Disulfiram/Copper Induces Immunogenic Cell Death and Enhances CD47 Blockade in Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Reagents
2.3. Cell Viability Assay
2.4. Colony Formation Assays
2.5. Determination of Apoptosis, Surface CRT, ATP and HMGB1 Release
2.6. Immunoblotting
2.7. Immunofluorescence
2.8. Tumor Model and Treatment
2.9. Dendritic Cell Activation and Phagocytosis Assays
2.10. Flow Cytometry Staining
2.11. siRNA
2.12. RNA Extraction, RNA-seq and Data Analysis
2.13. Statistics
3. Results
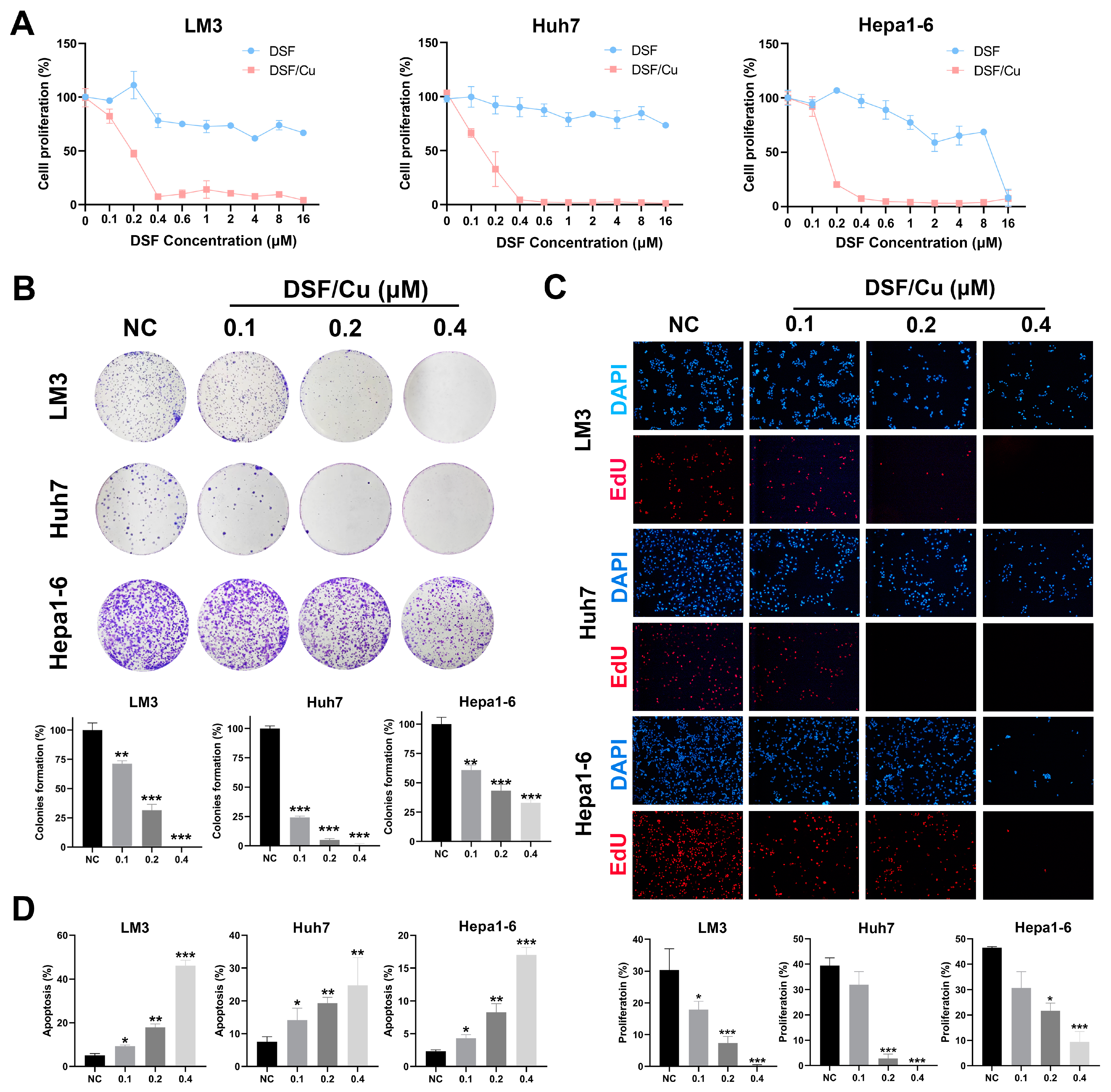
3.1. The Combination of DSF/Cu Inhibits Cell Proliferation and Induces Apoptosis of HCC Cells
3.2. DSF/Cu Induces ICD in HCC Cells and Enhances DC Activation
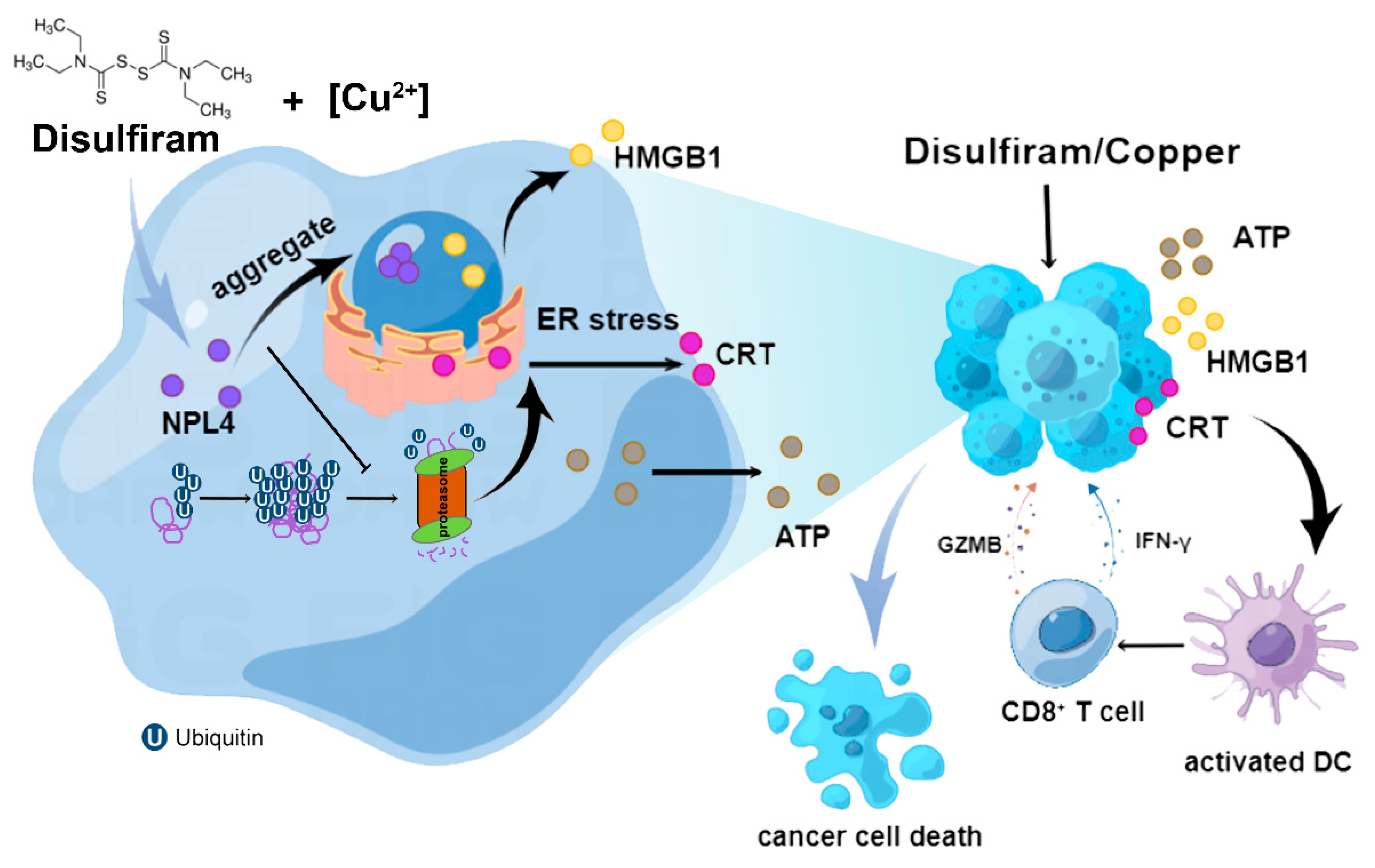
3.3. DSF/Cu Induces ER Stress through a Buildup of Poly-Ubiquitylated Proteins Mediated by NPL4 Aggregation in Cell-Nucleus
3.4. Inhibition of NPL4 Induces ICD in Hepatocellular Carcinoma
3.5. DSF/Cu induced Immune Memory and DC Activation In Vivo
3.6. Combination of DSF/Cu and Anti-CD47 Therapy Exhibited Great Antitumor Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yarchoan, M.; Xing, D.; Luan, L.; Xu, H.; Sharma, R.B.; Popovic, A.; Pawlik, T.M.; Kim, A.K.; Zhu, Q.; Jaffee, E.M.; et al. Characterization of the Immune Microenvironment in Hepatocellular Carcinoma. Clin. Cancer Res. 2017, 23, 7333–7339. [Google Scholar] [CrossRef]
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front. Immunol. 2019, 10, 168. [Google Scholar] [CrossRef]
- Crispe, I.N. Immune Tolerance in Liver Disease. Hepatology 2014, 60, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ramadori, P.; Pfister, D.; Seehawer, M.; Zender, L.; Heikenwalder, M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer 2021, 21, 541–557. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galassi, C.; Zitvogel, L.; Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol. 2022, 23, 487–500. [Google Scholar] [CrossRef]
- Aaes, T.L.; Vandenabeele, P. The intrinsic immunogenic properties of cancer cell lines, immunogenic cell death, and how these influence host antitumor immune responses. Cell Death Differ. 2021, 28, 843–860. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017, 17, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef]
- Ahmed, A.; Tait, S.W.G. Targeting Immunogenic Cell Death in Cancer. Mol. Oncol. 2020, 14, 2994–3006. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Chen, Y.; Wang, H.; Hua, Y.; Cai, Z. Immunogenic cell death in cancer therapy: Present and emerging inducers. J. Cell Mol. Med. 2019, 23, 4854–4865. [Google Scholar] [CrossRef]
- Yu, Z.; Guo, J.; Hu, M.; Gao, Y.; Huang, L. Icaritin Exacerbates Mitophagy and Synergizes with Doxorubicin to Induce Immunogenic Cell Death in Hepatocellular Carcinoma. ACS Nano 2020, 14, 4816–4828. [Google Scholar] [CrossRef]
- Zhu, H.; Shan, Y.; Ge, K.; Lu, J.; Kong, W.; Jia, C. Oxaliplatin induces immunogenic cell death in hepatocellular carcinoma cells and synergizes with immune checkpoint blockade therapy. Cell Oncol. 2020, 43, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Kepp, O.; Zitvogel, L.; Kroemer, G. Clinical evidence that immunogenic cell death sensitizes to PD-1/PD-L1 blockade. Oncoimmunology 2019, 8, e1637188. [Google Scholar] [CrossRef]
- D’Amico, L.; Menzel, U.; Prummer, M.; Müller, P.; Buchi, M.; Kashyap, A.; Haessler, U.; Yermanos, A.; Gébleux, R.; Briendl, M.; et al. A novel anti-HER2 anthracycline-based antibody-drug conjugate induces adaptive anti-tumor immunity and potentiates PD-1 blockade in breast cancer. J. Immunother. Cancer 2019, 7, 16. [Google Scholar] [CrossRef]
- Lu, C.; Li, X.; Ren, Y.; Zhang, X. Disulfiram: A novel repurposed drug for cancer therapy. Cancer Chemother. Pharmacol. 2021, 87, 159–172. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Wu, C.; Wang, L.; Chen, Z.-S.; Cui, W. The combination of disulfiram and copper for cancer treatment. Drug Discov. Today. 2020, 25, 1099–1108. [Google Scholar] [CrossRef]
- Skrott, Z.; Mistrik, M.; Andersen, K.K.; Friis, S.; Majera, D.; Gursky, J.; Oždian, T.; Bartkova, J.; Turi, Z.; Moudry, P.; et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 2017, 552, 194–199. [Google Scholar] [CrossRef]
- Skrott, Z.; Majera, D.; Gursky, J.; Buchtova, T.; Hajduch, M.; Mistrik, M.; Bartek, J. Disulfiram’s anti-cancer activity reflects targeting NPL4, not inhibition of aldehyde dehydrogenase. Oncogene 2019, 38, 6711–6722. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Yang, W.; Toprani, S.M.; Guo, W.; He, L.; DeLeo, A.B.; Ferrone, S.; Zhang, G.; Wang, E.; Lin, Z.; et al. Induction of immunogenic cell death in radiation-resistant breast cancer stem cells by repurposing anti-alcoholism drug disulfiram. Cell Commun. Signal. 2020, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- You, S.-Y.; Rui, W.; Chen, S.-T.; Chen, H.-C.; Liu, X.-W.; Huang, J. Process of immunogenic cell death caused by disulfiram as the anti-colorectal cancer candidate. Biochem. Biophys. Res. Commun. 2019, 513, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Sagar, V.; Vatapalli, R.; Lysy, B.; Pamarthy, S.; Anker, J.; Rodriguez, Y.; Han, H.; Unno, K.; Stadler, W.M.; Catalona, W.J.; et al. EPHB4 inhibition activates ER stress to promote immunogenic cell death of prostate cancer cells. Cell Death Dis. 2019, 10, 801. [Google Scholar] [CrossRef]
- Humeau, J.; Bezu, L.; Kepp, O.; Kroemer, G. EIF2α phosphorylation: A hallmark of both autophagy and immunogenic cell death. Mol. Cell Oncol. 2020, 7, 1776570. [Google Scholar] [CrossRef]
- Bouwstra, R.; van Meerten, T.; Bremer, E. CD47-SIRPα blocking-based immunotherapy: Current and prospective therapeutic strategies. Clin. Transl. Med. 2022, 12, e943. [Google Scholar] [CrossRef]
- Chao, M.P.; Jaiswal, S.; Weissman-Tsukamoto, R.; Alizadeh, A.A.; Gentles, A.J.; Volkmer, J.; Weiskopf, K.; Willingham, S.B.; Raveh, T.; Park, C.Y.; et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci. Transl. Med. 2010, 2, 63ra94. [Google Scholar] [CrossRef]
- de Oca, R.M.; Alavi, A.S.; Vitali, N.; Bhattacharya, S.; Blackwell, C.; Patel, K.; Seestaller-Wehr, L.; Kaczynski, H.; Shi, H.; Dobrzynski, E.; et al. Belantamab Mafodotin (GSK2857916) Drives Immunogenic Cell Death and Immune-mediated Antitumor Responses In Vivo. Mol. Cancer Ther. 2021, 20, 1941–1955. [Google Scholar] [CrossRef]
- Kepp, O.; Zitvogel, L.; Kroemer, G. Lurbinectedin: An FDA-approved inducer of immunogenic cell death for the treatment of small-cell lung cancer. Oncoimmunology 2020, 9, 1795995. [Google Scholar] [CrossRef] [PubMed]
- Costantini, S.; Capone, F.; Polo, A.; Bagnara, P.; Budillon, A. Valosin-Containing Protein (VCP)/p97: A Prognostic Biomarker and Therapeutic Target in Cancer. Int. J. Mol. Sci. 2021, 22, 10177. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Ruggiano, A.; Carvalho, P.; Rapoport, T.A. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 2014, 158, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.-S.; Yin, Y.-W.; Zhang, Y.-P.; Guo, P.-Y.; Li, W.; Liu, K.-L. Upregulation of NPL4 promotes bladder cancer cell proliferation by inhibiting DXO destabilization of cyclin D1 mRNA. Cancer Cell Int. 2019, 19, 149. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Yamada, Y.; Enokida, H.; Osako, Y.; Tsuruda, M.; Kuroshima, K.; Sakaguchi, T.; Sugita, S.; Tatarano, S.; Nakagawa, M. Targeting NPL4 via drug repositioning using disulfiram for the treatment of clear cell renal cell carcinoma. PLoS ONE 2020, 15, e0236119. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Nie, D.; Huang, Y.; Yu, X.; Chen, Z.; Zhong, M.; Liu, X.; Wang, X.; Sui, S.; Liu, Z.; et al. Anticancer effects of disulfiram in T-cell malignancies through NPL4-mediated ubiquitin-proteasome pathway. J. Leukoc. Biol. 2022. [Google Scholar] [CrossRef]
- Jiang, Z.; Sun, H.; Yu, J.; Tian, W.; Song, Y. Targeting CD47 for cancer immunotherapy. J. Hematol. Oncol. 2021, 14, 180. [Google Scholar] [CrossRef]
- Zhou, F.; Feng, B.; Yu, H.; Wang, D.; Wang, T.; Ma, Y.; Wang, S.; Li, Y. Tumor Microenvironment-Activatable Prodrug Vesicles for Nanoenabled Cancer Chemoimmunotherapy Combining Immunogenic Cell Death Induction and CD47 Blockade. Adv. Mater. 2019, 31, e1805888. [Google Scholar] [CrossRef]
- Greten, T.F.; Lai, C.W.; Li, G.; Staveley-O’Carroll, K.F. Targeted and Immune-Based Therapies for Hepatocellular Carcinoma. Gastroenterology 2019, 156, 510–524. [Google Scholar] [CrossRef]
- Rizzo, A.; Nannini, M.; Novelli, M.; Ricci, A.D.; Di Scioscio, V.; Pantaleo, M.A. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: A systematic review and meta-analysis. Ther. Adv. Med. Oncol. 2020, 12, 1758835920936932. [Google Scholar] [CrossRef]
- Yau, T.; Park, J.-W.; Finn, R.S.; Cheng, A.-L.; Mathurin, P.; Edeline, J.; Kudo, M.; Harding, J.J.; Merle, P.; Rosmorduc, O.; et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): A randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 2022, 23, 77–90. [Google Scholar] [CrossRef]
- Rizzo, A.; Ricci, A.D.; Gadaleta-Caldarola, G.; Brandi, G. First-line immune checkpoint inhibitor-based combinations in unresectable hepatocellular carcinoma: Current management and future challenges. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 1245–1251. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. IMbrave150 Investigators. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Ikeda, M.; Zhu, A.X.; Sung, M.W.; Baron, A.D.; Kudo, M.; Okusaka, T.; Kobayashi, M.; Kumada, H.; Kaneko, S.; et al. Phase Ib Study of Lenvatinib Plus Pembrolizumab in Patients With Unresectable Hepatocellular Carcinoma. J. Clin. Oncol. 2020, 38, 2960–2970. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Huang, H.; Pan, C.; Mei, Z.; Yin, S.; Zhou, L.; Zheng, S. Disulfiram/Copper Induces Immunogenic Cell Death and Enhances CD47 Blockade in Hepatocellular Carcinoma. Cancers 2022, 14, 4715. https://doi.org/10.3390/cancers14194715
Gao X, Huang H, Pan C, Mei Z, Yin S, Zhou L, Zheng S. Disulfiram/Copper Induces Immunogenic Cell Death and Enhances CD47 Blockade in Hepatocellular Carcinoma. Cancers. 2022; 14(19):4715. https://doi.org/10.3390/cancers14194715
Chicago/Turabian StyleGao, Xingxing, Hechen Huang, Caixu Pan, Zhibin Mei, Shengyong Yin, Lin Zhou, and Shusen Zheng. 2022. "Disulfiram/Copper Induces Immunogenic Cell Death and Enhances CD47 Blockade in Hepatocellular Carcinoma" Cancers 14, no. 19: 4715. https://doi.org/10.3390/cancers14194715
APA StyleGao, X., Huang, H., Pan, C., Mei, Z., Yin, S., Zhou, L., & Zheng, S. (2022). Disulfiram/Copper Induces Immunogenic Cell Death and Enhances CD47 Blockade in Hepatocellular Carcinoma. Cancers, 14(19), 4715. https://doi.org/10.3390/cancers14194715








