Imaging Immune Cells Using Fc Domain Probes in Mouse Cancer Xenograft Models
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
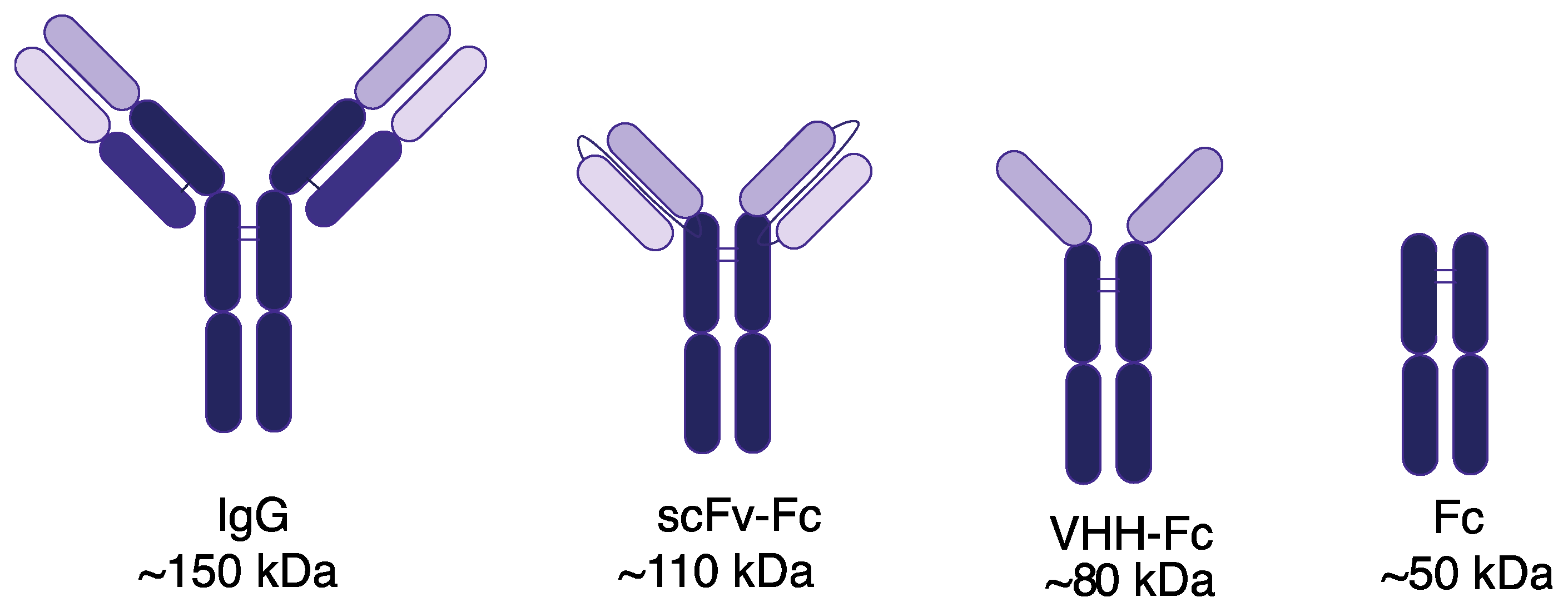
2.1. Cloning and Expression of VHH-Fc and Fc Domain
2.2. Tissue Culture
2.3. Protein Labeling and Flow Cytometry
2.4. Mouse Xenograft Models, Imaging, and Xenograft and Bone Marrow Flow Cytometry
3. Results
3.1. Fc Imaging Probe Expression and Fluorescent Labeling
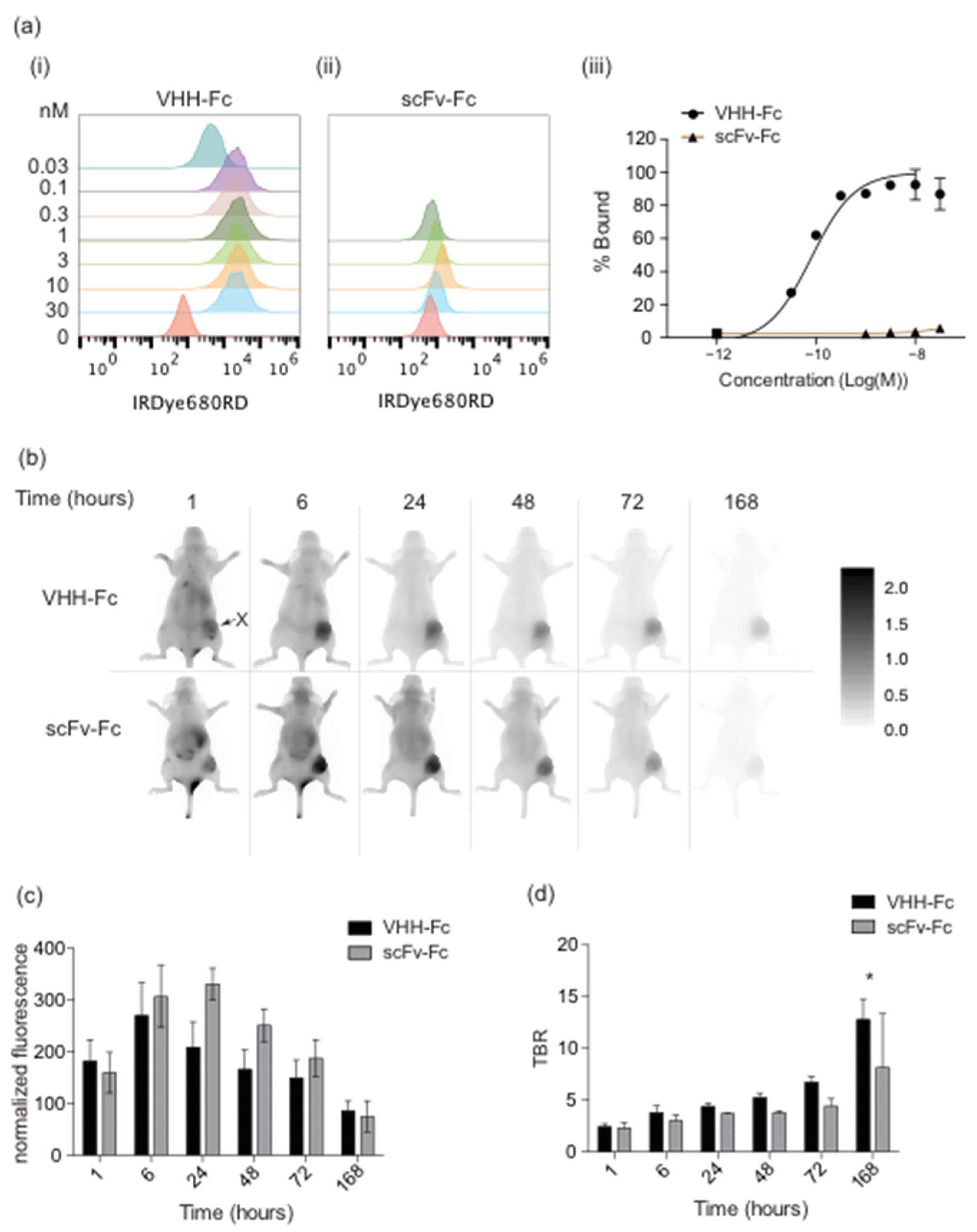
3.2. FaDu Cell Targeted VHH-Fc and MBP Binding scFv-Fc Imaging Probes Accumulate in FaDu Xenografts
3.3. Fc Domain Blocks Tumor Accumulation of FaDu Cell Line-Targeted VHH-Fc Probes
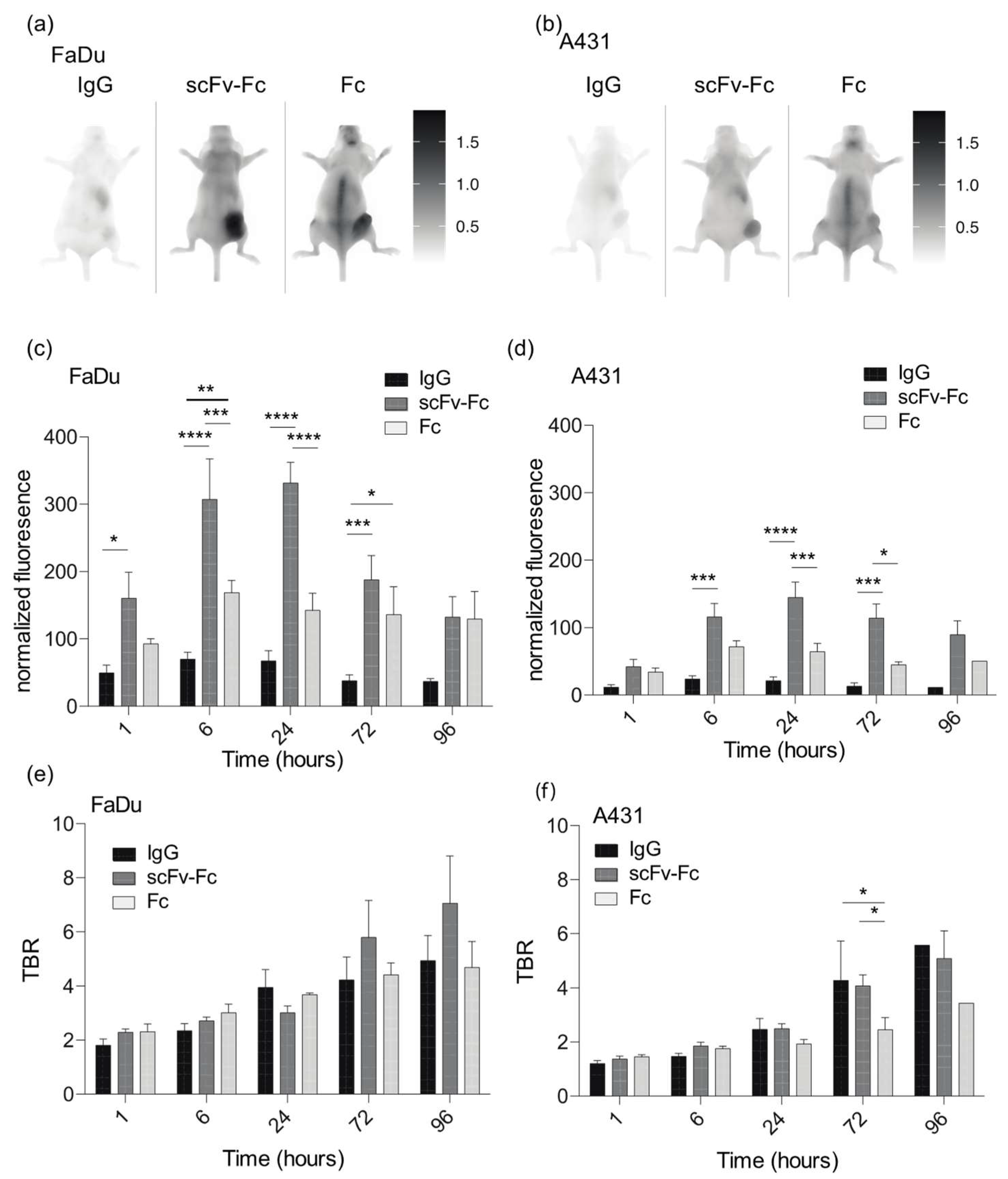
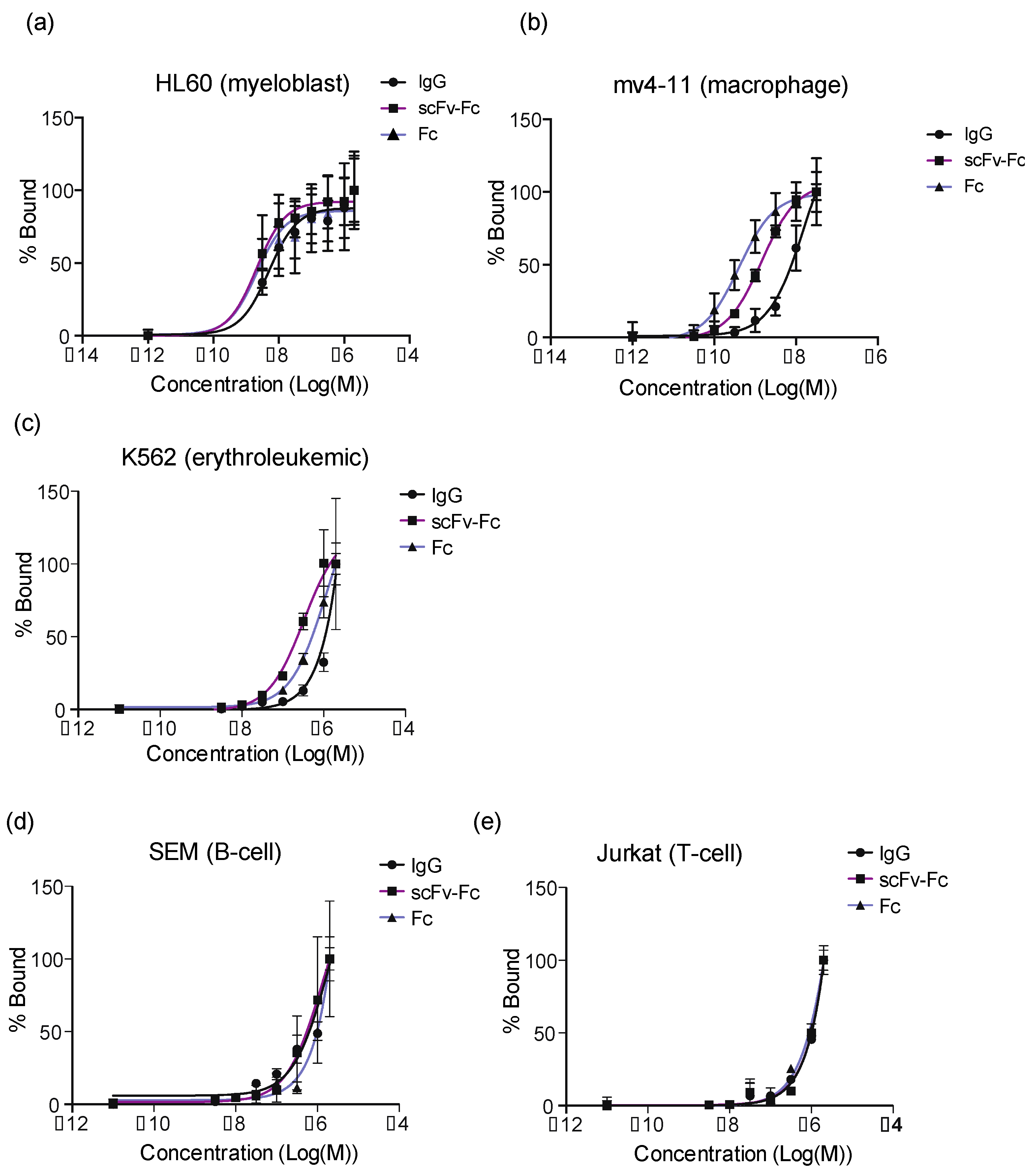
3.4. Accumulation of Fc-Containing Probes Depends on the Cell Origin of the Xenograft
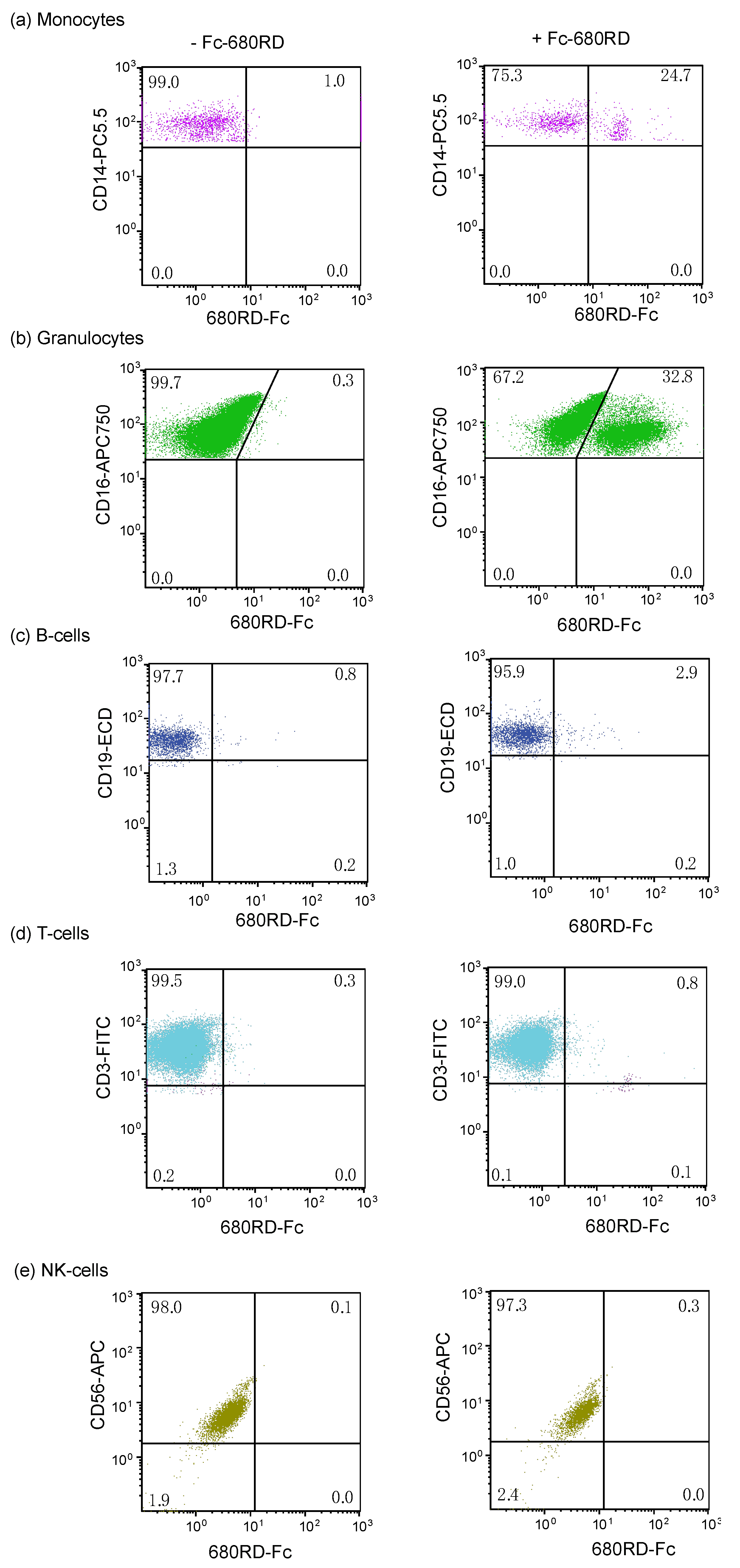
3.5. Analysis of Fc Domain Binding in Mouse Xenografts and Bone Marrow
3.6. Binding of Fc Domain Probe to Peripheral Blood
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karin, M.; Lawrence, T.; Nizet, V. Innate immunity gone awry: Linking microbial infections to chronic inflammation and cancer. Cell 2006, 124, 823–835. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Jeong, J.; Suh, Y.; Jung, K. Context drives diversification of monocytes and neutrophils in orchestrating the tumor microenvironment. Front. Immunol. 2019, 10, 1817. [Google Scholar] [CrossRef]
- Jensen, H.K.; Donskov, F.; Marcussen, N.; Nordsmark, M.; Lundbeck, F.; von der Maase, H. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J. Clin. Oncol. 2009, 27, 4709–4717. [Google Scholar] [CrossRef]
- Schiffmann, L.M.; Fritsch, M.; Gebauer, F.; Günther, S.D.; Stair, N.R.; Seeger, J.M.; Thangarajah, F.; Dieplinger, G.; Bludau, M.; Alakus, H.; et al. Tumour-infiltrating neutrophils counteract anti-vegf therapy in metastatic colorectal cancer. Br. J. Cancer 2018, 120, 69–78. [Google Scholar] [CrossRef]
- Zhuang, H.; Pourdehnad, M.; Lambright, E.S.; Yamamoto, A.J.; Lanuti, M.; Li, P.; Mozley, P.D.; Rossman, M.D.; Albelda, S.M.; Alavi, A. Dual time point 18F-FDG PET imaging for differentiating malignant from inflammatory processes. J. Nucl. Med. 2001, 42, 1412–1417. [Google Scholar] [PubMed]
- McCracken, M.N.; Radu, C.G. Targeted non-invasive imaging of the innate immune response. Proc. Natl. Acad. Sci. USA 2015, 112, 5868–5869. [Google Scholar] [CrossRef]
- Eisenblätter, M.; Ehrchen, J.; Varga, G.; Sunderkötter, C.; Heindel, W.; Roth, J.; Wall, A. In vivo optical imaging of cellular inflammatory response in granuloma formation using fluorescence-labeled macrophages. J. Nucl. Med. 2009, 50, 1676–1682. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, H.W.; Jeon, Y.H.; Hwang, M.-H.; Kim, J.-E.; Park, T.-I.; Ha, J.-H.; Lee, S.-W.; Ahn, B.-C.; Lee, J. Dual reporter gene imaging for tracking macrophage migration using the human sodium iodide symporter and an enhanced firefly luciferase in a murine inflammation model. Mol. Imaging Biol. 2013, 15, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Abreo, K.; Lieberman, L.M.; Moorthy, A.V. Distribution studies of 111in-oxine-labeled peritoneal mononuclear cells in tumor-bearing rats. Int. J. Nucl. Med. Biol. 1985, 12, 53–55. [Google Scholar] [CrossRef]
- Nahrendorf, M.; Zhang, H.; Hembrador, S.; Panizzi, P.; Sosnovik, D.E.; Aikawa, E.; Libby, P.; Swirski, F.; Weissleder, R. Nanoparticle PET-CT imaging of macrophages in inflammatory atherosclerosis. Circulation 2008, 117, 379–387. [Google Scholar] [CrossRef]
- Lee, H.W.; Gangadaran, P.; Kalimuthu, S.; Ahn, B.C. Advances in molecular imaging strategies for in vivo tracking of immune cells. Biomed. Res. Int. 2016, 2016, 1–10. [Google Scholar]
- Nair-Gill, E.; Wiltzius, S.M.; Wei, X.X.; Cheng, D.; Riedinger, M.; Radu, C.G.; Witte, O.N. PET probes for distinct metabolic pathways have different cell specificities during immune responses in mice. J. Clin. Investig. 2010, 120, 2005–2015. [Google Scholar] [CrossRef][Green Version]
- Radu, C.G.; Shu, C.J.; Shelly, S.M.; Phelps, M.E.; Witte, O.N. Positron emission tomography with computed tomography imaging of neuroinflammation in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2007, 104, 1937–1942. [Google Scholar] [CrossRef]
- Eshghi, N.; Lundeen, T.F.; Kuo, P.H. Dynamic adaptation of tumor immune response with nivolumab demonstrated by 18F-FDG PET/CT. Clin. Nucl. Med. 2018, 43, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Lugade, A.A.; Moran, J.P.; Gerber, S.A.; Rose, R.C.; Frelinger, J.G.; Lord, E.M. Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen-specific effector cells that traffic to the tumor. J. Immunol. 2005, 174, 7516–7523. [Google Scholar] [CrossRef]
- van Elsas, A.; Hurwitz, A.A.; Allison, J.P. Combination immunotherapy of B16 melanoma using anti–cytotoxic T lymphocyte–associated antigen 4 (ctla-4) and granulocyte/macrophage colony-stimulating factor (gm-csf)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med. 1999, 190, 355–366. [Google Scholar]
- Bernhard, W.; El-Sayed, A.; Barreto, K.; Gonzalez, C.; Hill, W.; Parada, A.C.; Fonge, H.; Geyer, C.R. Near infrared fluorescence imaging of EGFR expression in vivo using irdye800cw-nimotuzumab. Oncotarget 2018, 9, 6213–6227. [Google Scholar] [CrossRef]
- El-Sayed, A.; Bernhard, W.; Barreto, K.; Gonzalez, C.; Hill, W.; Pastushok, L.; Fonge, H.; Geyer, C.R. Evaluation of antibody fragment properties for near-infrared fluorescence imaging of her3-positive cancer xenografts. Theranostics 2018, 8, 4856–4869. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.; Collins, S.; Trujillo, J.; McCredie, K.; Ahern, M.; Tsai, S.; Gallo, R. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood 1979, 54, 713–733. [Google Scholar] [CrossRef]
- Breitman, T.R.; Selonick, S.E.; Collins, S.J. Induction of Differentiation of Human Promyelocytic Leukemia Cell Line (HL-60) by Retinoic Acid. Proc. Natl. Acad. Sci. USA 1980, 77, 2936–2940. [Google Scholar] [CrossRef] [PubMed]
- Xenaki, K.T.; Oliveira, S.; van Bergen En Henegouwen, P.M.P. Antibody or antibody fragments: Implications for molecular imaging and targeted therapy of solid tumors. Front. Immunol. 2017, 8, 1287. [Google Scholar] [CrossRef] [PubMed]
- Woof, J.M.; van Egmond, M.; Kerr, M.A. Chapter 13: Fc Receptors. In Mucosal Immunology, 3rd ed.; Mestecky, J., Lamm, M.E., Ogra, P., Strober, W., Bienenstock, J., McGhee, J., Mayer, L., Eds.; Academic Press: Cambridge, MA, USA, 2005; Volume 3, pp. 251–265. [Google Scholar]
- Pagès, F.; Galon, J.; Dieu-Nosjean, M.-C.; Tartour, E.; Sautès-Fridman, C.; Fridman, W.-H. Immune infiltration in human tumors: A prognostic factor that should not be ignored. Oncogene 2009, 29, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Zhang, H.; Zhou, J.; Wang, B.; Chen, Y.; Kong, Y.; Zeng, H. Peritumoural neutrophils negatively regulate adaptive immunity via the PD-L1/PD-1 signalling pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 1–11. [Google Scholar] [CrossRef]
- Wang, J.; Jia, Y.; Wang, N.; Zhang, X.; Tan, B.; Zhang, G.; Cheng, Y. The clinical significance of tumor-infiltrating neutrophils and neutrophil-to-cd8+ lymphocyte ratio in patients with resectable esophageal squamous cell carcinoma. J. Transl. Med. 2014, 12, 1–10. [Google Scholar] [CrossRef]
- Jensen, T.O.; Schmidt, H.; Møller, H.J.; Donskov, F.; Høyer, M.; Sjoegren, P.; Christensen, I.J.; Steiniche, T. Intratumoral neutrophils and plasmacytoid dendritic cells indicate poor prognosis and are associated with pstat3 expression in AJCC stage I/II melanoma. Cancer 2012, 118, 2476–2485. [Google Scholar] [CrossRef]
- Wislez, M.; Rabbe, N.; Marchal, J.; Milleron, B.; Crestani, B.; Mayaud, C.; Cadranel, J. Hepatocyte Growth Factor Production by Neutrophils Infiltrating Bronchiolaoalveolar Subtype Pulmonary Adenocarcinoma: Role in Tumor Progression and Death. Cancer Res. 2003, 63, 1405–1412. [Google Scholar]
- Seth, A.; Park, H.S.; Hong, K.S. Current perspective on in vivo molecular imaging of immune cells. Molecules 2017, 22, 881. [Google Scholar] [CrossRef]
- Kirschbaum, K.; Sonner, J.K.; Zeller, M.W.; Deumelandt, K.; Bode, J.; Sharma, R.; Krüwel, T.; Fischer, M.; Hoffmann, A.; da Silva, M.C.; et al. In vivo nanoparticle imaging of innate immune cells can serve as a marker of disease severity in a model of multiple sclerosis. Proc. Natl. Acad. Sci. USA 2016, 113, 13227–13232. [Google Scholar] [CrossRef]
- Kang, S.; Lee, H.W.; Jeon, Y.H.; Singh, T.D.; Choi, Y.J.; Park, J.Y.; Kim, J.S.; Lee, H.; Hong, K.S.; Lee, I.; et al. Combined fluorescence and magnetic resonance imaging of primary macrophage migration to sites of acute inflammation using near-infrared fluorescent magnetic nanoparticles. Mol. Imaging Biol. 2015, 17, 643–651. [Google Scholar] [CrossRef]
- Kgoebane, K.; Ally, M.M.; Duim-Beytell, M.C.; Suleman, F.E. The role of imaging in rheumatoid arthritis. S. Afr. J. Radiol. 2018, 22, 1316. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A. Imaging of acute disseminated encephalomyelitis. Neuroimaging Clin. N. Am. 2008, 18, 149–161. [Google Scholar] [CrossRef]
- Fox, A.; Mountford, J.; Braakhuis, A.; Harrison, L.C. Innate and adaptive immune responses to nonvascular xenografts: Evidence that macrophages are direct effectors of xenograft rejection. J. Immunol. 2001, 166, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Fischman, A.; Rubin, R.; White, J.; Locke, E.; Wilkinson, R.; Nedelman, M.; Callahan, R.J.; A Khaw, B.; Strauss, H.W. Localization of Fc and Fab Fragments of Nonspecific Polyclonal IgG at Focal Sites of Inflammation. J. Nucl. Med. 1990, 31, 1199–1205. [Google Scholar] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernhard, W.; Barreto, K.; El-Sayed, A.; DeCoteau, J.; Geyer, C.R. Imaging Immune Cells Using Fc Domain Probes in Mouse Cancer Xenograft Models. Cancers 2022, 14, 300. https://doi.org/10.3390/cancers14020300
Bernhard W, Barreto K, El-Sayed A, DeCoteau J, Geyer CR. Imaging Immune Cells Using Fc Domain Probes in Mouse Cancer Xenograft Models. Cancers. 2022; 14(2):300. https://doi.org/10.3390/cancers14020300
Chicago/Turabian StyleBernhard, Wendy, Kris Barreto, Ayman El-Sayed, John DeCoteau, and C. Ronald Geyer. 2022. "Imaging Immune Cells Using Fc Domain Probes in Mouse Cancer Xenograft Models" Cancers 14, no. 2: 300. https://doi.org/10.3390/cancers14020300
APA StyleBernhard, W., Barreto, K., El-Sayed, A., DeCoteau, J., & Geyer, C. R. (2022). Imaging Immune Cells Using Fc Domain Probes in Mouse Cancer Xenograft Models. Cancers, 14(2), 300. https://doi.org/10.3390/cancers14020300





