The Role of N6-Methyladenosine (m6A) Methylation Modifications in Hematological Malignancies
Abstract
:Simple Summary
Abstract
1. Introduction
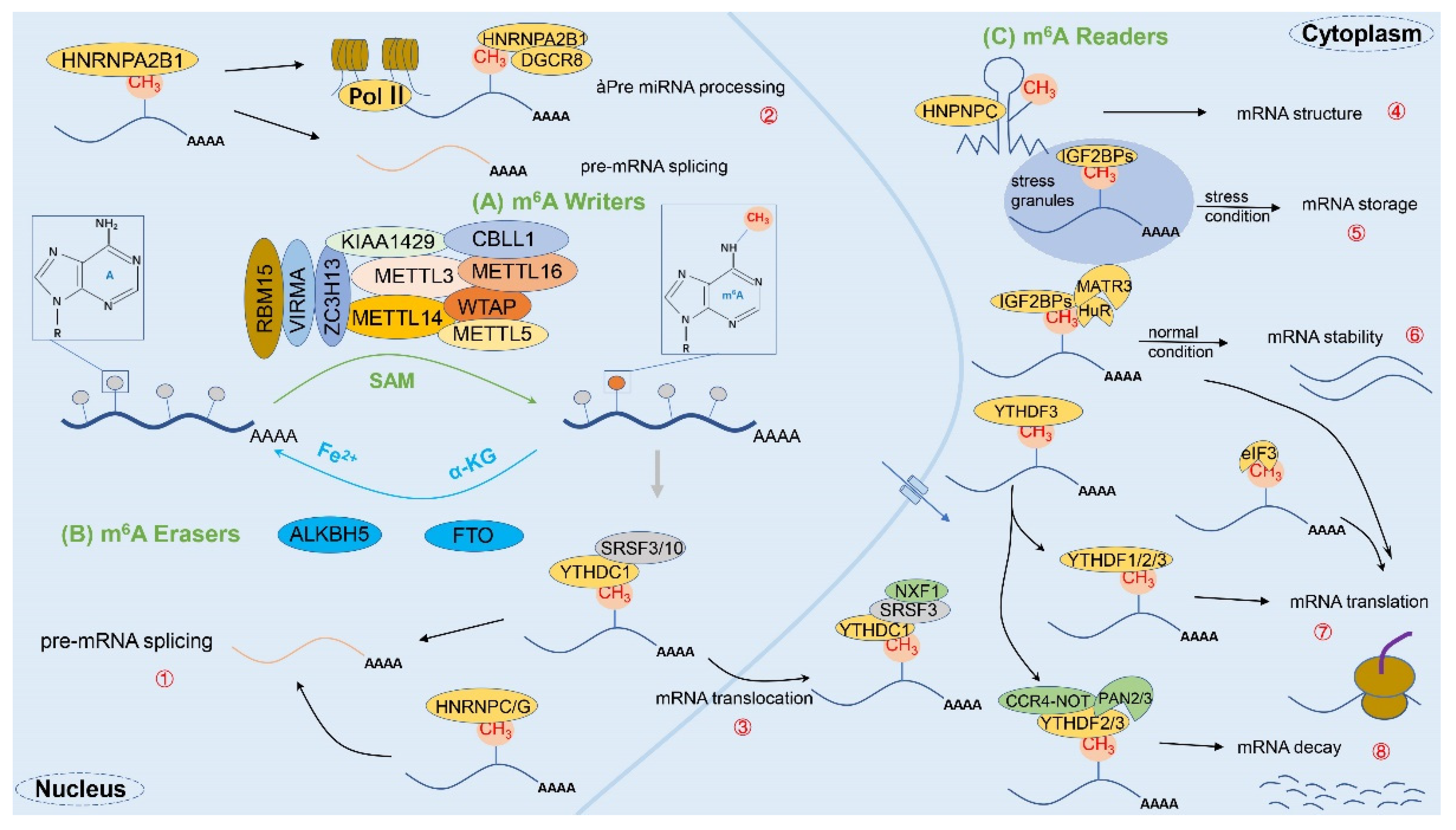
2. Biological Features of m6A Methylation Modifications
2.1. Overview of m6A Methylation
2.2. m6A-Related Enzymes
2.2.1. m6A Methyltransferase (Writers)
2.2.2. m6A Demethylases (Erasers)
2.2.3. m6A Binding Proteins (Readers)
2.3. m6A Modifications in Virus-Host Interaction
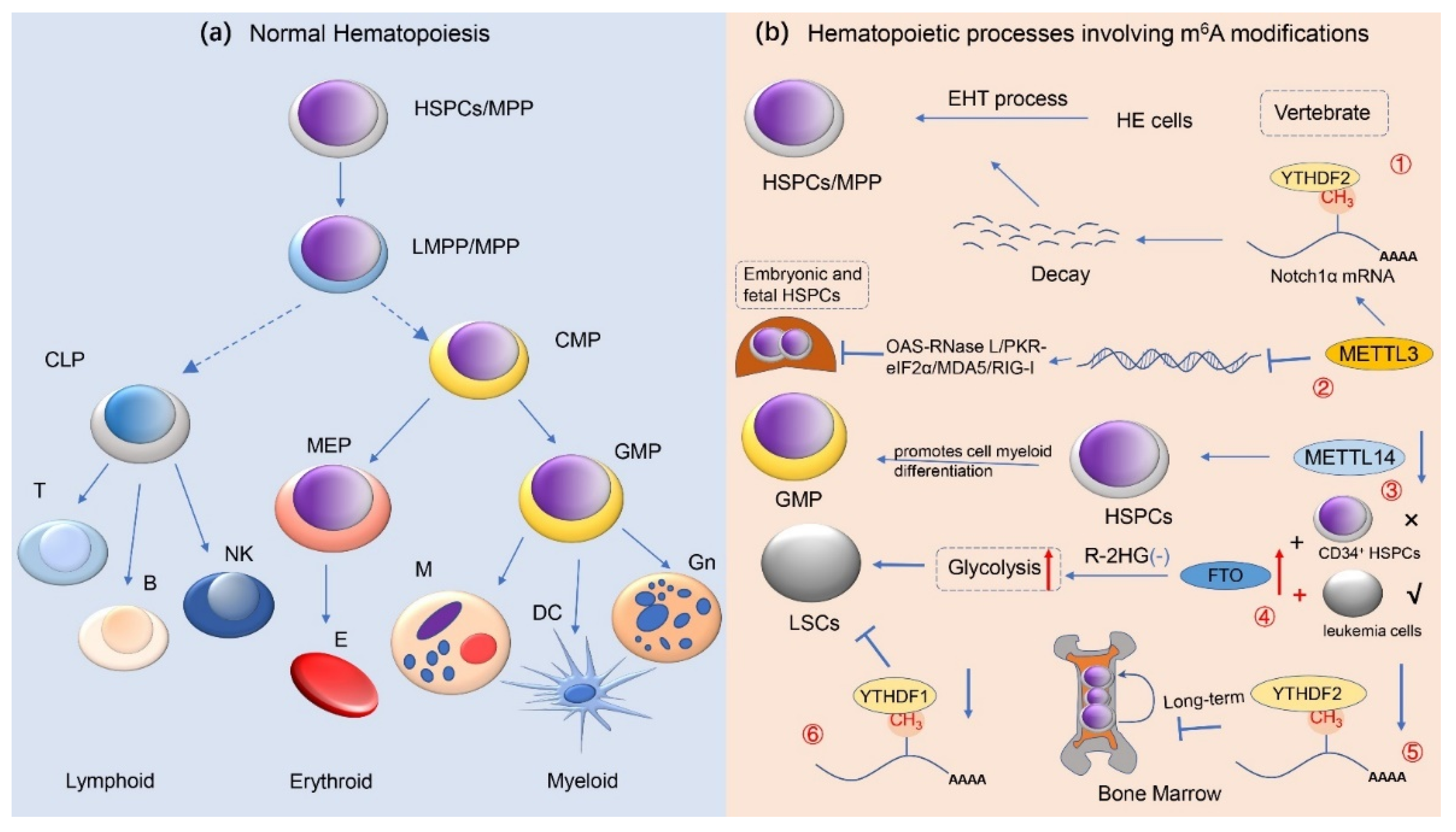
3. m6A Methylation Modification and Normal Hematopoietic Regulation
3.1. m6A Methylation Modification Regulates Hematopoietic Stem/Progenitor Cell Differentiation
3.2. m6A Methylation Modification Regulates the Differentiation of Bone Marrow Mesenchymal Stem Cells
3.3. m6A Methylation Modification Regulates Cellular Reprogramming
4. m6A Methylation Modifications and Hematological Malignancies
4.1. The Role of m6A Methylation Modifications in Acute Leukemia
4.1.1. RNA m6A Methylation and Acute Lymphoblastic Leukemia
4.1.2. RNA m6A Methylation and Acute Myeloid Leukemia
4.2. The Role of m6A Methylation Modifications in Chronic Leukemia
RNA m6A Methylation and Chronic Myeloid Leukemia
4.3. The Role of m6A Methylation Modifications in Lymphoma
4.3.1. RNA m6A Methylation and Diffuse Large B-Cell Lymphoma
4.3.2. RNA m6A Methylation and Other Types of Lymphoma
4.4. The Role of m6A Methylation Modifications in Multiple Myeloma
4.5. The Role of m6A Methylation Modifications in Myelodysplastic Syndromes
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [Green Version]
- Prakash, G.; Kaur, A.; Malhotra, P.; Khadwal, A.; Sharma, P.; Suri, V.; Varma, N.; Varma, S. Current Role of Genetics in Hematologic Malignancies. Indian J. Hematol. Blood Transfus. 2016, 32, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Mehrpouri, M.; Pourbagheri-Sigaroodi, A.; Bashash, D. The contributory roles of histone deacetylases (HDACs) in hematopoiesis regulation and possibilities for pharmacologic interventions in hematologic malignancies. Int. Immunopharmacol. 2021, 100, 108114. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Peng, Y.; Gao, A.; Du, C.; Herman, J.G. Epigenetic heterogeneity in cancer. Biomark. Res. 2019, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.A.; Issa, J.P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef] [Green Version]
- Roberti, A.; Valdes, A.F.; Torrecillas, R.; Fraga, M.F.; Fernandez, A.F. Epigenetics in cancer therapy and nanomedicine. Clin. Epigenet. 2019, 11, 81. [Google Scholar] [CrossRef]
- Liu, H.; Begik, O.; Lucas, M.C.; Ramirez, J.M.; Mason, C.E.; Wiener, D.; Schwartz, S.; Mattick, J.S.; Smith, M.A.; Novoa, E.M. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 2019, 10, 4079. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Pan, T. N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016, 23, 98–102. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Huang, H.; Chen, J. RNA N (6)-Methyladenosine Modification in Normal and Malignant Hematopoiesis. Adv. Exp. Med. Biol. 2019, 1143, 75–93. [Google Scholar] [CrossRef]
- Fong, C.Y.; Morison, J.; Dawson, M.A. Epigenetics in the hematologic malignancies. Haematologica 2014, 99, 1772–1783. [Google Scholar] [CrossRef] [Green Version]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Haruehanroengra, P.; Zheng, Y.Y.; Zhou, Y.; Huang, Y.; Sheng, J. RNA modifications and cancer. RNA Biol. 2020, 17, 1560–1575. [Google Scholar] [CrossRef]
- Wei, C.M.; Gershowitz, A.; Moss, B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 1975, 4, 379–386. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [Green Version]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Zhang, W.; Qian, Y.; Jia, G. The detection and functions of RNA modification m(6)A based on m(6)A writers and erasers. J. Biol. Chem. 2021, 297, 100973. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Dzhashiashvili, Y.; Shah, A.; Kunjamma, R.B.; Weng, Y.L.; Elbaz, B.; Fei, Q.; Jones, J.S.; Li, Y.I.; Zhuang, X.; et al. m(6)A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron 2020, 105, 293–309.e5. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Bodi, Z.; Bottley, A.; Archer, N.; May, S.T.; Fray, R.G. Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment. PLoS ONE 2015, 10, e0132090. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Narayan, P.; Ludwiczak, R.L.; Goodwin, E.C.; Rottman, F.M. Context effects on N6-adenosine methylation sites in prolactin mRNA. Nucleic Acids Res. 1994, 22, 419–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csepany, T.; Lin, A.; Baldick, C.J., Jr.; Beemon, K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 1990, 265, 20117–20122. [Google Scholar] [CrossRef]
- Narayan, P.; Rottman, F.M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science 1988, 242, 1159–1162. [Google Scholar] [CrossRef]
- Huang, J.; Yin, P. Structural Insights into N(6)-methyladenosine (m(6)A) Modification in the Transcriptome. Genom. Proteom. Bioinform. 2018, 16, 85–98. [Google Scholar] [CrossRef]
- Dai, D.; Wang, H.; Zhu, L.; Jin, H.; Wang, X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018, 9, 124. [Google Scholar] [CrossRef] [Green Version]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Ma, P.; Liu, Y.; Li, W.; Shu, Y. Multiple functions of m(6)A RNA methylation in cancer. J. Hematol. Oncol. 2018, 11, 48. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedi, R.K.; Huang, D.; Eberle, S.A.; Wiedmer, L.; Śledź, P.; Caflisch, A. Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem 2020, 15, 744–748. [Google Scholar] [CrossRef]
- Schöller, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 2018, 24, 499–512. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; He, L.; Liu, Z.; Ren, X.; Qi, L.; Wan, L.; Wang, W.; Tu, C.; Li, Z. Multifaceted Functions and Novel Insight Into the Regulatory Role of RNA N(6)-Methyladenosine Modification in Musculoskeletal Disorders. Front. Cell Dev. Biol. 2020, 8, 870. [Google Scholar] [CrossRef]
- Huang, J.; Dong, X.; Gong, Z.; Qin, L.Y.; Yang, S.; Zhu, Y.L.; Wang, X.; Zhang, D.; Zou, T.; Yin, P.; et al. Solution structure of the RNA recognition domain of METTL3-METTL14 N(6)-methyladenosine methyltransferase. Protein Cell 2019, 10, 272–284. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Zhuo, L.; Wang, J.; Zhang, Q.; Li, Q.; Li, G.; Yan, L.; Jin, T.; Pan, T.; Sui, X.; et al. METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 2020, 10, 1631–1646. [Google Scholar]
- Zheng, W.; Dong, X.; Zhao, Y.; Wang, S.; Jiang, H.; Zhang, M.; Zheng, X.; Gu, M. Multiple Functions and Mechanisms Underlying the Role of METTL3 in Human Cancers. Front. Oncol. 2019, 9, 1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [Green Version]
- Satterwhite, E.R.; Mansfield, K.D. RNA methyltransferase METTL16: Targets and function. Wiley Interdiscip. Rev. RNA 2021, e1681. [Google Scholar] [CrossRef]
- Ignatova, V.V.; Stolz, P.; Kaiser, S.; Gustafsson, T.H.; Lastres, P.R.; Sanz-Moreno, A.; Cho, Y.L.; Amarie, O.V.; Aguilar-Pimentel, A.; Klein-Rodewald, T.; et al. The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020, 34, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Gonçalves, V.; Lobo, J.; Guimarães-Teixeira, C.; Barros-Silva, D.; Guimarães, R.; Cantante, M.; Braga, I.; Maurício, J.; Oing, C.; Honecker, F.; et al. The component of the m(6)A writer complex VIRMA is implicated in aggressive tumor phenotype, DNA damage response and cisplatin resistance in germ cell tumors. J. Exp. Clin. Cancer Res. 2021, 40, 268. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, Y.; Yao, Y.; Xie, H.; Lu, G.; Du, C.; Cheng, J.; Zhou, J. VIRMA contributes to non-small cell lung cancer progression via N(6)-methyladenosine-dependent DAPK3 post-transcriptional modification. Cancer Lett. 2021, 522, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qi, X.; Liu, L.; Ma, S.; Liu, J.; Wu, J. Epigenetic Regulation of m(6)A Modifications in Human Cancer. Mol. Ther. Nucleic Acids 2020, 19, 405–412. [Google Scholar] [CrossRef]
- Gong, P.J.; Shao, Y.C.; Yang, Y.; Song, W.J.; He, X.; Zeng, Y.F.; Huang, S.R.; Wei, L.; Zhang, J.W. Analysis of N6-Methyladenosine Methyltransferase Reveals METTL14 and ZC3H13 as Tumor Suppressor Genes in Breast Cancer. Front. Oncol. 2020, 10, 578963. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Hao, Y.J.; Zhang, Y.; Li, M.M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16, 289–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Aas, A.; Isakson, P.; Bindesbøll, C.; Alemu, E.A.; Klungland, A.; Simonsen, A. Nucleocytoplasmic Shuttling of FTO Does Not Affect Starvation-Induced Autophagy. PLoS ONE 2017, 12, e0168182. [Google Scholar] [CrossRef]
- Wu, B.; Li, L.; Huang, Y.; Ma, J.; Min, J. Readers, writers and erasers of N(6)-methylated adenosine modification. Curr. Opin. Struct. Biol. 2017, 47, 67–76. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, L.H.; Wang, Y.; Xiao, Y.; Liu, J.; Zhang, W.; Yan, N.; Amu, G.; Tang, X.; Zhang, L.; et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. USA 2019, 116, 2919–2924. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, J.; Gu, Q.; Ma, Y.; Yang, Y.; Zhu, J.; Zhang, Q. The biological function of m6A demethylase ALKBH5 and its role in human disease. Cancer Cell Int. 2020, 20, 347. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Sheng, Y.; Zhu, A.C.; Robinson, S.; Jiang, X.; Dong, L.; Chen, H.; Su, R.; Yin, Z.; Li, W.; et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell 2020, 27, 64–80.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Wang, P.; Han, G.; Zhang, T.; Chang, J.; Yin, R.; Shan, Y.; Wen, J.; Xie, X.; et al. Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell 2020, 27, 81–97.e8. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Liao, S.; Sun, H.; Xu, C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genom. Proteom. Bioinform. 2018, 16, 99–107. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Gao, Y.; Pei, G.; Li, D.; Li, R.; Shao, Y.; Zhang, Q.C.; Li, P. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell Res. 2019, 29, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, K.; Zou, Z.; Qiu, M.; Tian, J.; Sieh, L.; Shi, H.; Zou, Y.; Wang, G.; Morrison, J.; et al. Genetic analyses support the contribution of mRNA N(6)-methyladenosine (m(6)A) modification to human disease heritability. Nat. Genet. 2020, 52, 939–949. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, J.N.; Wang, E.H.; Gong, C.J.; Lan, K.F.; Ding, X. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ 2020, 8, e10385. [Google Scholar] [CrossRef]
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R.; Ma, J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018, 9, 420. [Google Scholar] [CrossRef] [Green Version]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.I.; Shi, H.; Lyu, R.; Wylder, A.C.; Matuszek, Ż.; Pan, J.N.; He, C.; Parisien, M.; Pan, T. Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the Low-Complexity Protein hnRNPG. Mol. Cell 2019, 76, 70–81.e9. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell Mol. Life Sci. 2013, 70, 2657–2675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Kang, Y.; Wang, M.; Li, Y.; Xu, T.; Yang, W.; Song, H.; Wu, H.; Shu, Q.; Jin, P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018, 27, 3936–3950. [Google Scholar] [CrossRef]
- Wu, R.; Li, A.; Sun, B.; Sun, J.G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019, 29, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Orouji, E.; Peitsch, W.K.; Orouji, A.; Houben, R.; Utikal, J. Oncogenic Role of an Epigenetic Reader of m(6)A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers 2020, 12, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef]
- Imam, H.; Khan, M.; Gokhale, N.S.; McIntyre, A.B.R.; Kim, G.W.; Jang, J.Y.; Kim, S.J.; Mason, C.E.; Horner, S.M.; Siddiqui, A. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. USA 2018, 115, 8829–8834. [Google Scholar] [CrossRef] [Green Version]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Abreu, D.; Bordoni, A.; Zucca, E. Epidemiology of hematological malignancies. Ann. Oncol. 2007, 18 (Suppl. S1), i3–i8. [Google Scholar] [CrossRef]
- Peng, Z.; Gong, Y.; Wang, X.; He, W.; Wu, L.; Zhang, L.; Xiong, L.; Huang, Y.; Su, L.; Shi, P.; et al. METTL3-m(6)A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Mol. Ther. 2021. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Sun, H.; Guo, Y.; Zhou, Y.; Gu, X.; Jin, J.; Chen, X.; Wang, F.; Ma, H.; Guo, X.; et al. m(6) A reader protein YTHDF2 regulates spermatogenesis by timely clearance of phase-specific transcripts. Cell Prolif. 2021, e13164. [Google Scholar] [CrossRef]
- Yan, G.; Yuan, Y.; He, M.; Gong, R.; Lei, H.; Zhou, H.; Wang, W.; Du, W.; Ma, T.; Liu, S.; et al. m(6)A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol. Ther. Nucleic Acids 2020, 19, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gao, M.; He, J.; Wu, K.; Lin, S.; Jin, L.; Chen, Y.; Liu, H.; Shi, J.; Wang, X.; et al. The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 2021, 591, 322–326. [Google Scholar] [CrossRef]
- Mathoux, J.; Henshall, D.C.; Brennan, G.P. Regulatory Mechanisms of the RNA Modification m(6)A and Significance in Brain Function in Health and Disease. Front. Cell Neurosci. 2021, 15, 671932. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, S.; Xia, J.; Liu, F. Hematopoietic Hierarchy—An Updated Roadmap. Trends Cell Biol. 2018, 28, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006, 441, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.M.; Foroni, L. Core binding factor genes and human leukemia. Haematologica 2002, 87, 1307–1323. [Google Scholar]
- Kissa, K.; Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010, 464, 112–115. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Heng, J.; Ding, Y.; Xue, Y.; et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, Y.; Gao, S.; Zhang, C.; Chen, Y.; Li, W.; Yang, Y.G.; Zhou, Q.; Liu, F. Endothelial-specific m(6)A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 2018, 28, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Vasic, R.; Song, Y.; Teng, R.; Liu, C.; Gbyli, R.; Biancon, G.; Nelakanti, R.; Lobben, K.; Kudo, E.; et al. m(6)A Modification Prevents Formation of Endogenous Double-Stranded RNAs and Deleterious Innate Immune Responses during Hematopoietic Development. Immunity 2020, 52, 1007–1021.e8. [Google Scholar] [CrossRef]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 2018, 22, 191–205.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qing, Y.; Dong, L.; Gao, L.; Li, C.; Li, Y.; Han, L.; Prince, E.; Tan, B.; Deng, X.; Wetzel, C.; et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m(6)A/PFKP/LDHB axis. Mol. Cell 2021, 81, 922–939.e9. [Google Scholar] [CrossRef]
- Li, Z.; Qian, P.; Shao, W.; Shi, H.; He, X.C.; Gogol, M.; Yu, Z.; Wang, Y.; Qi, M.; Zhu, Y.; et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018, 28, 904–917. [Google Scholar] [CrossRef]
- Paris, J.; Morgan, M.; Campos, J.; Spencer, G.J.; Shmakova, A.; Ivanova, I.; Mapperley, C.; Lawson, H.; Wotherspoon, D.A.; Sepulveda, C.; et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 2019, 25, 137–148.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zuo, H.; Liu, J.; Wen, F.; Gao, Y.; Zhu, X.; Liu, B.; Xiao, F.; Wang, W.; Huang, G.; et al. Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018, 28, 1035–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mapperley, C.; van de Lagemaat, L.N.; Lawson, H.; Tavosanis, A.; Paris, J.; Campos, J.; Wotherspoon, D.; Durko, J.; Sarapuu, A.; Choe, J.; et al. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J. Exp. Med. 2021, 218. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Wei, J.; Yu, F.; Xu, H.; Yu, C.; Wu, Q.; Liu, Y.; Li, L.; Cui, X.-L.; Gu, X.; et al. A Critical Role of Nuclear m6A Reader YTHDC1 in Leukemogenesis by Regulating MCM Complex-Mediated DNA Replication. Blood 2021, 138, 2838–2852. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Yao, Y.; Bi, Z.; Wu, R.; Zhao, Y.; Liu, Y.; Liu, Q.; Wang, Y.; Wang, X. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m(6)A-YTHDF2-dependent manner. FASEB J. 2019, 33, 7529–7544. [Google Scholar] [CrossRef]
- Cen, S.; Li, J.; Cai, Z.; Pan, Y.; Sun, Z.; Li, Z.; Ye, G.; Zheng, G.; Li, M.; Liu, W.; et al. TRAF4 acts as a fate checkpoint to regulate the adipogenic differentiation of MSCs by activating PKM2. EBioMedicine 2020, 54, 102722. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xie, L.; Wang, M.; Xiong, Q.; Guo, Y.; Liang, Y.; Li, J.; Sheng, R.; Deng, P.; Wang, Y.; et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018, 9, 4772. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Choorapoikayil, S.; Kers, R.; Herbomel, P.; Kissa, K.; den Hertog, J. Pivotal role of Pten in the balance between proliferation and differentiation of hematopoietic stem cells in zebrafish. Blood 2014, 123, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zeng, H.M.; Xue, Y.J.; Lu, A.D.; Jia, Y.P.; Zuo, Y.X.; Zhang, L.P. The gene expression level of m6A catalytic enzymes is increased in ETV6/RUNX1-positive acute lymphoblastic leukemia. Int. J. Lab. Hematol. 2021, 43, e89–e91. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Chang, L.; Liu, C.; Chen, X.; Zhu, X. The study of METTL3 and METTL14 expressions in childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Mol. Genet. Genom. Med. 2019, 7, e00933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, H.T.; Liu, L.; Cui, L.N.; Ma, H.Y.; Shen, L.Y. ALKBH5-mediated m6A-demethylation of USP1 regulated T-cell acute lymphoblastic leukemia cell glucocorticoid resistance by Aurora B. Mol. Carcinog. 2021, 60, 644–657. [Google Scholar] [CrossRef]
- Cheng, Y.M.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.J.; Luo, H.Z.; Nguyen, D.T.; Mo, S.L.; Barin, E.; et al. N-6-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021, 39, 958–972.e8. [Google Scholar] [CrossRef]
- Chen, Z.; Shao, Y.L.; Wang, L.L.; Lin, J.; Zhang, J.B.; Ding, Y.; Gao, B.B.; Liu, D.H.; Gao, X.N. YTHDF2 is a potential target of AML1/ETO-HIF1α loop-mediated cell proliferation in t(8;21) AML. Oncogene 2021, 40, 3786–3798. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.D.; Xie, X.Q.; Han, G.Q.; Zhang, T.T.; Li, Y.S.; Li, Y.C.; Yin, R.; Wang, Q.F.; Zhang, T.; Wang, P.P.; et al. YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m(6)A-dependent manner. Blood 2021, 138, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Naren, D.; Yan, T.; Gong, Y.; Huang, J.; Zhang, D.; Sang, L.; Zheng, X.; Li, Y. High Wilms’ tumor 1 associating protein expression predicts poor prognosis in acute myeloid leukemia and regulates m(6)A methylation of MYC mRNA. J. Cancer Res. Clin. Oncol. 2021, 147, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yu, M.; Li, C.; Wang, Y.; Li, X.; Ulrich, B.; Su, R.; Dong, L.; Weng, H.; Huang, H.; et al. miR-550-1 functions as a tumor suppressor in acute myeloid leukemia via the hippo signaling pathway. Int. J. Biol. Sci. 2020, 16, 2853–2867. [Google Scholar] [CrossRef]
- Sorci, M.; Ianniello, Z.; Cruciani, S.; Larivera, S.; Ginistrelli, L.C.; Capuano, E.; Marchioni, M.; Fazi, F.; Fatica, A. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018, 9, 796. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Du, Y.; Hou, Y.; Zhao, M.; Li, J.; Du, Y.; Zhang, L.; Chen, C.; Yang, H.; Yan, F.; et al. Saikosaponin D exhibits anti-leukemic activity by targeting FTO/m(6)A signaling. Theranostics 2021, 11, 5831–5846. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691.e10. [Google Scholar] [CrossRef] [Green Version]
- Ianniello, Z.; Sorci, M.; Ceci Ginistrelli, L.; Iaiza, A.; Marchioni, M.; Tito, C.; Capuano, E.; Masciarelli, S.; Ottone, T.; Attrotto, C.; et al. New insight into the catalytic -dependent and -independent roles of METTL3 in sustaining aberrant translation in chronic myeloid leukemia. Cell Death Dis. 2021, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.Y.; Zhao, C.; Zhong, F.M.; Qin, T.Y.; Wen, F.; Li, M.Y.; Liu, J.; Huang, B.; Wang, X.Z. m(6)A Modification of lncRNA NEAT1 Regulates Chronic Myelocytic Leukemia Progression via miR-766-5p/CDKN1A Axis. Front. Oncol. 2021, 11, 679634. [Google Scholar] [CrossRef]
- Lai, X.; Wei, J.; Gu, X.Z.; Yao, X.M.; Zhang, D.S.; Li, F.; Sun, Y.Y. Dysregulation of LINC00470 and METTL3 promotes chemoresistance and suppresses autophagy of chronic myelocytic leukaemia cells. J. Cell. Mol. Med. 2021, 25, 4248–4259. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Li, M.; Hong, H.; Xu, Q.; He, Z.; Peng, Z. Expression of N(6)-methyladenosine (m(6)A) regulators correlates with immune microenvironment characteristics and predicts prognosis in diffuse large cell lymphoma (DLBCL). Bioengineered 2021, 12, 6115–6133. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Fu, Y.; Wang, Y.; Wang, J. The m6A Methyltransferase METTL3 Is Functionally Implicated in DLBCL Development by Regulating m6A Modification in PEDF. Front. Genet. 2020, 11, 955. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Hou, Y.; Chen, Q.; Chen, J.; Li, Y.; Zhang, E.; Gu, H.; Xu, R.; Liu, Y.; Cao, W.; et al. RNA demethylase ALKBH5 promotes tumorigenesis in multiple myeloma via TRAF1-mediated activation of NF-κB and MAPK signaling pathways. Oncogene 2021. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Fan, G.; Li, Q.; Su, Q.; Zhang, X.; Xue, X.; Wang, Z.; Qian, C.; Jin, Z.; Li, B.; et al. IDH2 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in multiple myeloma. Oncogene 2021, 40, 5393–5402. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Tang, X.; Tang, C.; Hua, Z.; Ke, M.; Wang, C.; Zhao, J.; Gao, S.; Jurczyszyn, A.; Janz, S.; et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J. Hematol. Oncol. 2021, 14, 54. [Google Scholar] [CrossRef]
- Chissoe, S.L.; Bodenteich, A.; Wang, Y.F.; Wang, Y.P.; Burian, D.; Clifton, S.W.; Crabtree, J.; Freeman, A.; Iyer, K.; Jian, L.; et al. Sequence and analysis of the human ABL gene, the BCR gene, and regions involved in the Philadelphia chromosomal translocation. Genomics 1995, 27, 67–82. [Google Scholar] [CrossRef]
- Liu, X.; Huang, L.; Huang, K.; Yang, L.; Yang, X.; Luo, A.; Cai, M.; Wu, X.; Liu, X.; Yan, Y.; et al. Novel Associations Between METTL3 Gene Polymorphisms and Pediatric Acute Lymphoblastic Leukemia: A Five-Center Case-Control Study. Front. Oncol. 2021, 11, 635251. [Google Scholar] [CrossRef]
- Luo, A.; Yang, L.; Li, M.; Cai, M.; Huang, A.; Liu, X.; Yang, X.; Yan, Y.; Wang, X.; Wu, X.; et al. Genetic Variants in METTL14 are Associated with the Risk of Acute Lymphoblastic Leukemia in Southern Chinese Children: A Five-Center Case-Control Study. Cancer Manag. Res. 2021, 13, 9189–9200. [Google Scholar] [CrossRef]
- Dohner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.H.; Dai, Y.F.; Huang, W.H.; Zhong, Q.F.; Zhu, P.; Zhang, W.J.; Wu, Z.H.; Lin, Q.; Zhu, H.Y.; Cui, L.Z.; et al. Prognostic Value of MicroRNA-20b in Acute Myeloid Leukemia. Front. Oncol. 2021, 10, 3467. [Google Scholar] [CrossRef]
- Shallis, R.M.; Wang, R.; Davidoff, A.; Ma, X.M.; Zeidan, A.M. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 2019, 36, 70–87. [Google Scholar] [CrossRef] [PubMed]
- Kelly, L.M.; Gilliland, D.G. Genetic’s of myeloid leukemias. Annu. Rev. Genom. Hum. Genet. 2002, 3, 179–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.M.; Lu, Z.K.; Yu, K.K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Pan, Z.P.; Wang, B.; Hou, D.Y.; You, R.L.; Wang, X.T.; Xie, W.H.; Huang, H.F. METTL3 mediates bone marrow mesenchymal stem cell adipogenesis to promote chemoresistance in acute myeloid leukaemia. FEBS Open Bio 2021, 11, 1659–1672. [Google Scholar] [CrossRef]
- Bansal, H.; Yihua, Q.; Iyer, S.P.; Ganapathy, S.; Proia, D.A.; Penalva, L.O.; Uren, P.J.; Suresh, U.; Carew, J.S.; Karnad, A.B.; et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 2014, 28, 1171–1174. [Google Scholar] [CrossRef] [Green Version]
- Raffel, G.D.; Mercher, T.; Shigematsu, H.; Williams, I.R.; Cullen, D.E.; Akashi, K.; Bernard, O.A.; Gilliland, D.G. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc. Natl. Acad. Sci. USA 2007, 104, 6001–6006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Renda, M.J.; Wang, L.; Cheng, E.C.; Niu, C.; Morris, S.W.; Chi, A.S.; Krause, D.S. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell Biol. 2007, 27, 3056–3064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 2018, 172, 90–105.e23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elcheva, I.A.; Wood, T.; Chiarolanzio, K.; Chim, B.; Wong, M.; Singh, V.; Gowda, C.P.; Lu, Q.; Hafner, M.; Dovat, S.; et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 2020, 34, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Mauro, M.J.; Cortes, J.E.; Minami, H.; Rea, D.; DeAngelo, D.J.; Breccia, M.; Goh, Y.T.; Talpaz, M.; Hochhaus, A.; et al. Asciminib in Chronic Myeloid Leukemia after ABL Kinase Inhibitor Failure. N. Engl. J. Med. 2019, 381, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- Vetrie, D.; Helgason, G.V.; Copland, M. The leukaemia stem cell: Similarities, differences and clinical prospects in CML and AML. Nat. Rev. Cancer 2020, 20, 158–173. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef]
- Höglund, M.; Sandin, F.; Simonsson, B. Epidemiology of chronic myeloid leukaemia: An update. Ann. Hematol. 2015, 94 (Suppl. S2), S241–S247. [Google Scholar] [CrossRef]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Trela, E.; Glowacki, S.; Błasiak, J. Therapy of chronic myeloid leukemia: Twilight of the imatinib era? ISRN Oncol. 2014, 2014, 596483. [Google Scholar] [CrossRef] [Green Version]
- Perrotti, D.; Turturro, F.; Neviani, P. BCR/ABL, mRNA translation and apoptosis. Cell Death Differ. 2005, 12, 534–540. [Google Scholar] [CrossRef]
- Prabhu, S.; Saadat, D.; Zhang, M.; Halbur, L.; Fruehauf, J.P.; Ong, S.T. A novel mechanism for Bcr-Abl action: Bcr-Abl-mediated induction of the eIF4F translation initiation complex and mRNA translation. Oncogene 2007, 26, 1188–1200. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.Q.; Zeng, H.M.; Zheng, R.S.; Zhang, S.W.; He, J. Cancer incidence and mortality in china, 2007. Chin. J. Cancer Res. 2012, 24, 1–8. [Google Scholar] [CrossRef] [Green Version]
- An, L.; Li, X.; Yang, J. MicroRNA-211 attenuates cell proliferation in T-cell lymphoblastic lymphoma through targeting TCF12. Leuk. Res. 2021, 110, 106653. [Google Scholar] [CrossRef]
- Ma, H.; Shen, L.; Yang, H.; Gong, H.; Du, X.; Li, J. m6A methyltransferase Wilms’ tumor 1-associated protein facilitates cell proliferation and cisplatin resistance in NK/T cell lymphoma by regulating dual-specificity phosphatases 6 expression via m6A RNA methylation. IUBMB Life 2021, 73, 108–117. [Google Scholar] [CrossRef]
- Wu, G.; Suo, C.; Yang, Y.; Shen, S.; Sun, L.; Li, S.T.; Zhou, Y.; Yang, D.; Wang, Y.; Cai, Y.; et al. MYC promotes cancer progression by modulating m(6) A modifications to suppress target gene translation. EMBO Rep. 2021, 22, e51519. [Google Scholar] [CrossRef]
- Xia, T.L.; Li, X.; Wang, X.; Zhu, Y.J.; Zhang, H.; Cheng, W.; Chen, M.L.; Ye, Y.; Li, Y.; Zhang, A.; et al. N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep. 2021, 22, e50128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; He, X.; Hu, J.; Yang, P.; Liu, C.; Wang, J.; An, R.; Zhen, J.; Pang, M.; Hu, K.; et al. Dysregulation of N(6)-methyladenosine regulators predicts poor patient survival in mantle cell lymphoma. Oncol. Lett. 2019, 18, 3682–3690. [Google Scholar] [CrossRef] [Green Version]
- Terpos, E.; Ntanasis-Stathopoulos, I. Multiple Myeloma: Clinical Updates From the American Society of Hematology Annual Meeting 2018. Clin. Lymphoma Myeloma Leuk. 2019, 19, e324–e336. [Google Scholar] [CrossRef] [PubMed]
- Pratanwanich, P.N.; Yao, F.; Chen, Y.; Koh, C.W.Q.; Wan, Y.K.; Hendra, C.; Poon, P.; Goh, Y.T.; Yap, P.M.L.; Chooi, J.Y.; et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 2021, 39, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Adès, L.; Itzykson, R.; Fenaux, P. Myelodysplastic syndromes. Lancet 2014, 383, 2239–2252. [Google Scholar] [CrossRef]
- Fei, D.L.; Zhen, T.; Durham, B.; Ferrarone, J.; Zhang, T.; Garrett, L.; Yoshimi, A.; Abdel-Wahab, O.; Bradley, R.K.; Liu, P.; et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc. Natl. Acad. Sci. USA 2018, 115, E10437–E10446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaguchi, H.; Nakanishi, K.; Kojima, S. Inherited bone marrow failure syndromes in 2012. Int. J. Hematol. 2013, 97, 20–29. [Google Scholar] [CrossRef] [PubMed]

| Cancer Type | m6A Modifiers | Patients/Cell Lines | Role of m6A in Cancer | Functions | Mechanism | References |
|---|---|---|---|---|---|---|
| ALL | Writers/Erasers | In ETV6/RUNX1-positive ALL patients | oncogene | High levels of m6A “writers” (METTL3, METTL14, WTAP) and m6A “erasers” (FTO and ALKBH5) mRNA expression prior to induction therapy resulted in a high disease burden in ALL patients | Not available | [117] |
| METTL3/METTL14 | In childhood ETV6/RUNX1-positive P-ALL | oncogene | The decreased levels of METTL3 and METTL14 indicate a possible role in the pathogenesis and progression of E/R-positive P-ALL. | Not available | [118] | |
| ALKBH5 | In glucocorticoid (GC)-resistant T-ALL patients/CEM-C1 cells/mice | oncogene | Inhibition of ALKBH5-mediated m6A modification decreased USP1 expression, and downregulation of USP1 ameliorated GC resistance in T-ALL by suppressing Aurora B expression and elevating GR levels | ALKBH5/USP1/Aurora B | [119] | |
| AML | YTHDC1 | In human AML cells/LSCs in vivo in mice | oncogene | YTHDC1 is overexpressed in AML, and it contributes to the proliferation and survival of human HSPCs/AML cells, as well as the self-renewal of leukemia stem cells (LSCs) in mice | YTHDC1/ MCM4 | [108] |
| YTHDC1 | In AML cells | oncogene | nYACs maintain mRNA stability, as well as AML cell survival and undifferentiated state; moreover, nYACs protect m6A-mRNA from degradation by PAXT complex and exosome-related RNAs | YTHDC1-m6A condensates (nYACs) | [120] | |
| YTHDF2 | In leukemic cells in vitro and in mice/in AML patients | oncogene | Overexpressed in t (8; 21)-type AML patients; when YTHDF2 is knocked down, it inhibits tumor cell proliferation in vitro and in mice | AML1/ETO-HIF1α loop/YTHDF2/TNFRSF1b | [121] | |
| YTHDF2 | In mouse and human AML | oncogene | YTHDF2 contributes to the initiation of AML disease as well as proliferation and maintains the overall functional integrity of LSCs | YTHDF2/ TNFR2 | [105] | |
| YBX1/IGF2BPs | Primary human and mouse AML cells in vitro and in vivo | oncogene | Expression of YBX1 is markedly upregulated in myeloid leukemia cells, and YBX1 deficiency greatly induces apoptosis and promotes differentiation while reducing proliferation and impairing leukemic competence of primary human and mouse AML cells in vitro and in vivo | YBX1/MYC/BCL2 (mRNA) | [122] | |
| METTL3 | In MOLM-13 cell lines/recipient mice in vivo | oncogene | METTL3 is highly expressed in AML cells as well as promotes AML cell proliferation and inhibits cell differentiation; m6A modification elevates the translation levels of c-MYC, BCL2, and PTEN gene mRNAs in AML cells | METTL3/c-MYC/BCL2/PTEN (mRNA) | [114] | |
| METTL3 | In AML cells and immunodeficient mice | oncogene | In AML cells, METTL3 promotes tumor cell proliferation and inhibits cell differentiation; downregulation of METTL3 results in the inability of immunodeficient mice to develop leukemia. | CEBPZ/ METTL3/ SP1 | [123] | |
| METTL14 | In normal HSPCs and AML cells | oncogene | METTL14 is overexpressed in AML cells and can block the differentiation of normal myeloid cells and promote malignant hematopoiesis via m6A modifications | SPI1-METTL14-MYB/MYC | [102] | |
| WTAP | In AML patients/WTAP knockout AML cells | oncogene | WTAP promotes AML cell proliferation, tumorigenesis, and inhibits cell differentiation. In addition, WTAP causes chemoresistance in AML cells | WTAP/MYC mRNA | [124] | |
| WTAP | In AML patients or in AML cells in vitro in vivo | oncogene | miR-550-1 leads to a further decrease in WWTR1 stability by downregulating the expression level of WTAP, which ultimately disrupts AML cell proliferation and tumorigenesis | miR-550-1/WTAP/ WWTR1 | [125] | |
| WTAP | In different AML cell lines, e.g., K562 cell line | oncogene | Under the regulation of functional METTL3, the expression of WTAP is upregulated and promotes the proliferation of AML cells | METTL3/WTAP | [126] | |
| FTO | In vitro, in mice, primary patient cells, and TKI-resistant cells | oncogene | SsD inhibits AML cell proliferation and promotes apoptosis and cell cycle arrest via targeting FTO/m6A signaling both in vitro and in vivo | Not available | [127] | |
| FTO | In AMLs | oncogene | FTO enhances leukemia oncogene-mediated cell transformation and leukemogenesis and suppresses all-trans retinoic acid (ATRA)-induced AML cell differentiation and apoptosis | FTO/ASB2, RARA | [128] | |
| FTO | In (R-2HG-sensitive) leukemia cells | oncogene | R-2HG abrogated FTO/m6A/YTHDF2-mediated post-transcriptional upregulation of PFKP and LDHB (two key glycolytic genes) expression, thereby attenuating aerobic glycolysis in leukemia | FTO/m6A/PFKP/LDHB axis | [103] | |
| FTO | In human AML cell lines and AML patients | oncogene | FTO inhibitors, namely FB23 and FB23-2, inhibit proliferation and promote differentiation/apoptosis in human AML cells and primary cells | Not available | [129] | |
| ALKBH5 | In human AML LSCs | oncogene | By regulating the chromatin state of the ALKBH5 locus, the expression of ALKBH5 can be elevated, thereby maintaining leukemogenesis in human AML | KDM4C, MYB, Pol II /ALKBH5/AXL Signaling Axis | [66] | |
| ALKBH5 | In human AML/in LSCs/LICs | oncogene | ALKBH5 not only facilitates the proliferation of AML cells, but also contributes to the self-renewal of leukemic stem/initiating cells (LSCs/LICs) | ALKBH5/TACC3 | [65] | |
| CML | METTL3 | In CML patients/CML cell lines | oncogene | Depletion of METTL3 strongly impairs the translation efficiency of mRNA and contributes to the proliferation of CML cells | METTL3/PES1 protein | [130] |
| METTL3 | PBMCs and CML cell lines | oncogene | Overexpression of NEAT1 inhibits cell viability and promotes apoptosis in CML cells | METTL3/NEAT1/miR-766-5p/CDKN1A axis | [131] | |
| METTL3 | In a mouse model, and in KCL22 and K562 cells | oncogene | Dysregulation of METTL3 promotes chemoresistance and inhibits autophagy in CML cells | LINC00470/METTL3/PTEN mRNA | [132] | |
| DLBCL | m6A regulators | In DLBCL patients | oncogene | In patients with DLBCL, high-risk m6A indicates worse survival when grouped according to prognostic characteristics | Not available | [133] |
| METTL3 | In DLBCL tissues and cell lines | oncogene | METTL3 promotes tumor cell proliferation | METTL3/ PEDF | [134] | |
| WTAP | In xenograft DLBCL models | oncogene | piRNA-30473 facilitates the proliferation of DLBCL cells and induces cell cycle arrest via upregulating WTAP | piRNA-30473/WTAP/HK2 m6A | [135] | |
| MM | ALKBH5 | in MM cells, xenograft models or patients | oncogene | ALKBH5 deficiency induces apoptosis and inhibits the growth of MM cells in vitro | ALKBH5/ TRAF1/NF-κB and MAPK | [136] |
| FTO | in CD138+ cells from MM | oncogene | IDH2 promotes the growth of myeloma cells in vitro by targeting FTO to regulate the m6A RNA level of MM | IDH2/FTO/WNT7B/Wnt | [137] | |
| HNRNPA2B1 | in MM patients and in MM cells | oncogene | Overexpression of HNRNPA2B1 promotes the proliferation of MM cells in vitro and in vivo | HNRNPA2B1/ILF3 mRNA/AKT3 | [138] | |
| MDS | YTHDC1 | In MDS cells | oncogene | Causes abnormalities in hematopoietic function | YTHDC1/SRSF3 or SRSF10 | [74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Peng, H. The Role of N6-Methyladenosine (m6A) Methylation Modifications in Hematological Malignancies. Cancers 2022, 14, 332. https://doi.org/10.3390/cancers14020332
Zhao Y, Peng H. The Role of N6-Methyladenosine (m6A) Methylation Modifications in Hematological Malignancies. Cancers. 2022; 14(2):332. https://doi.org/10.3390/cancers14020332
Chicago/Turabian StyleZhao, Yan, and Hongling Peng. 2022. "The Role of N6-Methyladenosine (m6A) Methylation Modifications in Hematological Malignancies" Cancers 14, no. 2: 332. https://doi.org/10.3390/cancers14020332
APA StyleZhao, Y., & Peng, H. (2022). The Role of N6-Methyladenosine (m6A) Methylation Modifications in Hematological Malignancies. Cancers, 14(2), 332. https://doi.org/10.3390/cancers14020332






