burden and suffer from drug-related adverse reactions, such as bone marrow suppression and digestive system symptoms, without survival benefits. Furthermore, some lose the opportunity for PARPi benefits. Finding a method that more accurately reflects the individual efficacy of PARPis and exploring other clinical and molecular biomarkers are critical for precision treatment with PARPis.
Another phenomenon that caught our attention was the expanding population benefit from PARPis. The Ambition (KGOG 3045) clinical study supported the treatment effect of olaparib in combination with antiangiogenic agents or immunotherapy (PD-L1) on platinum-resistant advanced ovarian cancer with HRD+ [21]. Even more encouraging, preliminary data from a multicenter, prospective, phase 2, single-arm clinical study (NANT) confirmed the efficacy and safety of niraparib monotherapy in neoadjuvant therapy for advanced ovarian cancer patients who bore unresectable lesions [22,23]. These positive results have led us to consider the use of PARPis as a first-line treatment and whether PARPis can replace platinum-based chemotherapy for treating ovarian cancer patients after surgery.
Patient-derived xenografts (PDXs) can simulate tumor development, evolution, and drug response. They are considered one of the best preclinical models because they retain the histopathological, genetic, and tumor microenvironment characteristics of the original tissue [24]. As such, this study re-established PDX models derived from 40 ovarian cancer patients randomly selected from our established PDX library [25] and conducted niraparib, olaparib, and paclitaxel and cisplatin sensitivity tests to evaluate whether PDXs can provide a more accurate personalized efficacy of PARPis, to search for other clinical and molecular markers of PARPis, and to explore the possibility of niraparib as a first-line treatment.
2. Materials and Methods
2.1. Patients
All patients in this study were from the Department of Obstetrics and Gynecology of Peking Union Medical College Hospital. Informed consent was obtained, and all procedures followed the ethical principles of the Institutional Review Board of Peking Union Medical College Hospital [25]. Clinical information, including PARPi history, therapeutic type (maintenance/treatment), treatment scenario (first-line/second-line), and prognostic information, was updated. Since the median follow-up time was less than two years, the primary endpoint was progression-free survival (PFS), and the secondary endpoint was overall survival (OS).
2.2. Establishment of the PDX Models and Performance of Drug Sensitivity Tests
After recovering cryopreserved tumor tissue and implanting a 3 mm × 3 mm × 3 mm tumor mass into female NOD-Prkdcem1Idmo-Il2rgem2Idmo (NPI) mice aged six to eight weeks, we monitored tumor volume and body weight regularly. All procedures were conducted following the ‘Guiding Principles in the Care and Use of Animals’ (China).
Pathological characteristics and the lymphocyte ratio were examined before tissue cryopreservation [25]. All the tumor tissues of PDX models were compared with the original tumor tissues for the consistency of pathology and characteristic proteins. The tumor lymphocyte ratio was detected before cryopreservation of tumor tissues in each PDX model in order to avoid the change of ovarian cancer tumor tissues into lymphoma. Leukocytes were identified as CD19+ and CD45+ (CD19 antibody and PE anti-human CD45 antibody), and samples containing less than 1% leukocytes were qualified for further mouse-to-mouse transplantation.
Then, a drug sensitivity test was performed when the tumor grew to 200 mm3. Clinically used olaparib (Lynparza, AstraZeneca AB, Cambridge, UK), niraparib (Zejula, GlaxoSmithKline, Bingford, UK), paclitaxel (Anzatax, Hospira Australia Pty Ltd., Melbourne, Australia), and carboplatin (Paraplatin, MYERS SQUIBB AE, Dubai, United Arab Emirates) were adopted and compared with normal saline (NS), 0.5% methylcellulose (0.5% MC) or 10% HP-β-CD PBS as a vehicle depending on the dissolved reagent.
PDX models generated from the same patient were randomly divided into experimental and control groups (four repeated in each) and received one of the following treatments: (1) niraparib 50 mg/kg (qd) or olaparib 75 mg/kg (qd) via intragastric administration; (2) paclitaxel 30 mg/kg (every four days × 8 cycles) and carboplatin 25 mg/kg (every five days × 6 cycles) via intravenous injection; or (3) the same volume and approach of the corresponding vehicle. Mouse body weight and tumor volume were recorded every three days. The administration was continued until the mice could no longer tolerate the drug, the tumor reached four times the initial volume, or the administration persisted for more than 150 days. Then, tumor tissue was harvested after the mice were euthanized.
2.3. Response Evaluation
The best response (%) calculated based on the Modified Response Evaluation Criteria in Solid Tumors (mRECIST) was used to evaluate PARPi efficacy (Table S1). PFS and OS in the PDXs were defined as the interval between the start of administration and when the tumor had doubled and quadrupled in size, respectively [24]. As long as more than half of the PDX models in the experimental group achieved stable disease (SD), partial response (PR), or complete response (CR) and the best response or survival in the experimental group was better than that in the vehicle group, a patient was considered to respond to PARPis.
2.4. WES of Patient Samples and Data Analysis
Tumor and normal tissues were used for whole exon sequencing (WES). The DNA extraction, gene library establishment, and quality control procedures were the same as those in our previous article [25]. Paired tumor and normal tissues were used to detect somatic mutations. The data were analyzed by TCGA-MC3, a scalable open-source scientific approach for mutation calling of tumor exomes using multiple genomic pipelines. All mutations with at least two callers were maintained and annotated by VEP to ensure accuracy, followed by filtering of noncoding regions, mutations of types 2 and 3 in the ClinVar (201912) database, and mutations whose frequencies were more significant than 1% among East Asian populations in the ExAC and gnomAD databases to obtain final somatic mutations. The HaplotypeCaller module of GATK was used to detect germline mutations, which were further filtered by the officially recommended hard-filter criteria of SNP/INDEL and the remaining filter conditions, such as somatic mutations. GSITIC2.0 was used to calculate significantly changed regions (Q-value = 0.01) to determine the copy number variation (CNV) status. DeconstructSigs (R package) based on nonnegative matrix decomposition was employed to extract 96 somatic mutation patterns, map them onto 30 features, and correlate them to data in the COSMIC (version 2) database to predict mutation-driving factors. The HRDscore algorithm, which considers genome heterozygous deletions (LOH), telomere allele imbalances (TAI), and large fragment migrations (LST), was built by Precision Scientific Co. (Beijing).
2.5. RNA Extraction and RT-qPCR
Tissue was crushed, and total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The mRNA was reverse transcribed into cDNA using the PrimeScript cDNA Synthesis Kit (Takara Bio, Inc., Kusatsu shi, Japan). Applied Biosystems SYBR Green Master Mix (Thermo Fisher Scientific, Inc., US) was employed for qPCR. GAPDH was used as the internal reference. NCBI Primer Blast was used to design the following primers: GAPDH forward 5′-ACCCAGAAGACTGTGGATGG-3′, reverse 5′-TCTAGACGGCAGGTCAGGTC-3′; KRAS forward 5′-ACAGGCTCAGGACTTAGCAA-3′, reverse 5′-AAGGCATCATCAACACCCAGA-3′; ATK1 forward 5′-CTGCACAAACGAGGGGAGTA-3′, reverse 5′-TCACGTTGGTCCACATCCTG-3′.
2.6. Protein Extraction and Immunoblotting
Tissue was crushed in RIPA lysis buffer (Beyotime Institute of Biotechnology) with PMSF (1%) (Beyotime Institute of Biotechnology) and a cocktail (1%) (Roche, Basel, Switzerland). This was followed by rotation at 4 °C for 60 min and centrifugation at 12,000 rpm at 4 °C for 15 min. Then, the supernatant was obtained. The protein concentration was determined using a bicinchoninic acid assay kit (LABLEAD). SDS−PAGE loading buffer (Beyotime Institute of Biotechnology) was added to the protein samples, and then the samples were boiled at 100 °C for 10 min. The 25 µg protein sample was subjected to SDS−PAGE electrophoresis (4% concentrated gel and 10% separated gel). Then, it was transferred to a PVDF membrane (EMD Millipore, Burlington, MA, USA), which was ultimately incubated in 5% skim milk at room temperature for one hour. The sealed PVDF membrane was then incubated with the following primary antibodies overnight at 4 °C: rabbit anti-actin monoclonal antibody (mAb) (1:1000; ABclonal, cat. no. A2319), rabbit anti-KRAS polyclonal antibody (pAb) (1:1000; ABclonal, cat. no. A1190) and rabbit anti-AKT1 mAb (1:1000; ABclonal, Wuhan, China, cat. no. A20799). After elution in TBST (LABLEAD), the membrane was incubated in HRP-conjugated goat anti-rabbit IgG (H + L) secondary antibodies (1:5000; ABclonal, cat. no. A5014) at room temperature for one hour. Enhanced chemiluminescence (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to detect the signal.
2.7. Statistics
Comparisons between the treatment and vehicle groups were performed by Student’s t-tests or Mann−Whitney U tests (measurement data) and chi-squared tests or Fisher’s exact tests (ranked data). The kappa coefficient value was used for consistency analysis, McNemar’s test was used for paired classification data analysis, and the log-rank test and Kaplan–Meier method were used for survival analysis. All data statistics were calculated by SPSS 26.0 (IBM Corporation, Armonk, NY, USA). A p value less than 0.05 was considered statistically significant.
3. Result
3.1. Patient Characteristics and PARPi Sensitivity Test
Forty patients were enrolled, and the detailed clinical information is depicted in Table S2. All patients except patient 34 (P34) underwent niraparib sensitivity testing (177 PDXs in the experimental group and 181 PDXs in the vehicle group), and olaparib sensitivity testing was performed for 27 patients (109 PDXs in the experimental group and 104 PDXs in the vehicle group). A total of 26 patients underwent both sensitivity tests simultaneously. The experimental group achieved superior drug efficacy compared to the vehicle group. This was indicated by the fact that the niraparib group induced a significantly higher response rate (59.89% vs. 33.15%, p < 0.001) and median best response rate (47.34% vs. 109.62%, p = 0.001) compared to the vehicle group, and a similar pattern was observed in the olaparib cohort (72.48% vs. 33.65%, p < 0.001; 37.06% vs. 113.52%, p = 0.001, respectively) (Figure S1).
3.2. PDX Is a More Accurate Indicator of the Individualized Efficacy of PARPis
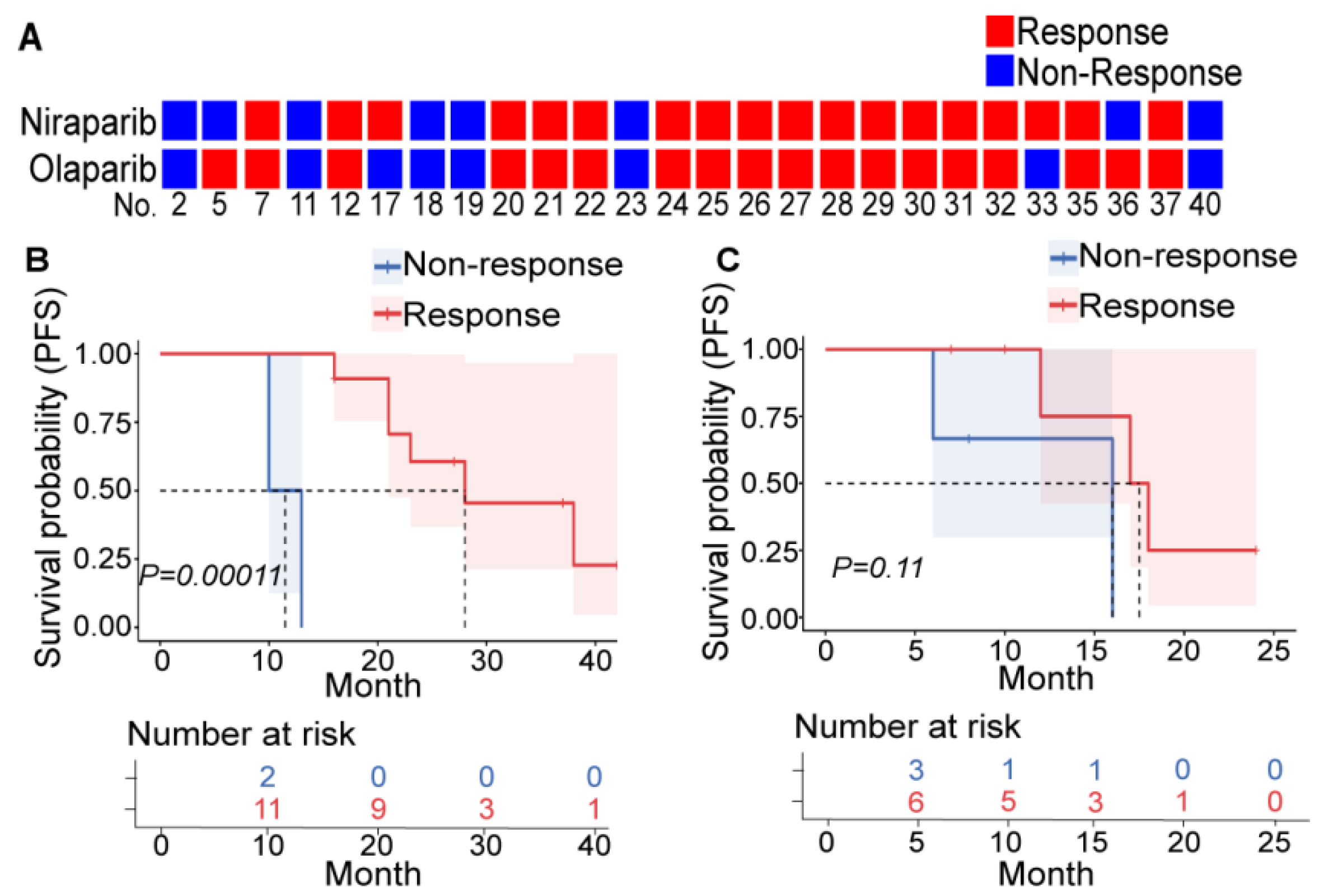
Among the 26 patients who underwent both niraparib and olaparib sensitivity tests, only 4 patients had different efficacy (no significant difference, p = 1.00 and high consistency, Kappa index = 0.675, p = 0.001) (Figure 1A, Table S3). In addition, all four patients with clear cell carcinoma (CC) were resistant to PARPis. BRCAmut and HRD+ are relatively reliable indicators of PARPi sensitivity in primary patients with HGSOC, and platinum sensitivity seemed to be a decent indicator in relapsing patients (Table S4, and tumor volume changes of typical patients are depicted in Figure S2). These findings were consistent with clinical experience and confirmed the repeatability and veracity of PDXs for evaluating PARPi efficacy.
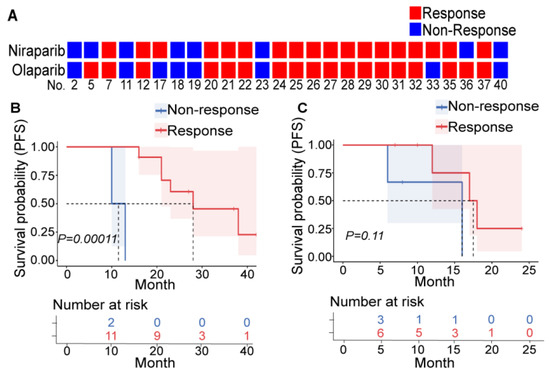
Figure 1.
PDXs can reflect PARPi efficacy. (A) Results of the niraparib (upper row) and olaparib (lower row) susceptibility tests in PDXs derived from 26 patients. Each square represents a response to a drug (red: nonresponder; blue: responder), and each column represents a patient (the patient number is at the bottom). Patients who were treated with PARPis as first-line (B) or second-line (C) maintenance therapy were divided into the responder (red) and nonresponder (blue) groups according to the PDX drug sensitivity test, and the differences in PFS between these two groups were compared. The upper picture depicts the Kaplan–Meier curve and log-rank value between the responder and nonresponder groups. The lower table shows the number of patients who did not reach the endpoint at the corresponding time points in each group.
BRCAmut, HRD+, and platinum sensitivity are not accurate indicators of PARPi response since approximately half of BRCAwt, HRD-, and platinum-resistant primary patients respond to PARPis. Additionally, none of these indicators significantly increased the prediction effect for PARPi benefit (Table S4). The correlation between PARPi efficacy and BRCAmut and HRD+ was even weaker in relapsed patients (Table S4).
Furthermore, we explored the evaluation efficacy of PDX in patients. Patients who were clinically treated with PARPis as first-line (13 patients) and second-line (9 patients) maintenance therapy were included (Table S5). They were divided into two groups according to the PARPi response suggested by PDXs. In first-line maintenance patients, the PARPi-effective group had significantly longer PFS than the PARPi-ineffective group (Figure 1B). However, this was not observed in patients with second-line maintenance (Figure 1C) due to a small number of enrolled patients and various second-line treatment regimens. This suggests that PDXs can screen patients with more significant survival benefits from PARPis among the population meeting the existing administration indicators.
3.3. Using PDX to Detect Novel Molecular Indicators of PARPis
Patients with WES results and similar sensitivity to olaparib and niraparib were divided into a PARPi-effective group (12 patients) and a PARPi-ineffective group (8 patients), and the genome characteristics of these groups were compared.
3.3.1. CNVs
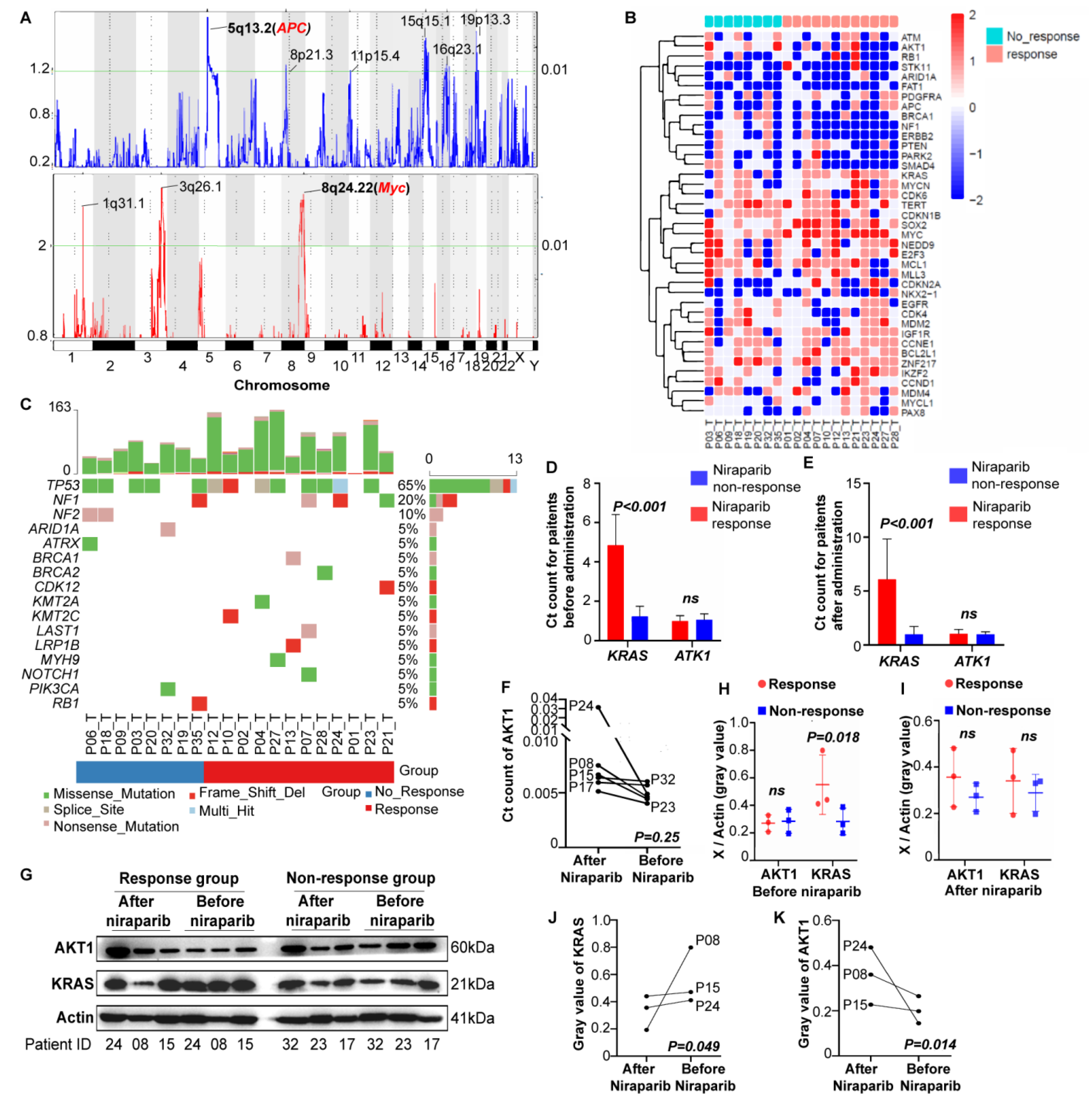
The effective group showed enriched Myc amplification and APC deletion (Figure 2A and Figure S3A). In addition, AKT1 deficiency was significantly enriched in the effective group, and KRAS deficiency was significantly enriched in the ineffective group (Tables S6 and S7). The distribution of CNVs in the two groups was slightly different. The chemotherapeutic resistance-related genes (mainly AKT1 and ERBB1) and EGFR tyrosine kinase inhibitor-related genes were decreased in the effective group, and multiple tumor driver genes (mainly Myc) were increased in the ineffective group (Figure 2B).

Figure 2.
Finding novel molecular biomarkers of PARPis. (A) The copy number variations (CNVs) in the olaparib and niraparib response groups. The bottom of the figure indicates the chromosome, the left number is the GISTIC Gscore (CNV frequency multiplied by CNV amplitude), and the right number is the q-value. The upper figure shows gene deletions (blue), and the lower figure shows gene enrichments (red). The detailed loci of any statistically significant CNVs (p < 0.01) are labeled. (B) The cluster analysis of CNVs in the responder and nonresponder groups. (C) Driver mutation analysis in the responder and nonresponder groups. The RNA (D–F) and protein (G–K) levels of KRAS and AKT1 were measured in the PDX tumor tissue before and after niraparib administration in the responder and nonresponder groups (three in each). Differences in KRAS and AKT1 RNA levels between the responder and nonresponder groups before (A) and after (B) niraparib treatment. (C) Changes in AKT1 RNA levels before and after niraparib treatment in the same patient. (D) The immunoblotting result, followed by the analysis of its gray values. Differences in KRas and Akt1 protein levels between the responder and nonresponder groups before (E) and after (F) niraparib treatment. Changes in Akt1 (G) and KRas (H) levels before and after niraparib treatment in the same patient from the responder group.
3.3.2. Gene Mutations
The number of functional loss sites in the effective group was significantly increased compared with that in the ineffective group (92.25 ± 51.40 vs. 56.63 ± 20.65, p = 0.047) (Figure S3B). Then, we analyzed driving mutations in epithelial ovarian cancer. TP53 mutations were the most common (63%), followed by NF1 (20%) and NF2 (10%) mutations. Additionally, these mutations were enriched in the effective group in general, especially BRCA1/2 mutations (Figure 2C). Nevertheless, no significant difference in gene mutations was detected (Table S8). In the signature analysis, only age was identified as significantly different between the effective group (lower) and the ineffective group (higher) (Figure S3C). Mutual exclusion and coexistence examination of mutated genes highlighted a coexisting trend of BRCA1-LRP1B and NOTCH1-LATS1 mutations in the effective group, but the differences were not significant (Figure S3D,H).
3.3.3. Verification by RNA and Protein
Patients in the niraparib responder (P08, P15, P24) and nonresponder (P17, P23, P32) groups were randomly selected to explore the effects of KRAS and AKT1 on PARPi efficacy. In both the pre-niraparib and post-niraparib tissues, the RNA levels of KRAS in the responder group were significantly higher than those in the nonresponder group, but AKT1 showed no significant difference (before treatment, p = 0.618; after treatment, p = 0.389) (Figure 2D,E). Therefore, we further explored the change in AKT1 expression after treatment. After niraparib treatment, the AKT1 level showed an increasing trend, although there was no statistical significance (Figure 2F). The protein analysis showed similar results. The KRAS level before treatment in the responder group was significantly higher than that in the nonresponder group, but there was no significant difference after treatment (p = 0.402) or in the AKT1 level (before treatment, p = 0.125; after treatment, p = 0.055) (Figure 2G–I). Similarly, AKT1 levels significantly increased after niraparib treatment, while KRAS levels significantly decreased after niraparib treatment in the responder group (Figure 2J,K).
These experiments suggest that tumor tissues with high KRAS expression are responsive to niraparib and that the enrichment of AKT1 during treatment might lead to drug resistance.
3.4. Using PDX to Detect Novel Clinical Indicators of PARPis
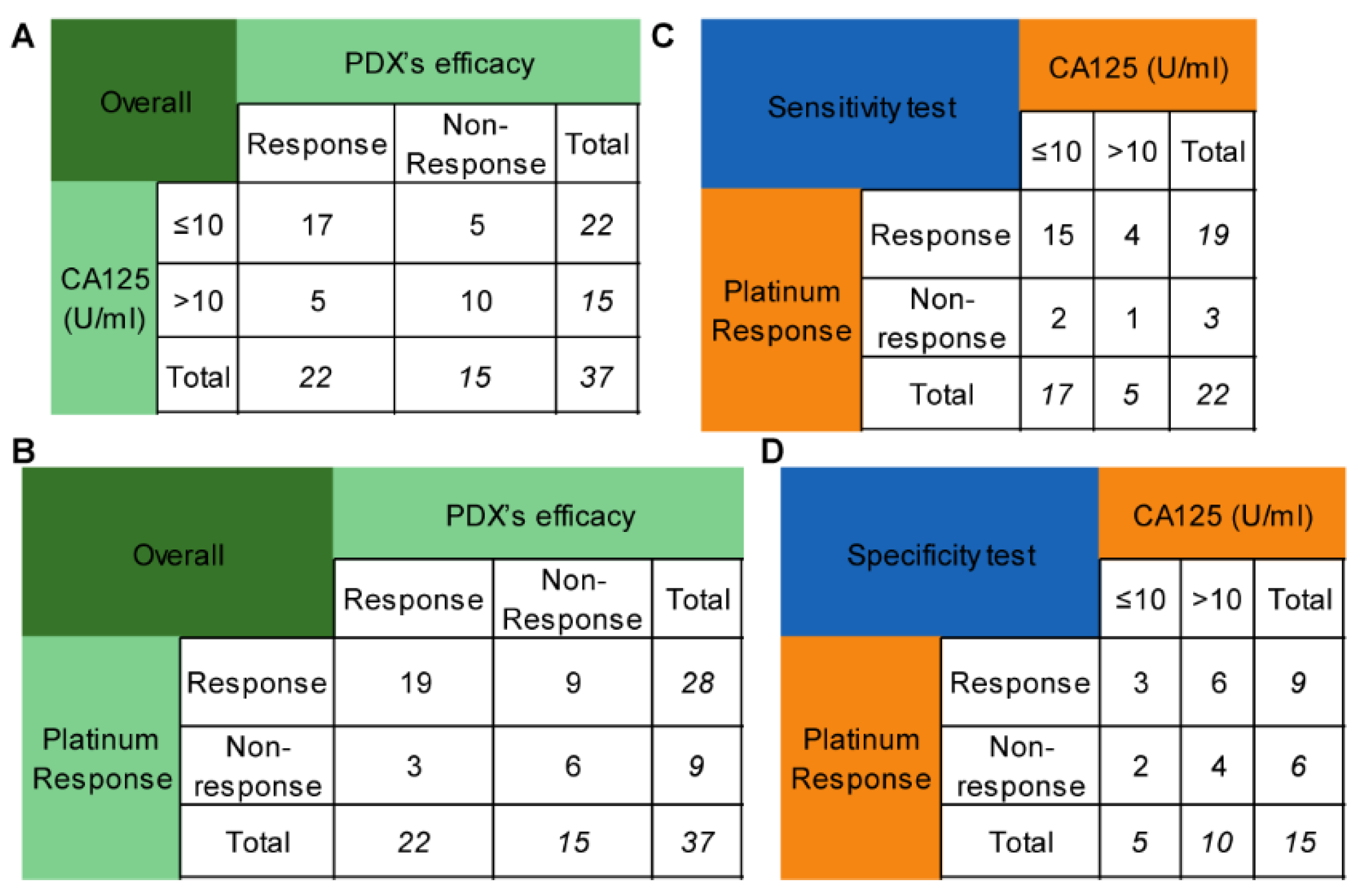
Platinum sensitivity is the clinical indicator for niraparib when it is used as maintenance therapy. However, it takes at least six months to determine the platinum response in naive patients, which can delay the administration. The degree of CA125 reduction during chemotherapy may partly reflect the sensitivity to platinum-based chemotherapy. As a result, this study investigated whether the lowest CA125 level (truncated at 10 U/mL) during chemotherapy could replace platinum sensitivity as a predictor of PARPi efficacy. Seventeen out of twenty-two patients with CA125 levels of no more than 10 U/mL were platinum-sensitive, while 5 out of 15 patients with CA125 levels less than 10 U/mL were platinum-resistant. The sensitivity and specificity of CA125 for predicting the efficacy of PARPis were 77.27% and 66.67% (Figure 3A), respectively, while those of platinum sensitivity were 86.36% and 40.00% (Figure 3B), respectively. These values were not significantly different (p = 0.687, p = 0.289, respectively) (Figure 3C,D).
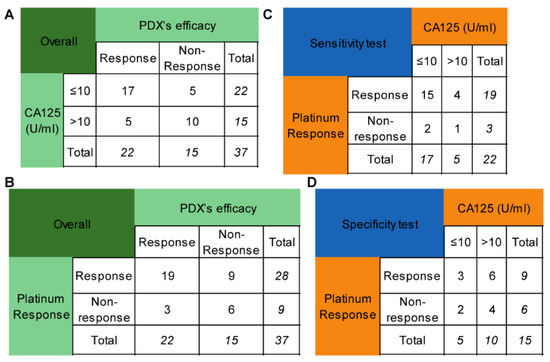
Figure 3.
Comparison of the CA125 level and platinum response in predicting niraparib efficacy. (A) The performance of the CA125 level (truncated at 10 U/mL) in predicting niraparib efficacy. (B) The performance of the platinum response in predicting niraparib efficacy. (C) Comparison of the sensitivity of the CA125 level and platinum response to predict niraparib efficacy. (D) Comparison of the specificity of the CA125 level and platinum response to predict niraparib efficacy.
3.5. Using PDX to Perform Clinical Trials
3.5.1. Reproducing NOVA, PRIMA, and SOLO I Trials
PDXs from five platinum-sensitive relapsing patients with HGSOC were employed to mimic the NOVA study. Both the PFS and OS of the niraparib group were significantly better than those of the control group (Figure S4A,B). Five platinum-sensitive primary patients with BRCAmut were employed to mimic the SOLO I study, and all PDXs in the experimental group had a longer PFS and OS than those in the vehicle group (Figure S4C,D). Seven platinum-sensitive advanced patients with residual lesions after surgery were employed to imitate the PRIMA study, including two patients with HRD+ and one with BRCAmut. All PDXs, HRD+ PDXs, and HRD- PDXs benefited from niraparib, as did the small number of BRCAmut patients who could not be analyzed effectively (Figure S4E–J).
3.5.2. Attempting a Prospective Trial to Determine Whether Niraparib Could Replace Chemotherapy as First-Line Treatment for Ovarian Cancer
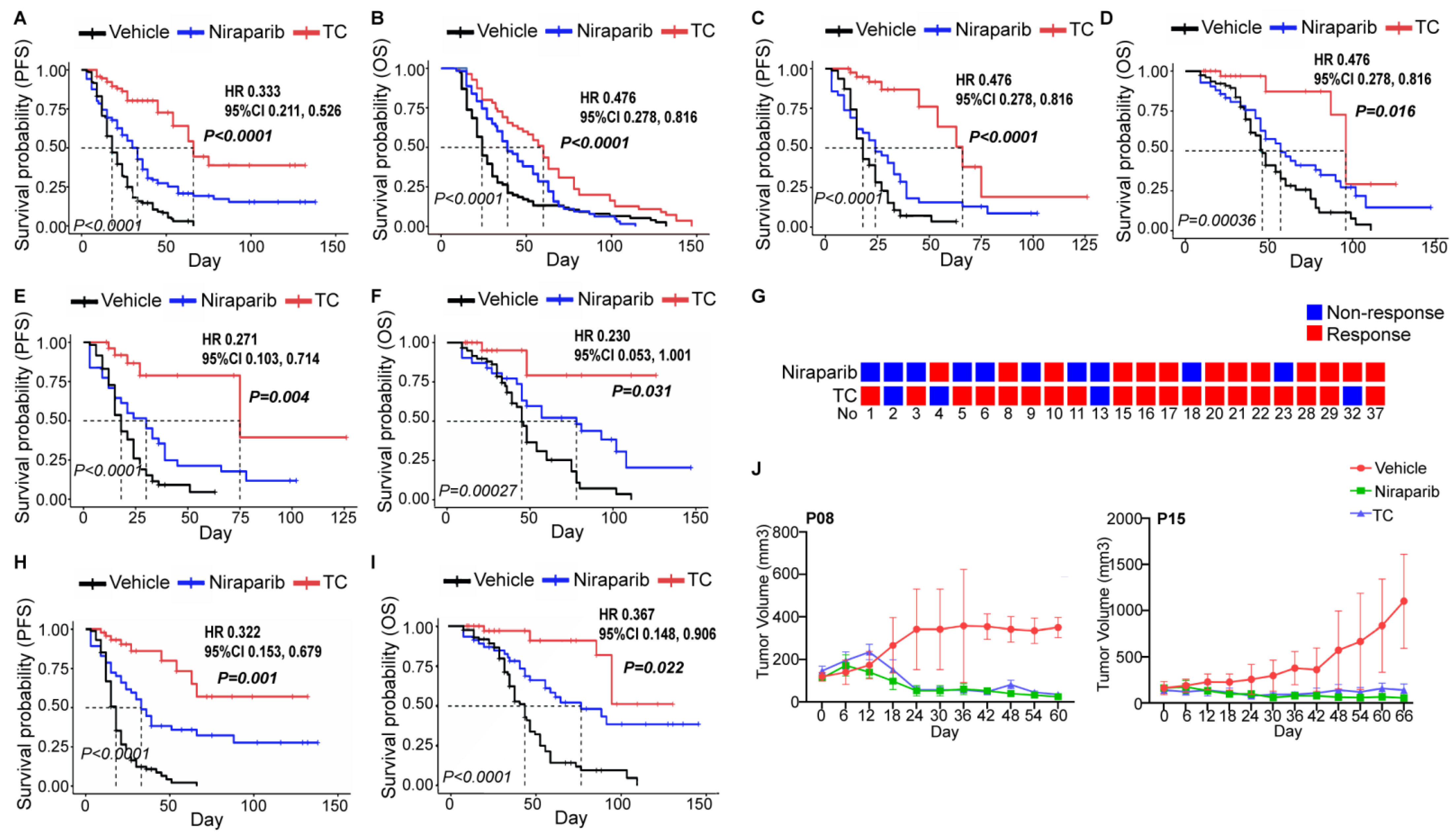
We further recruited PDXs established by 23 naive patients and compared the efficacy of the vehicle, niraparib, and TC chemotherapy. The efficacy evaluation indexes included best response (%), response category rate, PFS, and OS. The TC group exhibited a better treatment effect than the niraparib group among the general population (Figure 4A,B and Figure S5A,B) and patients in the HRD+ or BRCAmut subgroups (Figure 4C–F and Figure S5C–F). We noticed that 11 patients responded to both niraparib and TC (Figure 4G). Thus, we further explored this possibility in these patients and observed the same results (Figure 4H,I and Figure S5G,H). Nevertheless, niraparib and TC demonstrated a similar efficacy in several patients (P08 and P15) (Figure 4J), and two TC-resistant patients responded to niraparib (Figure 4G).

Figure 4.
Comparing the efficacies of niraparib and TC in PDX. The survival outcomes of the control, niraparib, and TC chemotherapy groups in PDXs of naive patients (A,B), the HRD+ subgroup (C,D), or the BRCA mutation subgroup (E,F) were analyzed. (G) Results of the niraparib and TC susceptibility tests evaluated by PDXs in 23 patients. (H,I). The survival outcomes of the control, niraparib, and TC chemotherapy groups in patients who simultaneously responded to niraparib and TC. (J) Tumor volume changes due to the vehicle (red), niraparib (green), and TC (blue) treatments in 2 patients (P08 and P15) who were responsive to niraparib but not TC. The italic p value in each Kaplan–Meier survival curve figure indicates the comparison of those three groups, and the p value (bold and italic), hazard ratio (HR), and 95% confidence interval (CI) represent the comparison between the niraparib and TC groups. TC, paclitaxel and carboplatin chemotherapy; PFS, progression-free survival; OS, overall survival.
Therefore, PDX can reproduce the clinical trials well, and niraparib has not yet been indicated as a replacement for TC chemotherapy in the first-line treatment of ovarian cancer in our preclinical trial.
4. Discussion
PDXs are considered one of the best preclinical models. In PDXs, the original tumor tissue is subcutaneously transplanted into immunodeficient mice, and PDXs retain their molecular and pathological characteristics, drug response, and tumor microenvironment despite host selection pressure [24,26,27,28]. The samples in the PDX model library used in this paper have been compared with the original tumor genome, transcriptome, proteome, and chemotherapy response data and have shown good consistency [25].
PDXs have been used to screen small-molecule targeted drugs for various tumors, and HGSOC-PDXs have been used to explore the PARPi response of patients with RAD51 or BRCA1 methylation and the therapeutic effect of PARPis combined with other drugs [29,30,31]. However, whether PDXs can accurately reflect PARPi efficacy has not been clearly indicated in experiments because few studies have reported consistent results on the correlation between BRCAmut and olaparib sensitivity [24,26]. In this study, the niraparib and olaparib sensitivity tests showed high consistency in PDXs established from the same patient, suggesting the repeatability of the PDX model for evaluating PARPi efficacy. Naive HGSOC patients with BRCAmut and HRD+ were more responsive to PARPis than platinum sensitivity. Additionally, platinum sensitivity seemed to be a more reliable indicator in relapsed patients, as identified by clinical experience and trials. Moreover, the PDX model reproduced the NOVA, PRIMA, and SOLO I trials. These results suggest that the PDX model can accurately predict the PARPi response.
The most prominent problem with PARPis in ovarian cancer is the lack of adequate application metrics, and BRCA mutation, HRD status, and platinum sensitivity are not perfect indicators for PARPis [10,14,15,16,17,32,33]. The mechanism of PARPis can partially explain this phenomenon. Although PARPis can lead to a ‘synthetic lethal’ interaction with HRD, the compensatory effects of other DNA damage repair pathways, BRCA1/2 recovery mutations, high expression of drug resistance pumps, and changes in tumor metabolism may cause acquired PARPi resistance [34]. In addition, other molecular defects in the homologous recombination (HR) pathway have the same killing effect (called ‘BRCAness’) [35]. Platinum agents lead to chromosomal cross-linking and kill tumor cells once nucleotide excision repair deficiency exists, indicating the presence of genomic instability and not HRD [12]. Therefore, we tested the accuracy of the current indicators and explored novel molecular and clinical indicators using PDXs.
In this experiment, we found that 20% of HRD+ and 40% of platinum-sensitive naive HGSOC patients had no response to PARPis, and approximately half of BRCAwt, HRD-, and platinum-resistant HGSOC patients responded to PARPis. In addition, we screened subpopulations with more significant survival benefits from patients clinically treated with PARPis as first-line maintenance therapy, demonstrating that the PDX model can effectively avoid the above limitations of clinical and molecular indications. Thus, the PDX model serves as a ‘black box’ that provides a more accurate and objective efficacy prediction than BRCAmut, HRD+, and platinum sensitivity.
PDX models feature patient-specific therapeutic effects and thus provide a possible tool for searching for molecular and clinical biomarkers of PARPis. Our research suggested that tumors with high KRAS expression were sensitive to PARPi treatment, and increased AKT1 expression after PARPi treatment may be associated with PARPi resistance.
The oncogene KRAS is associated with tumor development and drug resistance. Research has revealed that MEK inhibitors obstructing abnormally activated KRAS signaling pathways could trigger and amplify PARPi efficacy by increasing double-strand DNA breaks and activating the STING signaling pathway [36]. Although TP53 mutations are predominant in high-grade serous carcinoma and are the most common pathological type of epithelial ovarian cancer, KRAS mutations are more common in low-grade serous carcinoma and endometriosis-driven pathological types (clear cell or endometrioid carcinoma). PARPis are mainly used in the treatment of HGSOC patients, given that BRCA1/2 mutations mainly exist in HGSOC. The relationship between the KRAS expression level and PARPi efficacy in this study may provide a new direction for further preclinical trials of PARPi usage in such rare ovarian cancers and other malignancies with predominant KRAS mutations, such as colon cancer [37,38].
Through WES, ATK1 deletion mutations were enriched in the PARPi-sensitive group. We found that ATK1 was significantly enriched in tumors after PARPi application, and the residual tumor cells after PARPi administration were mostly drug-resistant cells after drug screening, which indicated that AKT1 enrichment might be related to PARPi resistance. However, there was no difference in AKT1 between the sensitive group and the drug resistance group, which may be caused by the insufficient number of tissues included. Additionally, PARPis activate AKT to form the phosphorylated ATM-Nemo-Akt-MTOR complex, located in the mitochondria, to protect cells from oxidative stress [39]. These two studies are consistent with our results, but further studies need to explore the underlying mechanisms in more detail.
Platinum sensitivity is the clinical indicator for PARPis, and the decrease in CA125 is closely correlated with platinum reactivity [8,40,41]. Therefore, adopting 10 U/mL as the cutoff value, this study showed that the minimum level of CA125 during chemotherapy could replace the platinum response as a predictor for the use of PARPis given that the two methods had similar sensitivity and specificity. This suggests that patients with CA125 less than 10 U/mL during chemotherapy can use PARPis as maintenance therapy immediately.
The other aspect that caught our attention was that PARPis have continuously expanded their application in ovarian cancer treatment, from posterior-line treatment to second-line maintenance to the current first-line maintenance [2,3,4,5,6,7]. Moreover, a study supported niraparib monotherapy as a neoadjuvant agent in advanced ovarian patients who cannot undergo optimal cytoreductive surgery [22,23]. Given this, we questioned whether PARPis had a similar first-line treatment effect to that of platinum-based chemotherapy in the PDX model. Unfortunately, TC consistently outperformed niraparib. However, it is undeniable that niraparib has shown similar efficacy to TC in several patients, and TC-resistant patients may benefit from niraparib. However, there is no approach to distinguish this subpopulation except the PDX susceptibility test. Therefore, clinical trials should be conducted with caution.
A limitation of this study is the small number of included patients. This made it challenging to analyze the molecular targets related to PARPi efficacy and identify subgroups that could benefit from niraparib as first-line treatment. The long time period that is required for model formation and drug sensitivity testing is often listed as a limitation to clinical application. However, the median PDX formation time was five months, and the median drug sensitivity testing time for patients lacking sensitivity (no effects on OS) was 23 days. That is, the PARPi maintenance efficacy results could be obtained within six months, which would not affect patient use of PARPis.
5. Conclusions
PDX models can better represent the personal therapeutic efficacy of PARPis in epithelial ovarian cancer than BRCA1/2mut, HRD+, and platinum sensitivity. High KRAS expression was correlated with PARPi sensitivity, and the accumulation of AKT1 during PARPi treatment might lead to PARPi resistance. A minimum CA125 of less than 10 U/mL during chemotherapy can be used as a clinical indicator of PARPi sensitivity. PARPis are not yet an alternative to platinum-based chemotherapy as a first-line treatment for ovarian cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14194649/s1, Figure S1: The experiment of niraparib and olapalib sensitivity tests in PDX models; Figure S2: Efficacy in PDXs from patients with different mutational characteristics; Figure S3: Genomic analysis of olaparib and niraparib response and non-response groups; Figure S4: Simulate the NOVA, SOLO I, and PRIMA clinical trials; Figure S5: Comparing efficacy between niraparib and TC in PDX; Table S1: Response Category; Table S2: Patients’ characteristics; Table S3: Drug efficacy of PDX; Table S4: The consistency of PARPi’s efficacy between PDX models and clinical guidelines recommendations; Table S5: Survival data of patients using PARPi as first- of second-line maintenance therapy; Table S6: The comparison of gene amplification between response and non-response groups; Table S7:The comparison of gene deletion between response and non-response groups; Table S8: The comparison of gene mutation between response and non-response groups.
Author Contributions
Data curation, J.C., Y.J. and Y.L.; formal analysis, J.C., H.W. and T.L.; funding acquisition, L.P.; investigation, J.C., Y.J. and Y.L.; methodology, L.P. and S.P.; resources, Y.G., W.W., Y.S., J.Y., Y.W., M.Q. and S.L.; supervision, L.P., Y.J., Y.L. and S.P.; writing—original draft, J.C.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by CAMS Innovation Fund for Medical Sciences (grant number CIFMS-2017-I2M-1-002), LE FUND (grant number KH-2020-LJJ-042), the Fund of National Key R&D Program of China (grant number 2016YFC1303700), the Fund of National Natural Science Foundation of China (grant numbers 30772315, 81572564, 31171416, and 31471383), and the Fund of Graduate Student Funding of Peking Union Medical College Hospital (grant number X344600).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Peking Union Medical College Hospital (protocol code S-072). The animal study protocol was approved by the Institutional Review Board of Peking Union Medical College Hospital.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information Files.
Acknowledgments
We are grateful to all the patients, animals, and unmentioned colleagues who participated in this study and the technical support and consultation from Beijing IDMO Co., Ltd. (Beijing, China), and Precision Scientific (Beijing) Co., Ltd. (Beijing, China).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Administration FAD. Highlights of Prescribing Information: Lynparza (Revised: January 2018). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/208558s001lbl.pdf (accessed on 15 July 2021).
- Administration FAD. Highlights of Presribing Information: Zejula (Revised: March 2017). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/208447s015s017lbledt.pdf (accessed on 15 July 2021).
- Del Campo, J.M.; Matulonis, U.A.; Malander, S.; Provencher, D.; Mahner, S.; Follana, P.; Waters, J.; Berek, J.S.; Woie, K.; Oza, A.; et al. Niraparib Maintenance Therapy in Patients With Recurrent Ovarian Cancer After a Partial Response to the Last Platinum-Based Chemotherapy in the ENGOT-OV16/NOVA Trial. J. Clin. Oncol. 2019, 37, 2968–2973. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.-M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- Hoppe, M.M.; Sundar, R.; Tan, D.S.P.; Jeyasekharan, A.D. Biomarkers for Homologous Recombination Deficiency in Cancer. J. Natl. Cancer Inst. 2018, 110, 704–713. [Google Scholar] [CrossRef]
- Pennington, K.P.; Walsh, T.; Harrell, M.I.; Lee, M.K.; Pennil, C.C.; Rendi, M.H.; Thornton, A.; Norquist, B.M.; Casadei, S.; Nord, A.S.; et al. Germline and Somatic Mutations in Homologous Recombination Genes Predict Platinum Response and Survival in Ovarian, Fallopian Tube, and Peritoneal Carcinomas. Clin. Cancer Res. 2014, 20, 764–775. [Google Scholar] [CrossRef]
- Ang, J.E.; Gourley, C.; Powell, C.B.; High, H.; Shapira-Frommer, R.; Castonguay, V.; De Greve, J.; Atkinson, T.; Yap, T.A.; Sandhu, S.; et al. Efficacy of Chemotherapy in BRCA1/2 Mutation Carrier Ovarian Cancer in the Setting of PARP Inhibitor Resistance: A Multi-Institutional Study. Clin. Cancer Res. 2013, 19, 5485–5493. [Google Scholar] [CrossRef] [PubMed]
- Baghmar, S.; Agarwal, A.; Gauda, C.; Qureshi, S.; Malik, P.S.; Vaibhav, V. PARP inhibitor in platinum resistant ovarian cancer: Single center real world experience. Ann. Oncol. 2019, 30, ix81–ix82. [Google Scholar] [CrossRef]
- Foo, T.; George, A.; Banerjee, S. PARP inhibitors in ovarian cancer: An overview of the practice-changing trials. Genes Chromosom. Cancer 2020, 60, 385–397. [Google Scholar] [CrossRef]
- Gogineni, V.; Morand, S.; Staats, H.; Royfman, R.; Devanaboyina, M.; Einloth, K.; Dever, D.; Stanbery, L.; Aaron, P.; Manning, L.; et al. Current Ovarian Cancer Maintenance Strategies and Promising New Developments. J. Cancer 2021, 12, 38–53. [Google Scholar] [CrossRef]
- Ruscito, I.; Bellati, F.; Ray-Coquard, I.; Mirza, M.R.; du Bois, A.; Gasparri, M.L.; Costanzi, F.; De Marco, M.P.; Nuti, M.; Caserta, D.; et al. Incorporating Parp-inhibitors in Primary and Recurrent Ovarian Cancer: A Meta-analysis of 12 phase II/III randomized controlled trials. Cancer Treat. Rev. 2020, 87, 102040. [Google Scholar] [CrossRef] [PubMed]
- Swisher, E.M.; Aghajanian, C.; O’Malley, D.M.; Fleming, G.F.; Kaufmann, S.H.; Levine, D.A.; Birrer, M.J.; Moore, K.N.; Spirtos, N.M.; Shahin, M.S.; et al. Impact of homologous recombination status and responses with veliparib combined with first-line chemotherapy in ovarian cancer in the Phase 3 VELIA/GOG-3005 study. Gynecol. Oncol. 2021, 164, 245–253. [Google Scholar] [CrossRef]
- Essel, K.; Behbakht, K.; Lai, T.; Hand, L.; Evans, E.; Dvorak, J.; Ding, K.; Konecny, G.; Moore, K. PARPi after PARPi in epithelial ovarian cancer. Gynecol. Oncol. Rep. 2021, 35, 100699. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Selle, F.; Scambia, G.; Asselain, B.; Marmé, F.; Lindemann, K.; Colombo, N.; Madry, R.; Glasspool, R.; Dubot, C.; et al. LBA33 Maintenance olaparib rechallenge in patients (pts) with ovarian carcinoma (OC) previously treated with a PARP inhibitor (PARPi): Phase IIIb OReO/ENGOT Ov-38 trial. Ann. Oncol. 2021, 32, S1308–S1309. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Kim, B.-G.; Kim, J.-W.; Park, E.; Joung, J.-G.; Kim, S.; Choi, C.H.; Kim, H.S.; on behalf of Korean Gynecologic Oncology Group (KGOG) investigators. Biomarker-guided targeted therapy in platinum-resistant ovarian cancer (AMBITION; KGOG 3045): A multicentre, open-label, five-arm, uncontrolled, umbrella trial. J. Gynecol. Oncol. 2022, 33, e45. [Google Scholar] [CrossRef]
- Zhou, D.; Liu, J.; Liu, R.; Li, H.; Huang, Y.; Ma, D.; Hong, L.; Gao, Q. Effectiveness and Safety of Niraparib as Neoadjuvant Therapy in Advanced Ovarian Cancer With Homologous Recombination Deficiency (NANT): Study Protocol for a Prospective, Multicenter, Exploratory, Phase 2, Single-Arm Study. Front. Oncol. 2022, 12, 852772. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, W.; Liu, R.; Shan, W.; Li, H.; Liu, J.; Xia, B.; He, S.; Xia, Y.; Wang, S.; et al. Effectiveness and safety of niraparib as neoadjuvant therapy in advanced ovarian cancer with homologous recombination deficiency: A prospective, multicenter, exploratory, phase 2 single-arm study (NANT). In Proceedings of the 2022 SGO Annual Meeting on Women’s Cancer, Phoenix, AZ, USA, 18–21 March 2022. [Google Scholar]
- Palmer, A.C.; Plana, D.; Gao, H.; Korn, J.M.; Yang, G.; Green, J.; Zhang, X.; Velazquez, R.; McLaughlin, M.E.; Ruddy, D.A.; et al. A Proof of Concept for Biomarker-Guided Targeted Therapy against Ovarian Cancer Based on Patient-Derived Tumor Xenografts. Cancer Res. 2020, 80, 4278–4287. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jin, Y.; Li, S.; Qiao, C.; Peng, X.; Li, Y.; Gu, Y.; Wang, W.; You, Y.; Yin, J.; et al. Patient-Derived Xenografts Are a Reliable Preclinical Model for the Personalized Treatment of Epithelial Ovarian Cancer. Front. Oncol. 2021, 11, 744256. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Honkala, A.; Malhotra, S.V.; Kummar, S.; Junttila, M.R. Harnessing the predictive power of preclinical models for oncology drug development. Nat. Rev. Drug Discov. 2022, 21, 99–114. [Google Scholar] [CrossRef]
- Woo, X.Y.; PDXNET Consortium; Giordano, J.; Srivastava, A.; Zhao, Z.-M.; Lloyd, M.W.; de Bruijn, R.; Suh, Y.-S.; Patidar, R.; Chen, L.; et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat. Genet. 2021, 53, 86–99. [Google Scholar] [CrossRef]
- Castroviejo-Bermejo, M.; Cruz, C.; Llop-Guevara, A.; Gutierrez-Enriquez, S.; Ducy, M.; Ibrahim, Y.H.; Gris-Oliver, A.; Pellegrino, B.; Bruna, A.; Guzmán, M.; et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol. Med. 2018, 10, e9172. [Google Scholar] [CrossRef]
- Kim, H.; Xu, H.; George, E.; Hallberg, D.; Kumar, S.; Jagannathan, V.; Medvedev, S.; Kinose, Y.; Devins, K.; Verma, P.; et al. Combining PARP with ATR inhibition overcomes PARP inhibitor and platinum resistance in ovarian cancer models. Nat. Commun. 2020, 11, 3726. [Google Scholar] [CrossRef]
- Sanij, E.; Hannan, K.M.; Xuan, J.; Yan, S.; Ahern, J.E.; Trigos, A.S.; Brajanovski, N.; Son, J.; Chan, K.T.; Kondrashova, O.; et al. CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 2020, 11, 2641. [Google Scholar] [CrossRef]
- Lee, C.K.; Friedlander, M.L.; Mph, A.T.; Ledermann, J.A.; Coleman, R.L.; Mirza, M.R.; Matulonis, U.A.; Pujade-Lauraine, E.; Bloomfield, R.; Goble, S.; et al. Molecular and clinical predictors of improvement in progression-free survival with maintenance PARP inhibitor therapy in women with platinum-sensitive, recurrent ovarian cancer: A meta-analysis. Cancer 2021, 127, 2432–2441. [Google Scholar] [CrossRef]
- Rosenthal, R.; McGranahan, N.; Herrero, J.; Taylor, B.S.; Swanton, C. deconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016, 17, 31. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.Y.; Wu, N.; Chen, Y.C.; Cheng, Q.; Wang, J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Mol. Cancer 2020, 19, 107. [Google Scholar] [CrossRef]
- da Cunha Colombo Bonadio, R.R.; Fogace, R.N.; Miranda, V.C.; Diz, M. Homologous recombination deficiency in ovarian cancer: A review of its epidemiology and management. Clinics 2018, 73 (Suppl. 1), e450s. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, X.; Fu, Y.; Guo, E.; Ye, Y.; Li, F.; Liu, S.; Xiao, R.; Liu, C.; Lu, F.; et al. MEK Inhibition Remodels the Immune Landscape of Mutant KRAS Tumors to Overcome Resistance to PARP and Immune Checkpoint Inhibitors. Cancer Res. 2021, 81, 2714–2729. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, P. Morphologic, Immunophenotypic, and Molecular Features of Epithelial Ovarian Cancer. Oncology 2016, 30, 166–176. [Google Scholar] [PubMed]
- Bulun, S.E.; Wan, Y.; Matei, D. Epithelial Mutations in Endometriosis: Link to Ovarian Cancer. Endocrinology 2019, 160, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Tapodi, A.; Bognar, Z.; Szabo, C.; Gallyas, F.; Sumegi, B.; Hocsak, E. PARP inhibition induces Akt-mediated cytoprotective effects through the formation of a mitochondria-targeted phospho-ATM-NEMO-Akt-mTOR signalosome. Biochem. Pharmacol. 2019, 162, 98–108. [Google Scholar] [CrossRef]
- Markman, M.; Federico, M.; Liu, P.Y.; Hannigan, E.; Alberts, D. Significance of early changes in the serum CA-125 antigen level on overall survival in advanced ovarian cancer. Gynecol. Oncol. 2006, 103, 195–198. [Google Scholar] [CrossRef]
- van Altena, A.M.; Kolwijck, E.; Spanjer, M.J.; Hendriks, J.C.; Massuger, L.F.; de Hullu, J.A. CA125 nadir concentration is an independent predictor of tumor recurrence in patients with ovarian cancer: A population-based study. Gynecol. Oncol. 2010, 119, 265–269. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).