Full Blood Count Trends for Colorectal Cancer Detection in Primary Care: Development and Validation of a Dynamic Prediction Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Data
2.2. Study Design
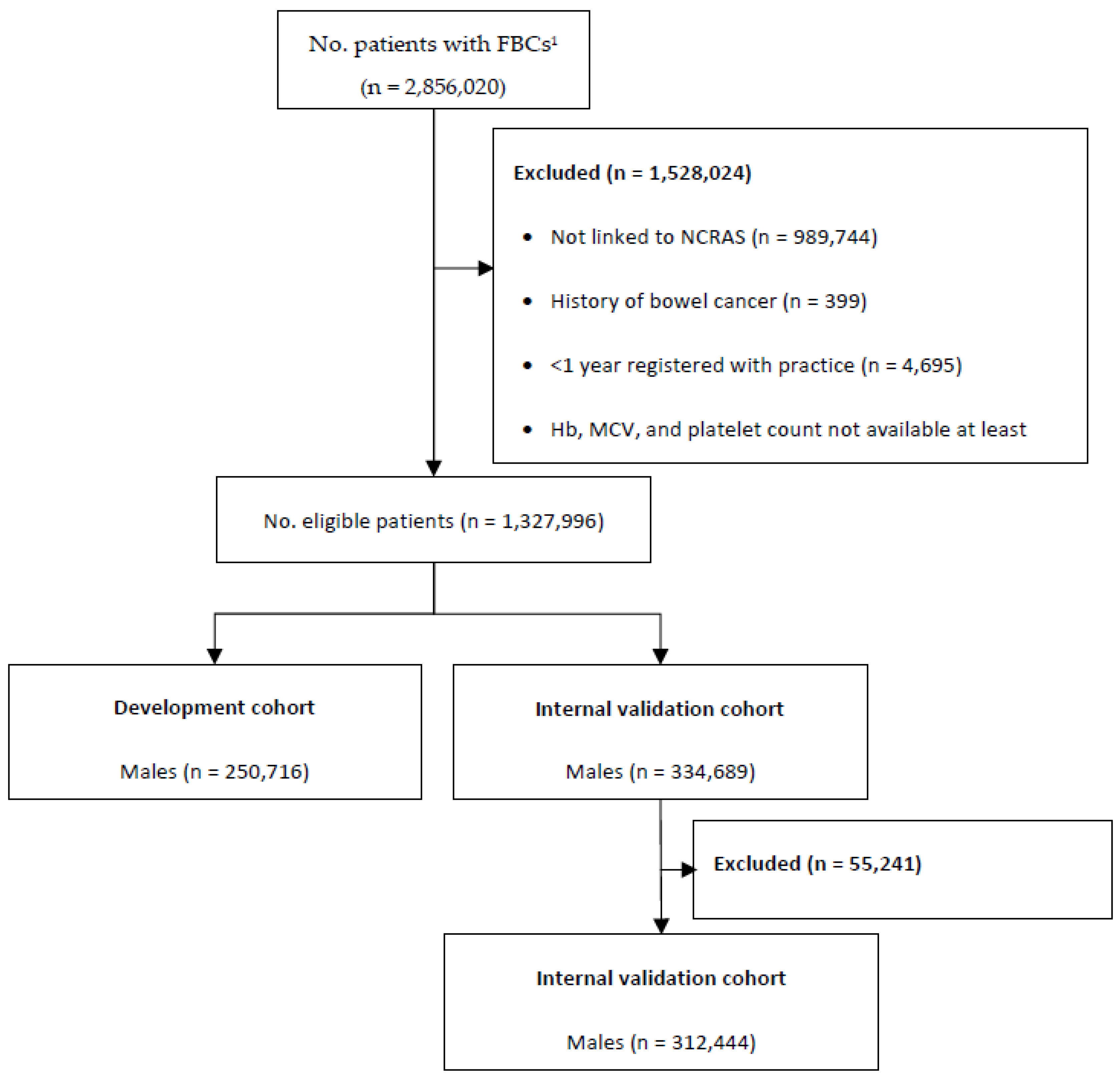
2.3. Participants
2.4. Outcome
2.5. Predictors
2.6. Missing Data
2.7. Sample Size
2.8. Model Development
2.9. Model Validation
2.10. Model Performance
2.11. Diagnostic Accuracy and Risk Thresholds
3. Results
3.1. Summary of Patient Data
3.2. Model Development
3.3. Model Performance
3.4. Performance in Subgroups (Validation Cohort)
3.5. Diagnostic Accuracy and Risk Thresholds (Validation Cohort)
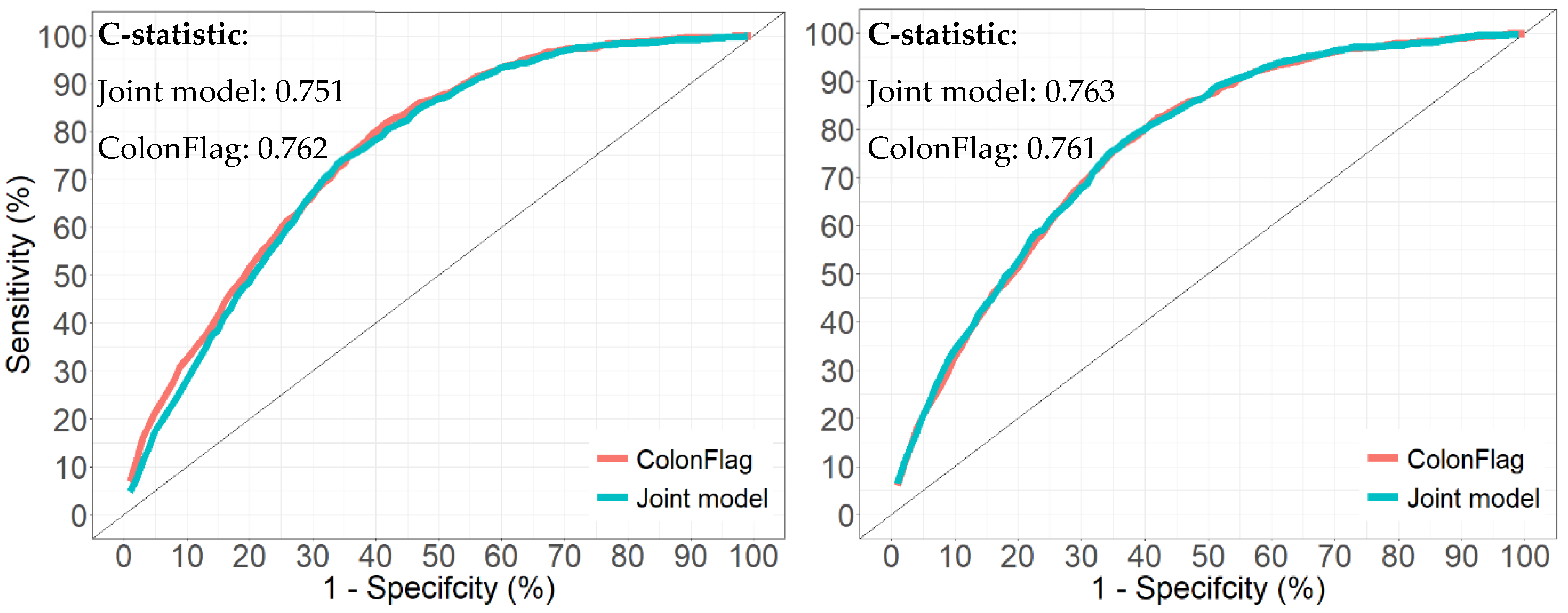
3.6. Comparison to the ColonFlag (Validation Cohort)
4. Discussion
4.1. Summary of Main Findings
4.2. Comparison with Existing Literature
4.3. Implications for Practice
4.4. Strengths and Limitations
4.4.1. Strengths
4.4.2. Limitations
5. Conclusions and Further Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Research UK. Bowel Cancer Incidence Statistics. 2020. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer#heading-Zero (accessed on 13 April 2022).
- Cancer Research UK. Bowel Cancer Mortality Statistics. 2021. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer#heading-One (accessed on 13 April 2022).
- Cancer Research UK. Bowel Cancer Survival by Stage at Diagnosis. 2019. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer/survival#heading-Three (accessed on 13 April 2022).
- Hewitson, P.; Glasziou, P.; Watson, E.; Towler, B.; Irwig, L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): An update. Am. J. Gastroenterol. 2008, 103, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Holt, T.; Birks, J.; Bankhead, C.; Nicholson, B.D.; Fuller, A.; Patnick, J. Do Changes in Full Blood Count Indices Predate Symptom Reporting in People with Undiagnosed Bowel Cancer? Retrospective Analysis Using Cohort and Case Control Designs. 2021. Available online: https://sapc.ac.uk/conference/2021/abstract/do-changes-full-blood-count-indices-predate-symptom-reporting-people (accessed on 13 April 2022).
- Hippisley-Cox, J.; Coupland, C. Identifying patients with suspected colorectal cancer in primary care: Derivation and validation of an algorithm. Br. J. Gen. Pract. 2012, 62, e29–e37. [Google Scholar] [CrossRef]
- MayoClinic. Complete Blood Count (CBC). 2020. Available online: https://www.mayoclinic.org/tests-procedures/complete-blood-count/about/pac-20384919 (accessed on 13 April 2022).
- Virdee, P.S.; Patnick, J.; Watkinson, P.; Birks, J.; Holt, T. Trends in the full blood count blood test and colorectal cancer detection: A longitudinal, case-control study of UK primary care patient data. NIHR Open Res. 2022, 2, 32. [Google Scholar] [CrossRef]
- The Royal Wolverhaptom NHS Trust. Haematology Normal Adult Reference Range. 2017. Available online: https://www.royalwolverhampton.nhs.uk/services/service-directory-a-z/pathology-services/departments/haematology/haematology-normal-adult-reference-ranges/ (accessed on 13 April 2022).
- York Teaching Hospital NHS Foundation Trust. Full Blood Count (FBC) Reference Ranges. 2021. Available online: https://www.yorkhospitals.nhs.uk/our-services/a-z-of-services/laboratory-medicine1/haematology-/ (accessed on 13 April 2022).
- Maidstone and Tunbridge Wells NHS Trust. Haematology Reference Ranges. 2020. Available online: https://www.mtw.nhs.uk/wp-content/uploads/2020/11/Haematology-reference-ranges.pdf (accessed on 13 April 2022).
- Gloucestershire Hospitals NHS Foundation Trust. Full Blood Count (FBC). 2013. Available online: https://www.gloshospitals.nhs.uk/our-services/services-we-offer/pathology/tests-and-investigations/full-blood-count-fbc/ (accessed on 13 April 2022).
- National Institute for Health and Care Excellence. Suspected Cancer Recognition and Referral. 2020. Available online: https://www.nice.org.uk/guidance/ng12/resources/suspected-cancer-recognition-and-referral-pdf-1837268071621 (accessed on 13 April 2022).
- World Health Organisation. Haemoglobin Concentrations for the Diagnosis of Anaemia and Assessment of Severity. 2011. Available online: https://apps.who.int/iris/bitstream/handle/10665/85839/WHO_NMH_NHD_MNM_11.1_eng.pdf?ua=1 (accessed on 13 April 2022).
- National Institute for Health and Care Excellence. Anaemia—Iron Deficiency. 2021. Available online: https://cks.nice.org.uk/topics/anaemia-iron-deficiency/ (accessed on 13 April 2022).
- Kinar, Y.; Kalkstein, N.; Akiva, P.; Levin, B.; Half, E.E.; Goldshtein, I.; Chodick, G.; Shalev, V. Development and validation of a predictive model for detection of colorectal cancer in primary care by analysis of complete blood counts: A binational retrospective study. J. Am. Med. Inform. Assoc. 2016, 23, 879–890. [Google Scholar] [CrossRef]
- Birks, J.; Bankhead, C.; Holt, T.A.; Fuller, A.; Patnick, J. Evaluation of a prediction model for colorectal cancer: Retrospective analysis of 2.5 million patient records. Cancer Med. 2017, 6, 2453–2460. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. BMJ 2015, 350, g7594. [Google Scholar] [CrossRef]
- Clinical Practice Research Datalink (CPRD). 2022. Available online: https://www.cprd.com/ (accessed on 13 April 2022).
- Virdee, P.S.; Marian, I.R.; Mansouri, A.; Elhussein, L.; Kirtley, S.; Holt, T.; Birks, J. The Full Blood Count Blood Test for Colorectal Cancer Detection: A Systematic Review, Meta-Analysis, and Critical Appraisal. Cancers 2020, 12, 2348. [Google Scholar] [CrossRef]
- Virdee, P.S.; Fuller, A.; Jacobs, M.; Holt, T.; Birks, J. Assessing data quality from the Clinical Practice Research Datalink: A methodological approach applied to the full blood count blood test. J. Big Data 2020, 7, 96. [Google Scholar] [CrossRef]
- Riley, R.D.; Ensor, J.; Snell, K.I.E.; Harrell, F.E., Jr.; Martin, G.P.; Reitsma, J.B.; Moons, K.G.M.; Collins, G.; van Smeden, M. Calculating the sample size required for developing a clinical prediction model. BMJ 2020, 368, m441. [Google Scholar] [CrossRef]
- Riley, R.D.; Snell, K.I.; Ensor, J.; Burke, D.L.; Harrell, F.E., Jr.; Moons, K.G.; Collins, G.S. Minimum sample size for developing a multivariable prediction model: PART II—binary and time-to-event outcomes. Stat. Med. 2019, 38, 1276–1296. [Google Scholar] [CrossRef]
- Wulfsohn, M.S.; Tsiatis, A.A. A joint model for survival and longitudinal data measured with error. Biometrics 1997, 53, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Diggle, P.; Dobson, A. Joint modelling of longitudinal measurements and event time data. Biostatistics 2000, 1, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Crowther, M.J. merlin—A unified modeling framework for data analysis and methods development in Stata. Stata J. 2020, 20, 763–784. [Google Scholar] [CrossRef]
- Elashoff, R.M.; Li, G.; Li, N. Joint Modeling of Longitudinal and Time-to-Event Data 2016; CRC Press: Boca Raton, FL, USA, 2016; ISBN 9781439807828. [Google Scholar]
- Hickey, G.L.; Philipson, P.; Jorgensen, A.; Kolamunnage-Dona, R. joineRML: A joint model and software package for time-to-event and multivariate longitudinal outcomes. BMC Med. Res. Methodol. 2018, 18, 50. [Google Scholar] [CrossRef]
- Rizopoulos, D. Joint Models for Longitudinal and Time-to-Event Data with Applications in R; Chapman and Hall/CRC; Routledge, London, UK, 2012; ISBN 9781439872864. [Google Scholar]
- Royston, P.; Sauerbrei, W. A new measure of prognostic separation in survival data. Stat. Med. 2004, 23, 723–748. [Google Scholar] [CrossRef]
- StatisticsHowTo. Brier Score: Definition, Examples. 2016. Available online: https://www.statisticshowto.com/brier-score/ (accessed on 13 April 2022).
- Steyerberg, E.W. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating, 1st ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Riley, R.D.; Windt, D.A.V.D.; Croft, P.; Moon, K.G.M. Prognosis Research in Healthcare: Concepts, Methods, and Impact; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Vergouwe, Y. Towards better clinical prediction models: Seven steps for development and an ABCD for validation. Eur. Heart J. 2014, 35, 1925–1931. [Google Scholar] [CrossRef]
- Van Calster, B.; van Smeden, M.; De Cock, B.; Steyerberg, E.W. Regression shrinkage methods for clinical prediction models do not guarantee improved performance: Simulation study. Stat. Methods Med. Res. 2020, 29, 3166–3178. [Google Scholar] [CrossRef]
- Panzuto, F.; Chiriatti, A.; Bevilacqua, S.; Giovannetti, P.; Russo, G.; Impinna, S.; Pistilli, F.; Capurso, G.; Annibale, B.; Fave, G.D. Symptom-based approach to colorectal cancer: Survey of primary care physicians in Italy. Dig. Liver Dis. 2003, 35, 869–875. [Google Scholar] [CrossRef]
- Bailey, S.E.R.; Abel, G.A.; Atkins, A.; Byford, R.; Davies, S.J.; Mays, J.; McDonald, T.J.; Miller, J.; Neck, C.; Renninson, J.; et al. Diagnostic performance of a faecal immunochemical test for patients with low-risk symptoms of colorectal cancer in primary care: An evaluation in the South West of England. Br. J. Cancer 2021, 124, 1231–1236. [Google Scholar] [CrossRef]
- Lee, J.K.; Liles, E.G.; Bent, S.; Levin, T.R.; Corley, D.A. Accuracy of fecal immunochemical tests for colorectal cancer: Systematic review and meta-analysis. Ann. Intern. Med. 2014, 160, 171. [Google Scholar] [CrossRef] [PubMed]
- Usher-Smith, J.A.; Walter, F.M.; Emery, J.D.; Win, A.K.; Griffin, S.J. Risk Prediction Models for Colorectal Cancer: A Systematic Review. Cancer Prev. Res. 2016, 9, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Wei, C.; Ensor, J.E.; Smolenski, D.J.; Amos, C.I.; Levin, B.; Berry, D.A. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control 2013, 24, 1207–1222. [Google Scholar] [CrossRef]
- Fitzgerald, M.; Saville, B.R.; Lewis, R.J. Decision curve analysis. JAMA 2015, 313, 409–410. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, L.; Vickers, A. Evaluation of prediction models for decision-making: Beyond calibration and discrimination. PLoS Med. 2013, 10, e1001491. [Google Scholar] [CrossRef]
- Localio, A.R.; Goodman, S. Beyond the usual prediction accuracy metrics: Reporting results for clinical decision making. Ann. Intern. Med. 2012, 157, 294–295. [Google Scholar] [CrossRef]
- Van Calster, B.; Wynants, L.; Verbeek, J.F.M.; Verbakel, J.Y.; Christodoulou, E.; Vickers, A.J.; Roobol, M.J.; Steyerberg, E.W. Reporting and Interpreting Decision Curve Analysis: A Guide for Investigators. Eur. Urol. 2018, 74, 796–804. [Google Scholar] [CrossRef]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef]
- Vickers, A.J.; Van Calster, B.; Steyerberg, E.W. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016, 352, i6. [Google Scholar] [CrossRef]
- Vickers, A.J.; van Calster, B.; Steyerberg, E.W. A simple, step-by-step guide to interpreting decision curve analysis. Diagn. Progn. Res. 2019, 3, 18. [Google Scholar] [CrossRef]
- Lab Tests Online. Full Blood Count (FBC). 2020. Available online: https://labtestsonline.org.uk/tests/full-blood-count-fbc (accessed on 4 August 2021).
- myDr. Full Blood Count (FBC). 2017. Available online: https://www.mydr.com.au/tests-investigations/full-blood-count-fbc/ (accessed on 4 August 2021).
- MedlinePlus. Complete Blood Count (CBC). 2020. Available online: https://medlineplus.gov/lab-tests/complete-blood-count-cbc/ (accessed on 4 August 2021).
- Gaskell, H.; Derry, S.; Moore, R.A. Is there an association between low dose aspirin and anemia (without overt bleeding)? Narrative review. BMC Geriatr. 2010, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Lassale, C.; Curtis, A.; Abete, I.; van der Schouw, Y.T.; Verschuren, W.M.M.; Lu, Y.; Bueno-de-Mesquita, H.B.A. Elements of the complete blood count associated with cardiovascular disease incidence: Findings from the EPIC-NL cohort study. Sci. Rep. 2018, 8, 3290. [Google Scholar] [CrossRef] [PubMed]
- Better Health Channel. Blood Count. 2019. Available online: https://www.betterhealth.vic.gov.au/health/conditionsandtreatments/blood-count (accessed on 4 August 2021).
- Arakawa, K.; Hosono, A.; Shibata, K.; Ghadimi, R.; Fuku, M.; Goto, C.; Imaeda, N.; Tokudome, Y.; Hoshino, H.; Marumoto, M.; et al. Changes in blood biochemical markers before, during, and after a 2-day ultramarathon. J. Sports Med. 2016, 7, 43–50. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boudjeltia, K.Z.; Faraut, B.; Stenuit, P.; Esposito, M.J.; Dyzma, M.; Brohee, D.; Ducobu, J.; Vanhaeverbeek, M.; Kerkhofs, M. Sleep restriction increases white blood cells, mainly neutrophil count, in young healthy men: A pilot study. Vasc. Health Risk Manag. 2008, 4, 1467–1470. [Google Scholar] [CrossRef]
- Liu, H.; Wang, G.; Luan, G.; and Liu, Q. Effects of sleep and sleep deprivation on blood cell count and hemostasis parameters in healthy humans. J. Thromb. Thrombolysis 2009, 28, 46–49. [Google Scholar] [CrossRef]
- Bain, B.J. A Beginner’s Guide to Blood Cells, 2nd ed.; Blackwell Pub.: Malden, MA, USA, 2004. [Google Scholar]
- Bain, B.J. Blood Cells: A Practical Guide, 5th ed.; John Wiley & Sons Ltd.: Chichester, UK, 2015. [Google Scholar]
- Akunov, A.; Sydykov, A.; Toktash, T.; Doolotova, A.; Sarybaev, A. Hemoglobin Changes After Long-Term Intermittent Work at High Altitude. Front. Physiol. 2018, 9, 1552. [Google Scholar] [CrossRef]
- Rowles, P.M.; Williams, E.S. Abnormal red cell morphology in venous blood of men climbing at high altitude. Br. Med. J. 1983, 286, 1396. [Google Scholar] [CrossRef][Green Version]
- Word Health Organisation. Guide to Cancer Early Diagnosis. 2020. Available online: https://www.who.int/cancer/publications/cancer_early_diagnosis/en/ (accessed on 5 August 2021).
- National Institute for Health and Care Excellence. What Investigations Should I Arrange to Confirm Iron Deficiency Anaemia? 2021. Available online: https://cks.nice.org.uk/topics/anaemia-iron-deficiency/diagnosis/investigations/ (accessed on 20 October 2021).
- Fitzmaurice, G.M.; Laird, N.M.; Ware, J.H. Applied Longitudinal Analysis, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Asar, O.; Ritchie, J.; Kalra, P.A.; and Diggle, P.J. Joint modelling of repeated measurement and time-to-event data: An introductory tutorial. Int. J. Epidemiol. 2015, 44, 334–344. [Google Scholar] [CrossRef]
- Roberts, S.; Osborne, M.; Ebden, M.; Reece, S.; Gibson, N.; Aigrain, S. Gaussian processes for time-series modelling. Philos. Trans. A Math. Phys. Eng. Sci. 2013, 371, 20110550. [Google Scholar] [CrossRef]
- Liu, X.; Rong, J.; Liu, X. Best linear unbiased prediction for linear combinationsin general mixed linear models. J. Multivar. Anal. 2008, 99, 1503–1517. [Google Scholar] [CrossRef]
- Philipson, P.; Hickey, G.L.; Crowther, M.J.; Kolamunnage-Dona, R. Faster Monte Carlo estimation of joint models for time-to-event and multivariate longitudinal data. Comput. Stat. Data Anal. (CSDA) 2020, 151, 107010. [Google Scholar] [CrossRef]
- Benoit, J.S.; Chan, W.; Doody, R.S. Joint coverage probability in a simulation study on Continuous-Time Markov Chain parameter estimation. J. Appl Stat. 2015, 42, 2531–2538. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Summary Statistic | Males | Females | ||
|---|---|---|---|---|
| Diagnosed | Not Diagnosed | Diagnosed | Not Diagnosed | |
| Development cohort: | ||||
| No. (%) | 865 (0.4%) | 249,851 (99.6%) | 677 (0.3%) | 246,018 (99.7%) |
| Mean age 1 (SD) | 70.9 (10.0) | 60.7 (13.0) | 73.2 (11.0) | 61.9 (14.6) |
| Age 1 range | 40–95 | 40–104 | 40–96 | 40–108 |
| Internal validation cohort: | ||||
| Males | Females | |||
| Diagnosed | Not diagnosed | Diagnosed | Not diagnosed | |
| No. (%) | 1,040 (0.3%) | 311,404 (99.7%) | 1,200 (0.3%) | 461,700 (99.7%) |
| Mean age 1 (SD) | 71.6 (10.2) | 60.6 (13.0) | 73.4 (11.2) | 61.7 (14.6) |
| Age 1 range | 40–95 | 40–109 | 40–98 | 40–107 |
| Variable | Males | Females |
|---|---|---|
| HR (95% CI) | HR (95% CI) | |
| Age 2 (years) 1 | 1.015 (1.013, 1.017) | 1.014 (1.012, 1.016) |
| Age 2 × log(Age) (years) 1 | 0.997 (0.997, 0.997) | 0.997 (0.997, 0.998) |
| Trend: haemoglobin (g/dL) 2 | 0.868 (0.824, 0.916) | 0.863 (0.805, 0.926) |
| Trend: mean cell volume (fL) 2 | 0.996 (0.983, 1.009) | 0.986 (0.972, 1.000) |
| Trend: platelets (1012/L) 2 | 1.001 (0.999, 1.002) | 1.002 (1.001, 1.003) |
| Baseline two-year survival 3 | 0.999941 | 0.9999618 |
| Performance Measure | Males | Females | ||
|---|---|---|---|---|
| Development | Validation | Development | Validation | |
| Brier score | 0.0034 | 0.0033 | 0.0027 | 0.0028 |
| RD2 | 0.28 | 0.30 | 0.31 | 0.34 |
| C-statistic | 0.739 (95% CI = 0.726–0.753) | 0.751 (95% CI = 0.739–0.764) | 0.753 (95% CI = 0.737–0.769) | 0.763 (95% CI = 0.753–0.775) |
| D-statistic | 1.27 (95% CI = 1.16–1.38) | 1.33 (95% CI = 1.23–1.43) | 1.38 (95% CI = 1.26–1.51) | 1.46 (95% CI = 1.37–1.55) |
| Calibration slope | 1.00 | 1.06 | 1.00 | 1.05 |
| Risk Centile | Risk Cut-Off | True Positives | False Positives | True Negatives | False Negatives | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|
| Males: | |||||||||
| 75% | 0.3670% | 600 | 77511 | 233893 | 440 | 57.69 | 75.11 | 0.77 | 99.81 |
| 80% | 0.4036% | 505 | 61984 | 249420 | 535 | 48.56 | 80.10 | 0.81 | 99.79 |
| 85% | 0.4406% | 401 | 46466 | 264938 | 639 | 38.56 | 85.08 | 0.86 | 99.76 |
| 90% | 0.4839% | 291 | 30954 | 280450 | 749 | 27.98 | 90.06 | 0.93 | 99.73 |
| 95% | 0.5525% | 180 | 15443 | 295961 | 860 | 17.31 | 95.04 | 1.15 | 99.71 |
| 99% | 0.7232% | 49 | 3076 | 308328 | 991 | 4.71 | 99.01 | 1.57 | 99.68 |
| Females: | |||||||||
| 75% | 0.2767% | 710 | 115018 | 346682 | 490 | 59.17 | 75.09 | 0.61 | 99.86 |
| 80% | 0.3043% | 614 | 91967 | 369733 | 586 | 51.17 | 80.08 | 0.66 | 99.84 |
| 85% | 0.3348% | 513 | 68922 | 392778 | 687 | 42.75 | 85.07 | 0.74 | 99.83 |
| 90% | 0.3747% | 397 | 45893 | 415807 | 803 | 33.08 | 90.06 | 0.86 | 99.81 |
| 95% | 0.4426% | 237 | 22909 | 438791 | 963 | 19.75 | 95.04 | 1.02 | 99.78 |
| 99% | 0.6446% | 75 | 4554 | 457146 | 1125 | 6.25 | 99.01 | 1.62 | 99.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virdee, P.S.; Patnick, J.; Watkinson, P.; Holt, T.; Birks, J. Full Blood Count Trends for Colorectal Cancer Detection in Primary Care: Development and Validation of a Dynamic Prediction Model. Cancers 2022, 14, 4779. https://doi.org/10.3390/cancers14194779
Virdee PS, Patnick J, Watkinson P, Holt T, Birks J. Full Blood Count Trends for Colorectal Cancer Detection in Primary Care: Development and Validation of a Dynamic Prediction Model. Cancers. 2022; 14(19):4779. https://doi.org/10.3390/cancers14194779
Chicago/Turabian StyleVirdee, Pradeep S., Julietta Patnick, Peter Watkinson, Tim Holt, and Jacqueline Birks. 2022. "Full Blood Count Trends for Colorectal Cancer Detection in Primary Care: Development and Validation of a Dynamic Prediction Model" Cancers 14, no. 19: 4779. https://doi.org/10.3390/cancers14194779
APA StyleVirdee, P. S., Patnick, J., Watkinson, P., Holt, T., & Birks, J. (2022). Full Blood Count Trends for Colorectal Cancer Detection in Primary Care: Development and Validation of a Dynamic Prediction Model. Cancers, 14(19), 4779. https://doi.org/10.3390/cancers14194779






