Meta-Analytic Comparison of Global RNA Transcriptomes of Acute and Chronic Myeloid Leukemia Cells Reveals Novel Gene Candidates Governing Myeloid Malignancies
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Data Extraction, Processing and Analysis
2.3. Gene Ontology
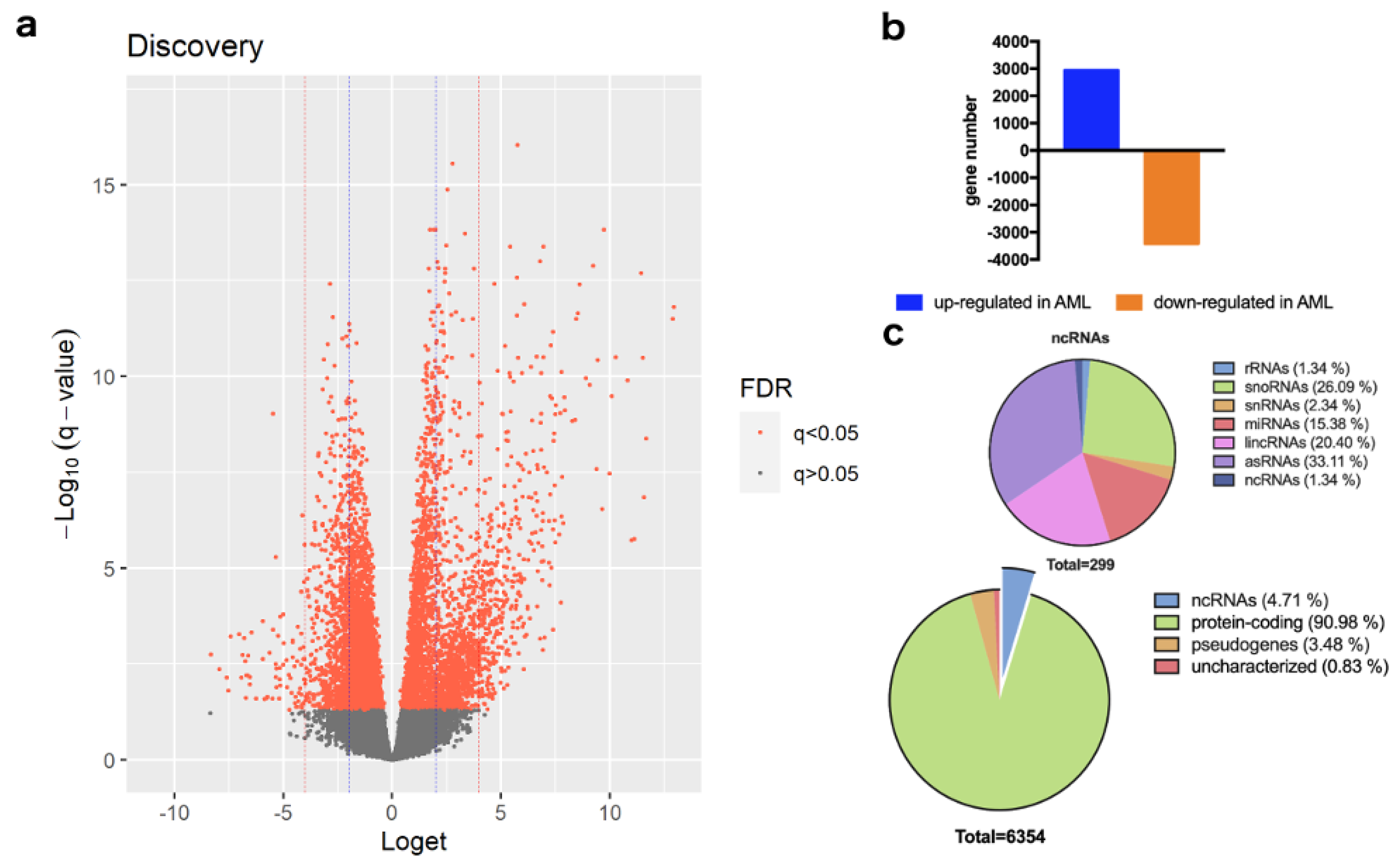
3. Results
Differentially Expressed Genes between AML and CML belong to Distinct Functional Groups
4. Discussion
4.1. Long Non-Coding RNAs and Pseudogenes
4.2. Pre-RNA Splicing and Processing
4.3. Oxidative Stress
4.4. Protein Glycosylation
4.5. Extracellular Vesicles and Secretory Granules
4.6. Migration of Neutrophils
4.7. Antimicrobial Immune Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Papaemmanuil, E.; Dohner, H.; Campbell, P.J. Genomic Classification in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 375, 900–901. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Vetrie, D.; Helgason, G.V.; Copland, M. The leukaemia stem cell: Similarities, differences and clinical prospects in CML and AML. Nat. Rev. Cancer 2020, 20, 158–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Pine, A.R.; Kotini, A.G.; Yuan, H.; Zamparo, L.; Starczynowski, D.T.; Leslie, C.; Papapetrou, E.P. Sequential CRISPR gene editing in human iPSCs charts the clonal evolution of myeloid leukemia and identifies early disease targets. Cell. Stem Cell 2021, 28, 1074–1089.e1077. [Google Scholar] [CrossRef]
- Lavallee, V.P.; Lemieux, S.; Boucher, G.; Gendron, P.; Boivin, I.; Armstrong, R.N.; Sauvageau, G.; Hebert, J. RNA-sequencing analysis of core binding factor AML identifies recurrent ZBTB7A mutations and defines RUNX1-CBFA2T3 fusion signature. Blood 2016, 127, 2498–2501. [Google Scholar] [CrossRef]
- Pabst, C.; Bergeron, A.; Lavallee, V.P.; Yeh, J.; Gendron, P.; Norddahl, G.L.; Krosl, J.; Boivin, I.; Deneault, E.; Simard, J.; et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016, 127, 2018–2027. [Google Scholar] [CrossRef]
- Lee, S.I.; Celik, S.; Logsdon, B.A.; Lundberg, S.M.; Martins, T.J.; Oehler, V.G.; Estey, E.H.; Miller, C.P.; Chien, S.; Dai, J.; et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat. Commun. 2018, 9, 42. [Google Scholar] [CrossRef]
- Assi, S.A.; Imperato, M.R.; Coleman, D.J.L.; Pickin, A.; Potluri, S.; Ptasinska, A.; Chin, P.S.; Blair, H.; Cauchy, P.; James, S.R.; et al. Subtype-specific regulatory network rewiring in acute myeloid leukemia. Nat. Genet. 2019, 51, 151–162. [Google Scholar] [CrossRef]
- Ehx, G.; Larouche, J.D.; Durette, C.; Laverdure, J.P.; Hesnard, L.; Vincent, K.; Hardy, M.P.; Theriault, C.; Rulleau, C.; Lanoix, J.; et al. Atypical acute myeloid leukemia-specific transcripts generate shared and immunogenic MHC class-I-associated epitopes. Immunity 2021, 54, 737–752.e710. [Google Scholar] [CrossRef]
- Li, S.Q.; Liu, J.; Zhang, J.; Wang, X.L.; Chen, D.; Wang, Y.; Xu, Y.M.; Huang, B.; Lin, J.; Li, J.; et al. Transcriptome profiling reveals the high incidence of hnRNPA1 exon 8 inclusion in chronic myeloid leukemia. J. Adv. Res. 2020, 24, 301–310. [Google Scholar] [CrossRef]
- SRA Toolkit. SRA Toolkit Development Team, NCBI, Bethesda, Maryland, USA. Available online: https://github.com/ncbi/sra-tools (accessed on 18 May 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Gorenjak, M.; Zupin, M.; Jezernik, G.; Skok, P.; Potocnik, U. Omics data integration identifies ELOVL7 and MMD gene regions as novel loci for adalimumab response in patients with Crohn’s disease. Sci. Rep. 2021, 11, 5449. [Google Scholar] [CrossRef] [PubMed]
- Krsteski, J.; Gorenjak, M.; But, I.; Pakiz, M.; Potocnik, U. Dysregulation of Synaptic Signaling Genes Is Involved in Biology of Uterine Leiomyoma. Genes 2021, 12, 1179. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed]
- Coutin, N. biogridr: BioGRID R API. 2015, Vancouver, Canada. Available online: https://github.com/npjc/biogridr (accessed on 18 May 2022).
- Dong, Y.; Shi, O.; Zeng, Q.; Lu, X.; Wang, W.; Li, Y.; Wang, Q. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp. Hematol. Oncol. 2020, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, Q.; Hou, M.; Peng, J.; Yang, X.; Xu, S. Magnitude and Temporal Trend of the Chronic Myeloid Leukemia: On the Basis of the Global Burden of Disease Study 2019. JCO Glob. Oncol. 2021, 7, 1429–1441. [Google Scholar] [CrossRef]
- Osman, A.E.G.; Deininger, M.W. Chronic Myeloid Leukemia: Modern therapies, current challenges and future directions. Blood Rev. 2021, 49, 100825. [Google Scholar] [CrossRef]
- Kantarjian, H.; Kadia, T.; DiNardo, C.; Daver, N.; Borthakur, G.; Jabbour, E.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F. Acute myeloid leukemia: Current progress and future directions. Blood Cancer J. 2021, 11, 41. [Google Scholar] [CrossRef]
- Cruz-Miranda, G.M.; Hidalgo-Miranda, A.; Barcenas-Lopez, D.A.; Nunez-Enriquez, J.C.; Ramirez-Bello, J.; Mejia-Arangure, J.M.; Jimenez-Morales, S. Long Non-Coding RNA and Acute Leukemia. Int. J. Mol. Sci. 2019, 20, 735. [Google Scholar] [CrossRef]
- Ng, M.; Heckl, D.; Klusmann, J.H. The Regulatory Roles of Long Noncoding RNAs in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 570. [Google Scholar] [CrossRef]
- Wurm, A.A.; Pina, C. Long Non-coding RNAs as Functional and Structural Chromatin Modulators in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 899. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Wang, X.; Chen, K.; Guo, G.; Chen, J.L. Noncoding RNAs and their functional involvement in regulation of chronic myeloid leukemia. Brief. Funct. Genom. 2016, 15, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Gabra, M.M.; Salmena, L. microRNAs and Acute Myeloid Leukemia Chemoresistance: A Mechanistic Overview. Front. Oncol. 2017, 7, 255. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.L.; Licht, J.D. Targeting Epigenetics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.L.; Li, L.; Jia, Y.X.; Zhang, B.Z.; Li, J.C.; Zhu, Y.H.; Li, M.Q.; He, J.Z.; Zeng, T.T.; Ban, X.J.; et al. LINC01554-Mediated Glucose Metabolism Reprogramming Suppresses Tumorigenicity in Hepatocellular Carcinoma via Downregulating PKM2 Expression and Inhibiting Akt/mTOR Signaling Pathway. Theranostics 2019, 9, 796–810. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, Z.; Zhang, S.; Chen, Y.; Zhou, B.; Li, G.; Sun, Q.; Zhou, D.; Ge, Y.; Yan, S.; et al. Down-regulation of Long Non-coding RNA LINC01554 in Hepatocellular Cancer and its Clinical Significance. J. Cancer 2020, 11, 3369–3374. [Google Scholar] [CrossRef]
- Li, L.; Huang, K.; Lu, Z.; Zhao, H.; Li, H.; Ye, Q.; Peng, G. Bioinformatics analysis of LINC01554 and its coexpressed genes in hepatocellular carcinoma. Oncol. Rep. 2020, 44, 2185–2197. [Google Scholar] [CrossRef]
- Luo, T.; Jiang, Y.; Yang, J. Long Noncoding RNA LINC01554 as a Novel Biomarker for Diagnosis and Prognosis Prediction of Epithelial Ovarian Cancer. Dis. Markers 2021, 2021, 1244612. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, J.; Deng, R.; Lin, C.; Huang, Y.; Yang, X.; Wang, C.; Yang, M.; He, Y.; Lu, J.; et al. G3BP2 regulated by the lncRNA LINC01554 facilitates esophageal squamous cell carcinoma metastasis through stabilizing HDGF transcript. Oncogene 2022, 41, 515–526. [Google Scholar] [CrossRef]
- Poliseno, L. Pseudogenes: Newly discovered players in human cancer. Sci. Signal. 2012, 5, re5. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Zhai, L.L.; Yin, J.Y.; Vanessa, M.E.; Zhou, J.; Zhang, J.; Tang, X.; Lin, J.; Qian, J.; Deng, Z.Q. Pseudogene BMI1P1 expression as a novel predictor for acute myeloid leukemia development and prognosis. Oncotarget 2016, 7, 47376–47386. [Google Scholar] [CrossRef]
- Li, L.; Zhao, W. The mutual regulatory loop between TPTEP1 and miR-1303 in leukemogenesis of acute myeloid leukemia. Cancer Cell. Int. 2021, 21, 260. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.L.; Zhou, J.; Zhang, J.; Tang, X.; Zhou, L.Y.; Yin, J.Y.; Vanessa, M.D.; Peng, W.; Lin, J.; Deng, Z.Q. Down-regulation of pseudogene Vimentin 2p is associated with poor outcome in de novo acute myeloid leukemia. Cancer Biomark. 2017, 18, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.Y.; Yin, J.Y.; Tang, Q.; Zhai, L.L.; Zhang, T.J.; Wang, Y.X.; Yang, D.Q.; Qian, J.; Lin, J.; Deng, Z.Q. High expression of dual-specificity phosphatase 5 pseudogene 1 (DUSP5P1) is associated with poor prognosis in acute myeloid leukemia. Int. J. Clin. Exp. Pathol. 2015, 8, 16073–16080. [Google Scholar]
- Jaju, R.J.; Haas, O.A.; Neat, M.; Harbott, J.; Saha, V.; Boultwood, J.; Brown, J.M.; Pirc-Danoewinata, H.; Krings, B.W.; Muller, U.; et al. A new recurrent translocation, t(5;11)(q35;p15.5), associated with del(5q) in childhood acute myeloid leukemia. The UK Cancer Cytogenetics Group (UKCCG). Blood 1999, 94, 773–780. [Google Scholar] [PubMed]
- Levanon, D.; Glusman, G.; Bangsow, T.; Ben-Asher, E.; Male, D.A.; Avidan, N.; Bangsow, C.; Hattori, M.; Taylor, T.D.; Taudien, S.; et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene 2001, 262, 23–33. [Google Scholar] [CrossRef]
- Lettnin, A.P.; Wagner, E.F.; Carrett-Dias, M.; Dos Santos Machado, K.; Werhli, A.; Canedo, A.D.; Trindade, G.S.; de Souza Votto, A.P. Silencing the OCT4-PG1 pseudogene reduces OCT-4 protein levels and changes characteristics of the multidrug resistance phenotype in chronic myeloid leukemia. Mol. Biol. Rep. 2019, 46, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Graubert, T.A.; Shen, D.; Ding, L.; Okeyo-Owuor, T.; Lunn, C.L.; Shao, J.; Krysiak, K.; Harris, C.C.; Koboldt, D.C.; Larson, D.E.; et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2011, 44, 53–57. [Google Scholar] [CrossRef]
- Inoue, D.; Bradley, R.K.; Abdel-Wahab, O. Spliceosomal gene mutations in myelodysplasia: Molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016, 30, 989–1001. [Google Scholar] [CrossRef]
- Larsson, C.A.; Cote, G.; Quintas-Cardama, A. The changing mutational landscape of acute myeloid leukemia and myelodysplastic syndrome. Mol. Cancer Res. 2013, 11, 815–827. [Google Scholar] [CrossRef]
- Crews, L.A.; Balaian, L.; Delos Santos, N.P.; Leu, H.S.; Court, A.C.; Lazzari, E.; Sadarangani, A.; Zipeto, M.A.; La Clair, J.J.; Villa, R.; et al. RNA Splicing Modulation Selectively Impairs Leukemia Stem Cell Maintenance in Secondary Human AML. Cell Stem Cell 2016, 19, 599–612. [Google Scholar] [CrossRef]
- Morgan, G.J.; Smith, M.T. Metabolic enzyme polymorphisms and susceptibility to acute leukemia in adults. Am. J. Pharm. 2002, 2, 79–92. [Google Scholar] [CrossRef]
- Smith, M.T.; Wang, Y.; Kane, E.; Rollinson, S.; Wiemels, J.L.; Roman, E.; Roddam, P.; Cartwright, R.; Morgan, G. Low NAD(P)H:quinone oxidoreductase 1 activity is associated with increased risk of acute leukemia in adults. Blood 2001, 97, 1422–1426. [Google Scholar] [CrossRef]
- Palande, K.K.; Beekman, R.; van der Meeren, L.E.; Beverloo, H.B.; Valk, P.J.; Touw, I.P. The antioxidant protein peroxiredoxin 4 is epigenetically down regulated in acute promyelocytic leukemia. PLoS ONE 2011, 6, e16340. [Google Scholar] [CrossRef]
- Bauer, S.; Abdgawad, M.; Gunnarsson, L.; Segelmark, M.; Tapper, H.; Hellmark, T. Proteinase 3 and CD177 are expressed on the plasma membrane of the same subset of neutrophils. J. Leukoc. Biol. 2007, 81, 458–464. [Google Scholar] [CrossRef]
- Jerke, U.; Rolle, S.; Dittmar, G.; Bayat, B.; Santoso, S.; Sporbert, A.; Luft, F.; Kettritz, R. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J. Biol. Chem. 2011, 286, 7070–7081. [Google Scholar] [CrossRef]
- Zhou, G.; Yu, L.; Fang, L.; Yang, W.; Yu, T.; Miao, Y.; Chen, M.; Wu, K.; Chen, F.; Cong, Y.; et al. CD177(+) neutrophils as functionally activated neutrophils negatively regulate IBD. Gut 2018, 67, 1052–1063. [Google Scholar] [CrossRef]
- Marino, S.F.; Jerke, U.; Rolle, S.; Daumke, O.; Kettritz, R. Competitively disrupting the neutrophil-specific receptor-autoantigen CD177:proteinase 3 membrane complex reduces anti-PR3 antibody-induced neutrophil activation. J. Biol. Chem. 2022, 298, 101598. [Google Scholar] [CrossRef]
- Passamonti, F.; Pietra, D.; Malabarba, L.; Rumi, E.; Della Porta, M.G.; Malcovati, L.; Bonfichi, M.; Pascutto, C.; Lazzarino, M.; Cazzola, M. Clinical significance of neutrophil CD177 mRNA expression in Ph-negative chronic myeloproliferative disorders. Br. J. Haematol. 2004, 126, 650–656. [Google Scholar] [CrossRef]
- Yoon, H.S.; Kim, H.Y.; Cho, K.A.; Kim, Y.H.; Woo, S.Y.; Kim, H.S.; Kang, J.L.; Ryu, K.H.; Park, J.W. Procollagen C-Endopeptidase Enhancer 2 Secreted by Tonsil-Derived Mesenchymal Stem Cells Increases the Oxidative Burst of Promyelocytic HL-60 Cells. Biology 2022, 11, 255. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Li, H.; Guan, F.; Li, X. Multiple Roles of Glycans in Hematological Malignancies. Front. Oncol. 2018, 8, 364. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Rangel, L.; Benitez-Guerrero, T.; Martinez-Vieyra, I.; Cisneros, B.; Martinez-Tovar, A.; Winder, S.J.; Cerecedo, D. A role for dystroglycan in the pathophysiology of acute leukemic cells. Life Sci. 2017, 182, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, B.; Verbavatz, J.M.; Jackson, C.L. GBF1 and Arf1 function in vesicular trafficking, lipid homoeostasis and organelle dynamics. Biol. Cell 2017, 109, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Caviston, J.P.; Holzbaur, E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell. Biol. 2009, 19, 147–155. [Google Scholar] [CrossRef]
- Chang, J.; Lee, S.; Blackstone, C. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J. Clin. Invest. 2014, 124, 5249–5262. [Google Scholar] [CrossRef]
- Caivano, A.; Laurenzana, I.; De Luca, L.; La Rocca, F.; Simeon, V.; Trino, S.; D’Auria, F.; Traficante, A.; Maietti, M.; Izzo, T.; et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumour Biol. 2015, 36, 9739–9752. [Google Scholar] [CrossRef]
- Tzoran, I.; Rebibo-Sabbah, A.; Brenner, B.; Aharon, A. Disease dynamics in patients with acute myeloid leukemia: New biomarkers. Exp. Hematol. 2015, 43, 936–943. [Google Scholar] [CrossRef]
- Hong, C.S.; Muller, L.; Whiteside, T.L.; Boyiadzis, M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front. Immunol. 2014, 5, 160. [Google Scholar] [CrossRef]
- Li, M.Y.; Zhao, C.; Chen, L.; Yao, F.Y.; Zhong, F.M.; Chen, Y.; Xu, S.; Jiang, J.Y.; Yang, Y.L.; Min, Q.H.; et al. Quantitative Proteomic Analysis of Plasma Exosomes to Identify the Candidate Biomarker of Imatinib Resistance in Chronic Myeloid Leukemia Patients. Front. Oncol. 2021, 11, 779567. [Google Scholar] [CrossRef]
- Kang, K.W.; Jung, J.H.; Hur, W.; Park, J.; Shin, H.; Choi, B.; Jeong, H.; Kim, D.S.; Yu, E.S.; Lee, S.R.; et al. The Potential of Exosomes Derived from Chronic Myelogenous Leukaemia Cells as a Biomarker. Anticancer Res. 2018, 38, 3935–3942. [Google Scholar] [CrossRef] [PubMed]
- Pando, A.; Reagan, J.L.; Quesenberry, P.; Fast, L.D. Extracellular vesicles in leukemia. Leuk. Res. 2018, 64, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Nehrbas, J.; Butler, J.T.; Chen, D.W.; Kurre, P. Extracellular Vesicles and Chemotherapy Resistance in the AML Microenvironment. Front. Oncol. 2020, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.H.; Sharifi, L.M.A.; Kakhharov, A.J.; Opulencia, M.J.C.; Alsaikhan, F.; Bokov, D.O.; Majdi, H.S.; Jawad, M.A.; Hammid, A.T.; Shalaby, M.N.; et al. Role of Acute Myeloid Leukemia (AML)-Derived exosomes in tumor progression and survival. Biomed. Pharmacother. 2022, 150, 113009. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Niazi, V.; Taheri, M. Contribution of extracellular vesicles in normal hematopoiesis and hematological malignancies. Heliyon 2021, 7, e06030. [Google Scholar] [CrossRef]
- Gargiulo, E.; Paggetti, J.; Moussay, E. Hematological Malignancy-Derived Small Extracellular Vesicles and Tumor Microenvironment: The Art of Turning Foes into Friends. Cells 2019, 8, 511. [Google Scholar] [CrossRef]
- Hedlund, M.; Nagaeva, O.; Kargl, D.; Baranov, V.; Mincheva-Nilsson, L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B cells. PLoS ONE 2011, 6, e16899. [Google Scholar] [CrossRef]
- Szajnik, M.; Czystowska, M.; Szczepanski, M.J.; Mandapathil, M.; Whiteside, T.L. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS ONE 2010, 5, e11469. [Google Scholar] [CrossRef]
- Viola, S.; Traer, E.; Huan, J.; Hornick, N.I.; Tyner, J.W.; Agarwal, A.; Loriaux, M.; Johnstone, B.; Kurre, P. Alterations in acute myeloid leukaemia bone marrow stromal cell exosome content coincide with gains in tyrosine kinase inhibitor resistance. Br. J. Haematol. 2016, 172, 983–986. [Google Scholar] [CrossRef]
- Corrado, C.; Saieva, L.; Raimondo, S.; Santoro, A.; De Leo, G.; Alessandro, R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. J. Cell. Mol. Med. 2016, 20, 1829–1839. [Google Scholar] [CrossRef]
- Wang, J.; Yu, M.; Guo, Q.; Ma, Q.; Hu, C.; Ma, Z.; Yin, X.; Li, X.; Wang, Y.; Pan, H.; et al. Prognostic significance of huntingtin interacting protein 1 expression on patients with acute myeloid leukemia. Sci. Rep. 2017, 7, 45960. [Google Scholar] [CrossRef]
- Kuckleburg, C.J.; Tilkens, S.B.; Santoso, S.; Newman, P.J. Proteinase 3 contributes to transendothelial migration of NB1-positive neutrophils. J. Immunol. 2012, 188, 2419–2426. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Grieshaber-Bouyer, R.; Wang, J.; Schmider, A.B.; Wilson, Z.S.; Zeng, L.; Halyabar, O.; Godin, M.D.; Nguyen, H.N.; Levescot, A.; et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood 2017, 130, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Cassatella, M.A.; Costantini, C.; Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011, 11, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Liew, P.X.; Kubes, P. The Neutrophil’s Role During Health and Disease. Physiol. Rev. 2019, 99, 1223–1248. [Google Scholar] [CrossRef]
- Jones, H.R.; Robb, C.T.; Perretti, M.; Rossi, A.G. The role of neutrophils in inflammation resolution. Semin. Immunol. 2016, 28, 137–145. [Google Scholar] [CrossRef]
- Rawat, K.; Syeda, S.; Shrivastava, A. Neutrophil-derived granule cargoes: Paving the way for tumor growth and progression. Cancer Metastasis Rev. 2021, 40, 221–244. [Google Scholar] [CrossRef]
- Bayat, B.; Werth, S.; Sachs, U.J.; Newman, D.K.; Newman, P.J.; Santoso, S. Neutrophil transmigration mediated by the neutrophil-specific antigen CD177 is influenced by the endothelial S536N dimorphism of platelet endothelial cell adhesion molecule-1. J. Immunol. 2010, 184, 3889–3896. [Google Scholar] [CrossRef]
- Yurchenko, V.; O’Connor, M.; Dai, W.W.; Guo, H.; Toole, B.; Sherry, B.; Bukrinsky, M. CD147 is a signaling receptor for cyclophilin B. Biochem. Biophys. Res. Commun. 2001, 288, 786–788. [Google Scholar] [CrossRef]
- Guo, L.L.; He, Z.C.; Yang, C.Q.; Qiao, P.T.; Yin, G.L. Epigenetic silencing of olfactomedin-4 enhances gastric cancer cell invasion via activation of focal adhesion kinase signaling. BMB Rep. 2015, 48, 630–635. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.; Xu, P. miR-590 accelerates lung adenocarcinoma migration and invasion through directly suppressing functional target OLFM4. Biomed. Pharmacother. 2017, 86, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Lei, X.; Zhang, L.; Fu, J. miR-103 regulates triple negative breast cancer cells migration and invasion through targeting olfactomedin 4. Biomed. Pharmacother. 2017, 89, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, H.; Wierusz-Wysocka, B.; Siekierka, H.; Szczepanik, A.; Klimas, R.; Wykretowicz, A. Polymorphonuclear neutrophils function in untreated patients with chronic myeloid leukemia. Oncology 1988, 45, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Wolach, B.; Gavrieli, R.; Manor, Y.; Lishner, M. Leukocyte function in chronic myeloproliferative disorders. Blood Cells. Mol. Dis. 1998, 24, 544–551. [Google Scholar] [CrossRef]
- Miyauchi, J.; Watanabe, Y.; Enomoto, Y.; Takeuchi, K. Lactoferrin-deficient neutrophil polymorphonuclear leucocytes in leukaemias: A semiquantitative and ultrastructural cytochemical study. J. Clin. Pathol. 1983, 36, 1397–1405. [Google Scholar] [CrossRef]
- Jenssen, H.; Hancock, R.E. Antimicrobial properties of lactoferrin. Biochimie 2009, 91, 19–29. [Google Scholar] [CrossRef]
- Bagheri, V. S100A12: Friend or foe in pulmonary tuberculosis? Cytokine 2017, 92, 80–82. [Google Scholar] [CrossRef]
- Kozlyuk, N.; Monteith, A.J.; Garcia, V.; Damo, S.M.; Skaar, E.P.; Chazin, W.J. S100 Proteins in the Innate Immune Response to Pathogens. Methods Mol. Biol. 2019, 1929, 275–290. [Google Scholar] [CrossRef]
- Rath, M.; Muller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37--A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta 2016, 1858, 546–566. [Google Scholar] [CrossRef]
- Guryanova, S.V.; Ovchinnikova, T.V. Immunomodulatory and Allergenic Properties of Antimicrobial Peptides. Int. J. Mol. Sci. 2022, 23, 2499. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Gupta, D. Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun. 2010, 16, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Giallongo, C.; Parrinello, N.; Tibullo, D.; La Cava, P.; Romano, A.; Chiarenza, A.; Barbagallo, I.; Palumbo, G.A.; Stagno, F.; Vigneri, P.; et al. Myeloid derived suppressor cells (MDSCs) are increased and exert immunosuppressive activity together with polymorphonuclear leukocytes (PMNs) in chronic myeloid leukemia patients. PLoS ONE 2014, 9, e101848. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Gui, X.; Kim, J.; Xie, L.; Chen, W.; Li, Z.; He, L.; Chen, Y.; Chen, H.; Luo, W.; et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 2018, 562, 605–609. [Google Scholar] [CrossRef]
- Takahashi, H.; Hatta, Y.; Iriyama, N.; Hasegawa, Y.; Uchida, H.; Nakagawa, M.; Makishima, M.; Takeuchi, J.; Takei, M. Induced differentiation of human myeloid leukemia cells into M2 macrophages by combined treatment with retinoic acid and 1alpha,25-dihydroxyvitamin D3. PLoS ONE 2014, 9, e113722. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.R.; Lee, J.H.; Hensel, J.A.; Sawant, A.B.; Davis, B.H.; Lee, C.M.; Deshane, J.S.; Ponnazhagan, S. Prostate cancer-derived cathelicidin-related antimicrobial peptide facilitates macrophage differentiation and polarization of immature myeloid progenitors to protumorigenic macrophages. Prostate 2016, 76, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zou, X.; Qi, G.; Tang, Y.; Guo, Y.; Si, J.; Liang, L. Roles and Mechanisms of Human Cathelicidin LL-37 in Cancer. Cell Physiol. Biochem. 2018, 47, 1060–1073. [Google Scholar] [CrossRef]
- Sharapova, T.N.; Ivanova, O.K.; Soshnikova, N.V.; Romanova, E.A.; Sashchenko, L.P.; Yashin, D.V. Innate Immunity Protein Tag7 Induces 3 Distinct Populations of Cytotoxic Cells That Use Different Mechanisms to Exhibit Their Antitumor Activity on Human Leukocyte Antigen-Deficient Cancer Cells. J. Innate Immun. 2017, 9, 598–608. [Google Scholar] [CrossRef]
- Mussai, F.; Egan, S.; Higginbotham-Jones, J.; Perry, T.; Beggs, A.; Odintsova, E.; Loke, J.; Pratt, G.; U, K.P.; Lo, A.; et al. Arginine dependence of acute myeloid leukemia blast proliferation: A novel therapeutic target. Blood 2015, 125, 2386–2396. [Google Scholar] [CrossRef]
- Azzaoui, I.; Uhel, F.; Rossille, D.; Pangault, C.; Dulong, J.; Le Priol, J.; Lamy, T.; Houot, R.; Le Gouill, S.; Cartron, G.; et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood 2016, 128, 1081–1092. [Google Scholar] [CrossRef]
- Yin, C.M.; Wong, J.H.; Xia, J.; Ng, T.B. Studies on anticancer activities of lactoferrin and lactoferricin. Curr. Protein Pept. Sci. 2013, 14, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Sashchenko, L.P.; Dukhanina, E.A.; Yashin, D.V.; Shatalov, Y.V.; Romanova, E.A.; Korobko, E.V.; Demin, A.V.; Lukyanova, T.I.; Kabanova, O.D.; Khaidukov, S.V.; et al. Peptidoglycan recognition protein tag7 forms a cytotoxic complex with heat shock protein 70 in solution and in lymphocytes. J. Biol. Chem. 2004, 279, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Dukhanina, E.A.; Kabanova, O.D.; Lukyanova, T.I.; Shatalov, Y.V.; Yashin, D.V.; Romanova, E.A.; Gnuchev, N.V.; Galkin, A.V.; Georgiev, G.P.; Sashchenko, L.P. Opposite roles of metastasin (S100A4) in two potentially tumoricidal mechanisms involving human lymphocyte protein Tag7 and Hsp70. Proc. Natl. Acad. Sci. USA 2009, 106, 13963–13967. [Google Scholar] [CrossRef] [PubMed]

| Accession Number | Phenotype | Number of Unique Blood Samples Analyzed | Study Description |
|---|---|---|---|
| GSE52656 | AML | 22 | Leucogene; mutations and gene expression in AML [5,6] |
| GSE108003 | AML | 12 | gene expression markers for drug sensitivity in AML [7] |
| GSE108266 | AML | 6 | chromatin landscape and gene expression in different AML subtypes [8] |
| GSE147524 | AML | 16 | tumor-specific antigens in AML [9] |
| GSE100026 | CML | 10 | gene expression in CML [10] |
| GSE100026 | Healthy | 5 | gene expression in CML [10] |
| Symbol | Gene Name | q-Value |
|---|---|---|
| PTMAP12 | prothymosin alpha pseudogene 12 | 9.20 × 10−17 |
| LOC644936 | actin beta pseudogene | 4.07 × 10−14 |
| RPS27AP20 | RPS27A pseudogene 20 | 2.64 × 10−13 |
| FAM133CP | family with sequence similarity 133, member A pseudogene | 3.83 × 10−13 |
| LINC01554 | long intergenic non-protein coding RNA 1554 | 2.53 × 10−12 |
| Go Annotation | Associated Genes * | p-Value | Specificity |
|---|---|---|---|
| Spliceosomal complex | CWC22, IK, ISY1, LSM6, PNN, PRPF3, PRPF38A, PRPF8, RBMX2, RNF113A, SLU7, SYF2, ZCRB1, ZRSR2 | 8.33 × 10−9 | CML |
| U2-type spliceosomal complex | CWC22, IK, ISY1, LSM6, PRPF3, PRPF38A, PRPF8, RBMX2, RNF113A, SYF2 | 3.29 × 10−7 | CML |
| U2-type precatalytic spliceosome | CWC22, IK, LSM6, PRPF3, PRPF38A, PRPF8, RBMX2, RNF113A | 1.45 × 10−6 | CML |
| mRNA processing | AKAP8L, CWC22, IK, ISY1, LSM6, PRPF3, PRPF38A, PRPF8, RBMX2, RNF113A, SLU7, SYF2, ZRSR2 | 3.57 × 10−5 | CML |
| Specific granule | CAMP, CRISP3, LTF, OLFM4 | 2.33 × 10−4 | CML |
| Catalytic step 2 spliceosome | CWC22, ISY1, PNN, PRPF8, SLU7, SYF2 | 3.92 × 10−3 | CML |
| Secretory granule | CA4, CAMP, CD177, CRISP3, LTF, OLFM4 | 4.55 × 10−3 | CML |
| Antimicrobial humoral immune response mediated by antimicrobial peptide | CAMP, LTF, PGLYRP1, S100A12 | 1.34 × 10−2 | CML |
| Reactive oxygen species metabolic process | AATF, CD177, GBF1, HP, HPR, PRG3 | 1.39 × 10−2 | CML |
| Glycoprotein metabolic process | ALG8, CHST14, EXT2, POMGNT1, POMT2, TMEM258, TMEM59 | 1.44 × 10−2 | AML |
| Superoxide anion generation | AATF, CD177, PRG3 | 1.53 × 10−2 | CML |
| Neutrophil migration | CD177, GBF1, OLFM4, PPIB | 1.57 × 10−2 | CML |
| Defense response to fungus | ARG1, LTF, S100A12 | 1.58 × 10−2 | CML |
| Regulation of reactive oxygen species metabolic process | AATF, CD177, HP, HPR | 1.65 × 10−2 | CML |
| Establishment of vesicle localization | GBF1, HTT, SPG11 | 1.99 × 10−2 | AML |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jurgec, S.; Jezernik, G.; Gorenjak, M.; Büdefeld, T.; Potočnik, U. Meta-Analytic Comparison of Global RNA Transcriptomes of Acute and Chronic Myeloid Leukemia Cells Reveals Novel Gene Candidates Governing Myeloid Malignancies. Cancers 2022, 14, 4681. https://doi.org/10.3390/cancers14194681
Jurgec S, Jezernik G, Gorenjak M, Büdefeld T, Potočnik U. Meta-Analytic Comparison of Global RNA Transcriptomes of Acute and Chronic Myeloid Leukemia Cells Reveals Novel Gene Candidates Governing Myeloid Malignancies. Cancers. 2022; 14(19):4681. https://doi.org/10.3390/cancers14194681
Chicago/Turabian StyleJurgec, Staša, Gregor Jezernik, Mario Gorenjak, Tomaž Büdefeld, and Uroš Potočnik. 2022. "Meta-Analytic Comparison of Global RNA Transcriptomes of Acute and Chronic Myeloid Leukemia Cells Reveals Novel Gene Candidates Governing Myeloid Malignancies" Cancers 14, no. 19: 4681. https://doi.org/10.3390/cancers14194681
APA StyleJurgec, S., Jezernik, G., Gorenjak, M., Büdefeld, T., & Potočnik, U. (2022). Meta-Analytic Comparison of Global RNA Transcriptomes of Acute and Chronic Myeloid Leukemia Cells Reveals Novel Gene Candidates Governing Myeloid Malignancies. Cancers, 14(19), 4681. https://doi.org/10.3390/cancers14194681







