The Mechanistic Roles of Sirtuins in Breast and Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
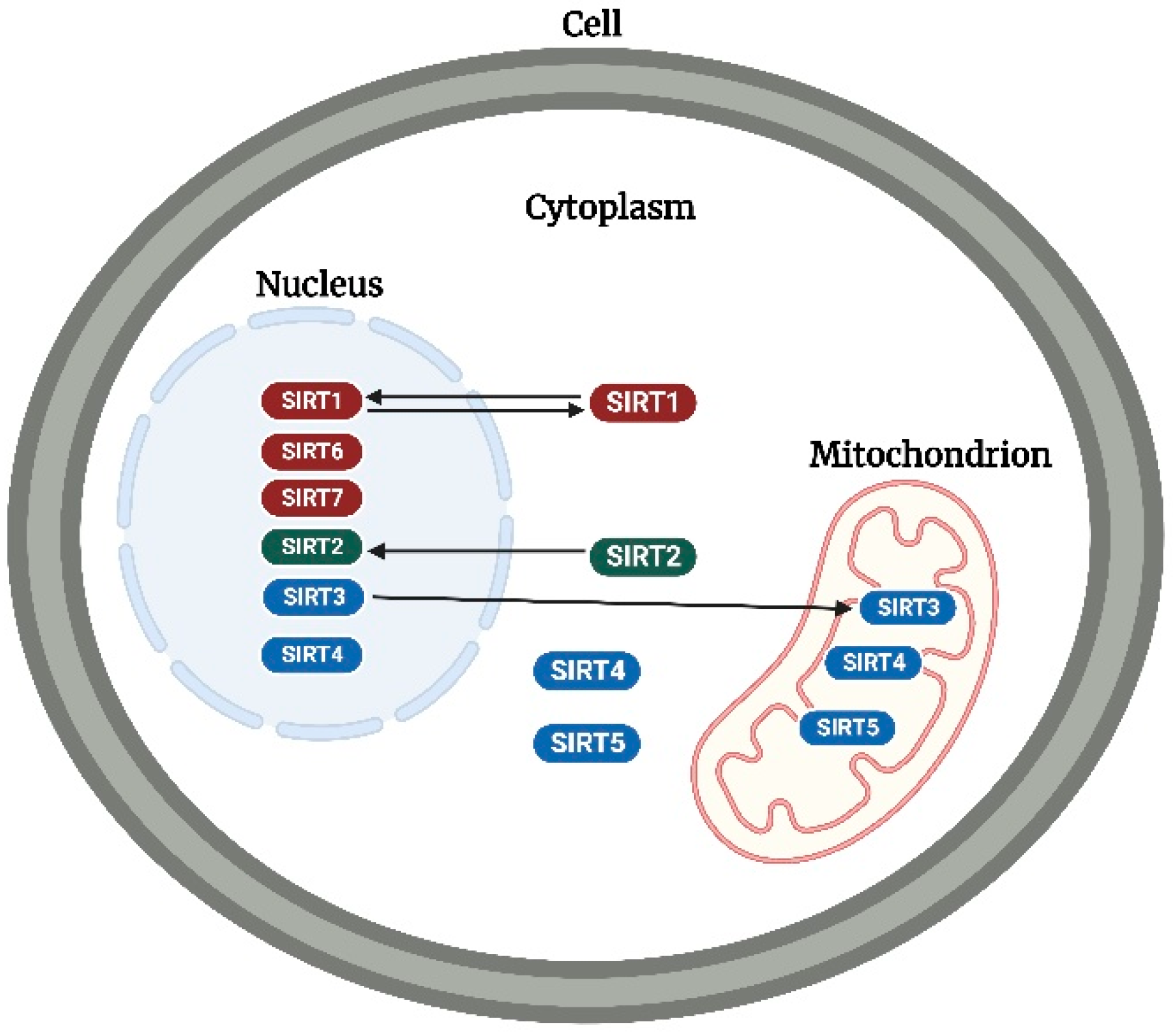
Overview of Sirtuins
2. Mechanistic Roles of Sirtuins in Breast and Prostate Cancer
2.1. Nuclear Sirtuins in Breast Cancer
2.1.1. SIRT1
2.1.2. SIRT6
2.1.3. SIRT7
2.2. Nuclear Sirtuins in Prostate Cancer
2.2.1. SIRT1
2.2.2. SIRT6
2.2.3. SIRT7
2.3. Cytoplasmic Sirtuins in Breast Cancer
SIRT2
2.4. Cytoplasmic Sirtuins in Prostate Cancer
SIRT2
2.5. Mitochondrial Sirtuins in Breast Cancer
2.5.1. SIRT3
2.5.2. SIRT4
2.5.3. SIRT5
2.6. Mitochondrial Sirtuins in Prostate Cancer
2.6.1. SIRT3
2.6.2. SIRT4
2.6.3. SIRT5
3. Mechanistic Role of Sirtuins Regulation by microRNAs (miRNAs) in Breast and Prostate Cancers
3.1. Overview of miRNA
3.2. SIRT1 Regulation by miRNAs in Breast Cancer
3.3. SIRT7 Regulation by miRNAs in Breast Cancer
3.4. SIRT1 Regulation by miRNAs in Prostate Cancer
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berdichevsky, A.; Viswanathan, M.; Horvitz, H.R.; Guarente, L.C. Elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell 2006, 125, 1165–1177. [Google Scholar] [PubMed] [Green Version]
- Bauer, J.H.; Morris, S.N.S.; Chang, C.; Flatt, T.; Wood, J.G.; Helfand, S.L. dSir2 and Dmp53 interact to mediate aspects of CR-dependent lifespan extension in D. melanogaster. Aging 2009, 1, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.-I.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef] [PubMed]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Denu, J.M. The Sir 2 family of protein deacetylases. Curr. Opin. Chem. Biol. 2005, 9, 431–440. [Google Scholar] [CrossRef]
- Sosnowska, B.; Mazidi, M.; Penson, P.; Gluba-Brzózka, A.; Rysz, J.; Banach, M. The sirtuin family members SIRT1, SIRT3 and SIRT6: Their role in vascular biology and atherogenesis. Atherosclerosis 2017, 265, 275–282. [Google Scholar] [CrossRef] [Green Version]
- Vassilopoulos, A.; Fritz, K.S.; Petersen, D.R.; Gius, D. The human sirtuin family: Evolutionary divergences and functions. Hum Genom. 2011, 5, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Haigis, M.C.; Mostoslavsky, R.; Haigis, K.M.; Fahie, K.; Christodoulou, D.C.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Karow, M.; Blander, G.; et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 2006, 126, 941–954. [Google Scholar] [CrossRef] [Green Version]
- Liszt, G.; Ford, E.; Kurtev, M.; Guarente, L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005, 280, 21313–21320. [Google Scholar] [CrossRef]
- Rauh, D.; Fischer, F.; Gertz, M.; Lakshminarasimhan, M.; Bergbrede, T.; Aladini, F.; Kambach, C.; Becker, C.F.W.; Zerweck, J.; Schutkowski, M.; et al. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathias, R.A.; Greco, T.M.; Oberstein, A.; Budayeva, H.G.; Chakrabarti, R.; Rowland, E.A.; Kang, Y.; Shenk, T.; Cristea, I.M. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 2014, 159, 1615–16125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorentino, F.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; et al. Therapeutic Potential and Activity Modulation of the Protein Lysine Deacylase Sirtuin 5. J. Med. Chem. 2022, 65, 9580–9606. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005, 16, 4623–4635. [Google Scholar] [CrossRef] [Green Version]
- Sacconnay, L.; Carrupt, P.-A.; Nurisso, A. Human sirtuins: Structures and flexibility. J. Struct. Biol. 2016, 196, 534–542. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Hu, H.; Hua, C.; Du, K.; Wei, T. Tissue distribution, subcellular localization, and enzymatic activity analysis of human SIRT5 isoforms. Biochem. Biophys. Res. Commun. 2018, 503, 763–769. [Google Scholar] [CrossRef]
- Iwahara, T.; Bonasio, R.; Narendra, V.; Reinberg, D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol. Cell. Biol. 2012, 32, 5022–5034. [Google Scholar] [CrossRef] [Green Version]
- Scher, M.B.; Vaquero, A.; Reinberg, D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007, 21, 920–928. [Google Scholar] [CrossRef]
- Ramadani-Muja, J.; Gottschalk, B.; Pfeil, K.; Burgstaller, S.; Rauter, T.; Bischof, H.; Waldeck-Weiermair, M.; Bugger, H.; Graier, W.F.; Malli, R. Visualization of Sirtuin 4 Distribution between Mitochondria and the Nucleus, Based on Bimolecular Fluorescence Self-Complementation. Cells 2019, 8, 1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldridge, M.J.G.; Pereira, J.M.; Impens, F.; Hamon, M.A. Active nuclear import of the deacetylase Sirtuin-2 is controlled by its C-terminus and importins. Sci. Rep. 2020, 10, 2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.-L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, W.; Zhang, X. Nucleus or cytoplasm? The mysterious case of SIRT1’s subcellular localization. Cell Cycle 2016, 15, 3337–3338. [Google Scholar] [CrossRef] [Green Version]
- Tanno, M.; Sakamoto, J.; Miura, T.; Shimamoto, K.; Horio, Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007, 282, 6823–6832. [Google Scholar] [CrossRef] [Green Version]
- Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793–798. [Google Scholar] [CrossRef]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef] [Green Version]
- George, J.; Ahmad, N. Mitochondrial Sirtuins in Cancer: Emerging Roles and Therapeutic Potential. Cancer Res. 2016, 76, 2500–2506. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, X.; Cui, Y.; Wei, Q.; Chen, S.; Wang, X. Effects of SIRT1 silencing on viability, invasion and metastasis of human glioma cell lines. Oncol. Lett. 2019, 17, 3701–3708. [Google Scholar] [CrossRef] [Green Version]
- Mei, Z.; Zhang, X.; Yi, J.; Huang, J.; He, J.; Tao, Y. Sirtuins in metabolism, DNA repair and cancer. J. Exp. Clin. Cancer Res. 2016, 35, 182. [Google Scholar] [CrossRef]
- Nagarajan, P.; Parthun, M.R. The flip side of sirtuins: The emerging roles of protein acetyltransferases in aging. Aging 2020, 12, 4673–4677. [Google Scholar] [CrossRef] [PubMed]
- Papa, L.; Germain, D. SirT3 Regulates a Novel Arm of the Mitochondrial Unfolded Protein Response. Mol. Cell. Biol. 2014, 34, 699–710, Erratum in Mol. Cell. Biol. 2017, 37, e00191–e00217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Xiang, H.; Liu, J.; Chen, Y.; He, R.-R.; Liu, B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics 2020, 10, 8315–8342. [Google Scholar] [CrossRef] [PubMed]
- Maissan, P.; Mooij, E.; Barberis, M. Sirtuins-Mediated System-Level Regulation of Mammalian Tissues at the Interface between Metabolism and Cell Cycle: A Systematic Review. Biology 2021, 10, 194. [Google Scholar] [CrossRef]
- Bosch-Presegué, L.; Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef]
- Bai, L.; Lin, G.; Sun, L.; Liu, Y.; Huang, X.; Cao, C.; Guo, Y.; Xie, C. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget 2016, 7, 40377–40386. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Hao, Y.; Zhao, Z.; Tong, Y. Sirtuin 6 contributes to migration and invasion of osteosarcoma cells via the ERK1/2/MMP9 pathway. FEBS Open Bio 2017, 7, 1291–1301. [Google Scholar] [CrossRef]
- Garcia-Peterson, L.M.; Wilking-Busch, M.J.; Ndiaye, M.A.; Philippe, C.G.A.; Setaluri, V.; Ahmad, N. Sirtuins in Skin and Skin Cancers. Ski. Pharm. Physiol. 2017, 30, 216–224. [Google Scholar] [CrossRef]
- Hu, J.; Jing, H.; Lin, H. Sirtuin inhibitors as anticancer agents. Future Med. Chem. 2014, 6, 945–966. [Google Scholar] [CrossRef] [Green Version]
- Kozako, T.; Suzuki, T.; Yoshimitsu, M.; Arima, N.; Honda, S.-I.; Soeda, S. Anticancer agents targeted to sirtuins. Molecules 2014, 19, 20295–20313. [Google Scholar] [CrossRef]
- McGlynn, L.M.; McCluney, S.; Jamieson, N.; Thomson, J.; MacDonald, A.I.; Oien, K.; Dickson, E.J.; Carter, C.R.; McKay, C.J.; Shiels, P.G. SIRT3 & SIRT7: Potential Novel Biomarkers for Determining Outcome in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0131344. [Google Scholar]
- Tan, Y.; Li, B.; Peng, F.; Gong, G.; Li, N. Integrative Analysis of Sirtuins and Their Prognostic Significance in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2020, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar]
- Risbridger, G.P.; Davis, I.D.; Birrell, S.N.; Tilley, W. Breast and prostate cancer: More similar than different. Nat. Rev. Cancer 2010, 10, 205–212. [Google Scholar] [CrossRef]
- Jørgensen, K.J.; Gøtzsche, P.C.; Kalager, M.; Zahl, P.-H. Breast Cancer Screening in Denmark: A Cohort Study of Tumor Size and Overdiagnosis. Ann. Intern. Med. 2017, 166, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tang, X.; Qian, M.; Liu, Z.; Meng, F.; Fu, L.; Wang, Z.; Zhu, W.-G.; Huang, J.-D.; Zhou, Z.; et al. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene 2018, 37, 6299–6315. [Google Scholar] [CrossRef] [Green Version]
- Latifkar, A.; Ling, L.; Hingorani, A.; Johansen, E.; Clement, A.; Zhang, X.; Hartman, J.; Fischbach, C.; Lin, H.; Cerione, R.A.; et al. Loss of Sirtuin 1 Alters the Secretome of Breast Cancer Cells by Impairing Lysosomal Integrity. Dev. Cell 2019, 49, 393–408.e7. [Google Scholar] [CrossRef]
- Mahmud, Z.; Gomes, A.R.; Lee, H.J.; Aimjongjun, S.; Jiramongkol, Y.; Yao, S.; Zona, S.; Alasiri, G.; Gong, G.; Yagüe, E.; et al. EP300 and SIRT1/6 Co-Regulate Lapatinib Sensitivity Via Modulating FOXO3-Acetylation and Activity in Breast Cancer. Cancers 2019, 11, 1067. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Qin, Q.; Chen, R.; Wei, C.; Mo, Q. SIRT1 promotes proliferation, migration, and invasion of breast cancer cell line MCF-7 by upregulating DNA polymerase delta1 (POLD1). Biochem. Biophys. Res. Commun. 2018, 502, 351–357. [Google Scholar] [CrossRef]
- Jin, X.; Wei, Y.; Xu, F.; Zhao, M.; Dai, K.; Shen, R.; Yang, S.; Zhang, N. SIRT1 promotes formation of breast cancer through modulating Akt activity. J. Cancer 2018, 9, 2012–2023. [Google Scholar] [CrossRef] [Green Version]
- Kuo, S.J.; Lin, H.-Y.; Chien, S.-Y.; Chen, D.-R. SIRT1 suppresses breast cancer growth through downregulation of the Bcl-2 protein. Oncol. Rep. 2013, 30, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holloway, K.R.; Barbieri, A.; Malyarchuk, S.; Saxena, M.; Nedeljkovic-Kurepa, A.; Mehl, M.C.; Wang, A.; Gu, X.; Pruitt, K. SIRT1 positively regulates breast cancer associated human aromatase (CYP19A1) expression. Mol. Endocrinol. 2013, 27, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Santolla, M.F.; Avino, S.; A Pellegrino, M.; De Francesco, E.M.; De Marco, P.; Lappano, R.; Vivacqua, A.; Cirillo, F.; Rigiracciolo, D.C.; Scarpelli, A.; et al. SIRT1 is involved in oncogenic signaling mediated by GPER in breast cancer. Cell Death Dis. 2015, 6, e1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, G.E.J.; Pandey, S.; Nedeljkovic-Kurepa, A.; Saxena, M.; Wang, A.; Pruitt, K. Frizzled 7 expression is positively regulated by SIRT1 and β-catenin in breast cancer cells. PLoS ONE 2014, 9, e98861. [Google Scholar] [CrossRef]
- Moore, R.L.; Faller, D.V. SIRT1 represses estrogen-signaling, ligand-independent ERα-mediated transcription, and cell proliferation in estrogen-responsive breast cells. J. Endocrinol. 2013, 216, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.-S.; Hyun, C.L.; Park, I.A.; Kim, J.Y.; Chung, Y.R.; Im, S.-A.; Lee, K.-H.; Moon, H.-G.; Ryu, H.S. SIRT1 induces tumor invasion by targeting epithelial mesenchymal transition-related pathway and is a prognostic marker in triple negative breast cancer. Tumour Biol. 2016, 37, 4743–4753. [Google Scholar] [CrossRef]
- Gollavilli, N.; Kanugula, A.K.; Koyyada, R.; Karnewar, S.; Neeli, P.K.; Kotamraju, S. AMPK inhibits MTDH expression via GSK3β and SIRT1 activation: Potential role in triple negative breast cancer cell proliferation. FEBS J. 2015, 282, 3971–3985. [Google Scholar] [CrossRef]
- Yao, Y.; Liu, T.; Wang, X.; Zhang, D. The Contrary Effects of Sirt1 on MCF7 Cells Depend on CD36 Expression Level. J. Surg. Res. 2019, 238, 248–254. [Google Scholar] [CrossRef]
- Elangovan, S.; Ramachandran, S.; Venkatesan, N.; Ananth, S.; Gnana-Prakasam, J.P.; Martin, P.M.; Browning, D.D.; Schoenlein, P.V.; Prasad, P.D.; Ganapathy, V.; et al. SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor α in breast cancer. Cancer Res. 2011, 71, 6654–6664. [Google Scholar] [CrossRef] [Green Version]
- Fiskus, W.; Coothankandaswamy, V.; Chen, J.; Ma, H.; Ha, K.; Saenz, D.T.; Krieger, S.S.; Mill, C.P.; Sun, B.; Huang, P.; et al. SIRT2 Deacetylates and Inhibits the Peroxidase Activity of Peroxiredoxin-1 to Sensitize Breast Cancer Cells to Oxidant Stress-Inducing Agents. Cancer Res. 2016, 76, 5467–5478. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Ni, T.K.; Wronski, A.; Glass, B.; Skibinski, A.; Beck, A.; Kuperwasser, C. The SIRT2 Deacetylase Stabilizes Slug to Control Malignancy of Basal-like Breast Cancer. Cell Rep. 2016, 17, 1302–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Vassilopoulos, A.; Wang, R.-H.; Lahusen, T.; Xiao, Z.; Xu, X.; Li, C.; Veenstra, T.D.; Li, B.; Yu, H.; et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011, 20, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Ozden, O.; Liu, G.; Song, H.Y.; Zhu, Y.; Yan, Y.; Zou, X.; Kang, H.-J.; Jiang, H.; Principe, D.R.; et al. SIRT2-Mediated Deacetylation and Tetramerization of Pyruvate Kinase Directs Glycolysis and Tumor Growth. Cancer Res. 2016, 76, 3802–3812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minten, E.V.; Kapoor-Vazirani, P.; Li, C.; Zhang, H.; Balakrishnan, K.; Yu, D.S. SIRT2 promotes BRCA1-BARD1 heterodimerization through deacetylation. Cell Rep. 2021, 34, 108921. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Liu, J.; Guo, M.; Gao, X.; Wu, X.; Bai, N.; Guo, W.; Li, N.; Yi, F.; Cheng, R.; et al. The NAD-dependent deacetylase SIRT2 regulates T cell differentiation involved in tumor immune response. Int. J. Biol. Sci. 2020, 16, 3075–3084. [Google Scholar] [CrossRef]
- Torrens-Mas, M.; Pons, D.G.; Sastre-Serra, J.; Oliver, J.; Roca, P. SIRT3 Silencing Sensitizes Breast Cancer Cells to Cytotoxic Treatments Through an Increment in ROS Production. J. Cell Biochem. 2017, 118, 397–406. [Google Scholar] [CrossRef]
- Zou, X.; Zhu, Y.; Park, S.-H.; Liu, G.; O’Brien, J.; Jiang, H.; Gius, D. SIRT3-Mediated Dimerization of IDH2 Directs Cancer Cell Metabolism and Tumor Growth. Cancer Res. 2017, 77, 3990–3999. [Google Scholar] [CrossRef] [Green Version]
- Pinterić, M.; Podgorski, I.I.; Sobočanec, S.; Hadžija, M.P.; Paradžik, M.; Dekanić, A.; Marinović, M.; Halasz, M.; Belužić, R.; Davidović, G.; et al. De novo expression of transfected sirtuin 3 enhances susceptibility of human MCF-7 breast cancer cells to hyperoxia treatment. Free Radic Res. 2018, 52, 672–684. [Google Scholar] [CrossRef] [Green Version]
- Podgorski, I.I.; Pinterić, M.; Marčinko, D.; Hadžija, M.P.; Filić, V.; Ciganek, I.; Pleše, D.; Balog, T.; Sobočanec, S. Combination of sirtuin 3 and hyperoxia diminishes tumorigenic properties of MDA-MB-231 cells. Life Sci. 2020, 254, 117812. [Google Scholar] [CrossRef]
- Pinterić, M.; Podgorski, I.I.; Hadžija, M.P.; Filić, V.; Paradžik, M.; Proust, B.L.J.; Dekanić, A.; Ciganek, I.; Pleše, D.; Marčinko, D.; et al. Sirt3 Exerts Its Tumor-Suppressive Role by Increasing p53 and Attenuating Response to Estrogen in MCF-7 Cells. Antioxidants 2020, 9, 294. [Google Scholar] [CrossRef] [Green Version]
- Zu, Y.; Chen, X.-F.; Li, Q.; Zhang, S.-T.; Si, L.-N. PGC-1α activates SIRT3 to modulate cell proliferation and glycolytic metabolism in breast cancer. Neoplasma 2021, 68, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Li, J.; Fu, L.; Gai, J.; Guan, J.; Li, Q. SIRT4 enhances the sensitivity of ER-positive breast cancer to tamoxifen by inhibiting the IL-6/STAT3 signal pathway. Cancer Med. 2019, 8, 7086–7097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, D.; Li, J.; Su, Y.; Liu, S.; Lei, Q.-Y.; Yin, M. Deacetylation of MTHFD2 by SIRT4 senses stress signal to inhibit cancer cell growth by remodeling folate metabolism. J. Mol. Cell Biol. 2022, 14, mjac020. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Liu, X.; Ren, Y.; Li, J.; Li, P.; Jiao, Q.; Meng, P.; Wang, F.; Wang, Y.; Wang, Y.-S.; et al. Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells. Theranostics 2020, 10, 9458–9476. [Google Scholar] [CrossRef]
- Polletta, L.; Vernucci, E.; Carnevale, I.; Arcangeli, T.; Rotili, D.; Palmerio, S.; Steegborn, C.; Nowak, T.; Schutkowski, M.; Pellegrini, L.; et al. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy 2015, 11, 253–270. [Google Scholar] [CrossRef] [Green Version]
- Greene, K.S.; Lukey, M.J.; Wang, X.; Blank, B.; Druso, J.E.; Lin, M.-C.J.; Stalnecker, C.A.; Zhang, C.; Abril, Y.N.; Erickson, J.W.; et al. SIRT5 stabilizes mitochondrial glutaminase and supports breast cancer tumorigenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 26625–26632. [Google Scholar] [CrossRef]
- Fernandez, I.; Abril, Y.N.; Hong, J.Y.; Yang, M.; Sadhukhan, S.; Lin, H.; Weiss, R. Abstract 4806: SIRT5 inhibition causes increased oxidative stress and impairs tumor progression and metastasis. Cancer Res. 2020, 80, 4806. [Google Scholar] [CrossRef]
- Sociali, G.; Grozio, A.; Caffa, I.; Schuster, S.; Becherini, P.; Damonte, P.; Sturla, L.; Fresia, C.; Passalacqua, M.; Mazzola, F.; et al. SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells. FASEB J. 2019, 33, 3704–3717. [Google Scholar] [CrossRef]
- Ioris, R.M.; Galié, M.; Ramadori, G.; Anderson, J.G.; Charollais, A.; Konstantinidou, G.; Brenachot, X.; Aras, E.; Goga, A.; Ceglia, N.; et al. SIRT6 Suppresses Cancer Stem-like Capacity in Tumors with PI3K Activation Independently of Its Deacetylase Activity. Cell Rep. 2017, 18, 1858–1868. [Google Scholar] [CrossRef]
- Becherini, P.; Caffa, I.; Piacente, F.; Damonte, P.; Vellone, V.G.; Passalacqua, M.; Benzi, A.; Bonfiglio, T.; Reverberi, D.; Khalifa, A.; et al. SIRT6 enhances oxidative phosphorylation in breast cancer and promotes mammary tumorigenesis in mice. Cancer Metab. 2021, 9, 6. [Google Scholar] [CrossRef]
- Tang, X.; Shi, L.; Xie, N.; Liu, Z.; Qian, M.; Meng, F.; Xu, Q.; Zhou, M.; Cao, X.; Zhu, W.-G.; et al. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat. Commun. 2017, 8, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Hu, Y.; Shao, L.; Zhuang, J.; Huo, Q.; He, S.; Chen, S.; Wang, J.; Xie, N. SIRT7 interacts with TEK (TIE2) to promote adriamycin induced metastasis in breast cancer. Cell Oncol. 2021, 44, 1405–1424. [Google Scholar] [CrossRef] [PubMed]
- Lovaas, J.D.; Zhu, L.; Chiao, C.Y.; Byles, V.; Faller, D.V.; Dai, Y. SIRT1 enhances matrix metalloproteinase-2 expression and tumor cell invasion in prostate cancer cells. Prostate 2013, 73, 522–530. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Li, J.; Zheng, F.; Ouyang, Y.; Chen, X.; Zhang, L.; Chen, Y.; Wang, L.; Mu, S.; Zhang, H. Effect of SIRT1 Gene on Epithelial-Mesenchymal Transition of Human Prostate Cancer PC-3 Cells. Med. Sci. Monit. 2016, 22, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Byles, V.; Zhu, L.; Lovaas, J.D.; Chmilewski, L.K.; Wang, J.; Faller, D.V.; Dai, Y. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene 2012, 31, 4619–4629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung-Hynes, B.; Nihal, M.; Zhong, W.; Ahmad, N. Role of sirtuin histone deacetylase SIRT1 in prostate cancer. A target for prostate cancer management via its inhibition? J. Biol. Chem. 2009, 284, 3823–3832. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Rivas, P.; Bedolla, R.; Thapa, D.; Reddick, R.L.; Ghosh, R.; Kumar, A.P. Dietary resveratrol prevents development of high-grade prostatic intraepithelial neoplastic lesions: Involvement of SIRT1/S6K axis. Cancer Prev. Res. 2013, 6, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Kokura, K.; Izumi, V.; Koomen, J.M.; Seto, E.; Chen, J.; Fang, J. MPP8 and SIRT1 crosstalk in E-cadherin gene silencing and epithelial-mesenchymal transition. EMBO Rep. 2015, 16, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Ngo, D.; Forman, L.W.; Qin, D.C.; Jacob, J.; Faller, D.V. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol. Endocrinol. 2007, 21, 1807–1821. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.; Liu, M.; Sauve, A.A.; Jiao, X.; Zhang, X.; Wu, X.; Powell, M.J.; Yang, T.; Gu, W.; Avantaggiati, M.L.; et al. Hormonal control of androgen receptor function through SIRT1. Mol. Cell Biol. 2006, 26, 8122–8135. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.B.; Thapa, D.; Munoz, A.R.; Hussain, S.S.; Yang, X.; Bedolla, R.G.; Osmulski, P.; Gaczynska, M.E.; Lai, Z.; Chiu, Y.-C.; et al. Androgen deprivation-induced elevated nuclear SIRT1 promotes prostate tumor cell survival by reactivation of AR signaling. Cancer Lett. 2021, 505, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Wang, N.; Chen, Q.; Xu, J.; Cheng, W.; Di, M.; Xia, W.; Gao, W.-Q. SIRT3 inhibits prostate cancer by destabilizing oncoprotein c-MYC through regulation of the PI3K/Akt pathway. Oncotarget 2015, 6, 26494–26507. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Quan, Y.; Xia, W. SIRT3 inhibits prostate cancer metastasis through regulation of FOXO3A by suppressing Wnt/β-catenin pathway. Exp. Cell Res. 2018, 364, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.; Singh, C.K.; Chhabra, G.; Nihal, M.; Iczkowski, K.A.; Ahmad, N. Abstract 539: Pro-proliferative function of the histone deacetylase SIRT3 in prostate cancer. Cancer Res. 2018, 78, 539. [Google Scholar] [CrossRef]
- Sawant Dessai, A.; Dominguez, M.P.; Chen, U.-I.; Hasper, J.; Prechtl, C.; Yu, C.; Katsuta, E.; Dai, T.; Zhu, B.; Jung, S.; et al. Transcriptional Repression of SIRT3 Potentiates Mitochondrial Aconitase Activation to Drive Aggressive Prostate Cancer to the Bone. Cancer Res. 2021, 81, 50–63. [Google Scholar] [CrossRef]
- Fu, W.; Li, H.; Fu, H.; Zhao, S.; Shi, W.; Sun, M.; Li, Y. The SIRT3 and SIRT6 Promote Prostate Cancer Progression by Inhibiting Necroptosis-Mediated Innate Immune Response. J. Immunol. Res. 2020, 2020, 8820355. [Google Scholar] [CrossRef]
- Csibi, A.; Fendt, S.-M.; Li, C.; Poulogiannis, G.; Choo, A.Y.; Chapski, D.J.; Jeong, S.M.; Dempsey, J.M.; Parkhitko, A.; Morrison, T.; et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 2013, 153, 840–854. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Li, Y.; Liu, T.; Hu, B.; Li, J.; Liu, C.; Liu, T.; Li, F. Mitochondrial PAK6 inhibits prostate cancer cell apoptosis via the PAK6-SIRT4-ANT2 complex. Theranostics 2020, 10, 2571–2586. [Google Scholar] [CrossRef]
- Guan, J.; Jiang, X.; Gai, J.; Sun, X.; Zhao, J.; Li, J.; Li, Y.; Cheng, M.; Du, T.; Fu, L.; et al. Sirtuin 5 regulates the proliferation, invasion and migration of prostate cancer cells through acetyl-CoA acetyltransferase 1. J. Cell. Mol. Med. 2020, 24, 14039–14049. [Google Scholar] [CrossRef]
- Choi, S.Y.; Jeon, J.M.; Na, A.Y.; Kwon, O.K.; Bang, I.H.; Ha, Y.-S.; Bae, E.J.; Park, B.-H.; Lee, E.H.; Kwon, T.G.; et al. SIRT5 Directly Inhibits the PI3K/AKT Pathway in Prostate Cancer Cell Lines. Cancer Genom. Proteom. 2022, 19, 50–59. [Google Scholar] [CrossRef]
- Kwon, O.K.; Bang, I.H.; Choi, S.Y.; Jeon, J.M.; Na, A.-Y.; Gao, Y.; Cho, S.S.; Ki, S.H.; Choe, Y.; Lee, J.N.; et al. SIRT5 Is the desuccinylase of LDHA as novel cancer metastatic stimulator in aggressive prostate cancer. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xie, Q.R.; Wang, B.; Shao, J.; Zhang, T.; Liu, T.; Huang, G.; Xia, W. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein. Cell 2013, 4, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wong, A.S.; Xia, W. Abstract 1151: SIRT6 induces EMT and promotes cancer cell invasion and migration in prostate cancer. Cancer Res. 2014, 74, 1151. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, R.; Song, L.-D.; Zhu, L.-F.; Zhan, J.-F. SIRT6 Promotes the Progression of Prostate Cancer via Regulating the Wnt/β-Catenin Signaling Pathway. J. Oncol. 2022, 2022, 2174758. [Google Scholar] [CrossRef] [PubMed]
- Haider, R.; Massa, F.; Kaminski, L.; Clavel, S.; Djabari, Z.; Robert, G.; Laurent, K.; Michiels, J.-F.; Durand, M.; Ricci, J.-E.; et al. Sirtuin 7: A new marker of aggressiveness in prostate cancer. Oncotarget 2017, 8, 77309–77316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, M.; Jiang, C.-Y.; Zhang, Y.; Zhao, J.; Han, B.-M.; Xia, S.-J. SIRT7 depletion inhibits cell proliferation and androgen-induced autophagy by suppressing the AR signaling in prostate cancer. J. Exp. Clin. Cancer Res. 2020, 39, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Villanova, L.; Tanaka, S.; Aonuma, M.; Roy, N.; Berber, E.; Pollack, J.R.; Michishita-Kioi, E.; Chua, K.F. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci. Rep. 2015, 5, 9841. [Google Scholar] [CrossRef] [Green Version]
- Deus, C.M.; Serafim, T.L.; Magalhães-Novais, S.; Vilaça, A.; Moreira, A.C.; Sardão, V.A.; Cardoso, S.M.; Oliveira, P.J. Sirtuin 1-dependent resveratrol cytotoxicity and pro-differentiation activity on breast cancer cells. Arch. Toxicol. 2017, 91, 1261–1278. [Google Scholar] [CrossRef]
- Kala, R.; Shah, H.N.; Martin, S.L.; Tollefsbol, T.O. Epigenetic-based combinatorial resveratrol and pterostilbene alters DNA damage response by affecting SIRT1 and DNMT enzyme expression, including SIRT1-dependent γ-H2AX and telomerase regulation in triple-negative breast cancer. BMC Cancer 2015, 15, 672. [Google Scholar] [CrossRef] [Green Version]
- Kugel, S.; Feldman, J.L.; Klein, M.A.; Silberman, D.M.; Sebastian, C.; Mermel, C.; Dobersch, S.; Clark, A.R.; Getz, G.; Denu, J.M.; et al. Identification of and Molecular Basis for SIRT6 Loss-of-Function Point Mutations in Cancer. Cell Rep. 2015, 13, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Li, C.; Zhang, X. The promoter methylation status and mRNA expression levels of CTCF and SIRT6 in sporadic breast cancer. DNA Cell Biol. 2014, 33, 581–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirumurthi, U.; Shen, J.; Xia, W.; LaBaff, A.M.; Wei, Y.; Li, C.-W.; Chang, W.-C.; Chen, C.-H.; Lin, H.-K.; Yu, D.; et al. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci. Signal 2014, 7, ra71. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, S.; Zhao, X.; Lai, S.; Yuan, Z.; Zhan, Y.; Ni, K.; Liu, Z.; Liu, L.; Xin, R.; et al. Clinicopathological and prognostic value of SIRT6 in patients with solid tumors: A meta-analysis and TCGA data review. Cancer Cell Int. 2022, 22, 84. [Google Scholar] [CrossRef] [PubMed]
- Aljada, A.; Saleh, A.M.; Alkathiri, M.; Shamsa, H.B.; Al-Bawab, A.; Nasr, A. Altered Sirtuin 7 Expression is Associated with Early Stage Breast Cancer. Breast Cancer 2015, 9, 3–8. [Google Scholar] [CrossRef]
- Huo, Q.; Li, Z.; Cheng, L.; Yang, F.; Xie, N. SIRT7 Is a Prognostic Biomarker Associated with Immune Infiltration in Luminal Breast Cancer. Front. Oncol. 2020, 10, 621. [Google Scholar] [CrossRef]
- Thiery, J. Epithelial—Mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Drescher, F.; Juárez, P.; Arellano, D.L.; Serafín-Higuera, N.; Olvera-Rodriguez, F.; Jiménez, S.; Licea-Navarro, A.F.; Fournier, P.G. TIE2 Induces Breast Cancer Cell Dormancy and Inhibits the Development of Osteolytic Bone Metastases. Cancers 2020, 12, 868. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.J.; Engers, R.; Florl, A.R.; Otte, A.P.; Muller, M.; Schulz, W.A. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol. Ther. 2007, 6, 1403–1412. [Google Scholar] [CrossRef] [Green Version]
- Kojima, K.; Ohhashi, R.; Fujita, Y.; Hamada, N.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem. Biophys. Res. Commun. 2008, 373, 423–428. [Google Scholar] [CrossRef]
- Nakane, K.; Fujita, Y.; Terazawa, R.; Atsumi, Y.; Kato, T.; Nozawa, Y.; Deguchi, T.; Ito, M. Inhibition of cortactin and SIRT1 expression attenuates migration and invasion of prostate cancer DU145 cells. Int. J. Urol. 2012, 19, 71–79. [Google Scholar] [CrossRef]
- Yang, Y.; Hou, H.; Haller, E.M.; Nicosia, S.V.; Bai, W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005, 24, 1021–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paik, J.H.; Kollipara, R.; Chu, G.; Ji, H.; Xiao, Y.; Ding, Z.; Miao, L.; Tothova, Z.; Horner, J.W.; Carrasco, D.R.; et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 2007, 128, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chan, C.-H.; Chen, K.; Guan, X.; Lin, H.-K.; Tong, Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 2012, 31, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Mens Health 2019, 37, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Rivas, P.; Yang, X.; Lai, Z.; Chen, Y.; Schadler, K.L.; Hu, M.; Reddick, R.L.; Ghosh, R.; Kumar, A.P. SIRT1 inhibition-induced senescence as a strategy to prevent prostate cancer progression. Mol. Carcinog. 2022, 61, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Noël, A.; Jost, M.; Maquoi, E. Matrix metalloproteinases at cancer tumor-host interface. Semin. Cell Dev. Biol. 2008, 19, 52–60. [Google Scholar] [CrossRef]
- Hay, N.; Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef] [Green Version]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Li, Q.; Yang, L.; Liu, L.; Cao, Q.; Li, Q. SMAD4 activates Wnt signaling pathway to inhibit granulosa cell apoptosis. Cell Death Dis. 2020, 11, 373. [Google Scholar] [CrossRef]
- Zhu, M.L.; Partin, J.V.; Bruckheimer, E.M.; Strup, S.E.; Kyprianou, N. TGF-beta signaling and androgen receptor status determine apoptotic cross-talk in human prostate cancer cells. Prostate 2008, 68, 287–295. [Google Scholar] [CrossRef]
- Shi, Y.; Han, J.J.; Tennakoon, J.B.; Mehta, F.F.; Merchant, F.; Burns, A.R.; Howe, M.K.; McDonnell, D.P.; Frigo, D.E. Androgens promote prostate cancer cell growth through induction of autophagy. Mol. Endocrinol. 2013, 27, 280–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGlynn, L.M.; Zino, S.; MacDonald, A.I.; Curle, J.; Reilly, J.E.; Mohammed, Z.M.A.; McMillan, D.C.; Mallon, E.; Payne, A.P.; Edwards, J.; et al. SIRT2: Tumour suppressor or tumour promoter in operable breast cancer? Eur. J. Cancer 2014, 50, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Zhou, M.; Yang, Y. Upregulated tumor sirtuin 2 expression correlates with reduced TNM stage and better overall survival in surgical breast cancer patients. Ir. J. Med. Sci. 2020, 189, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 2009, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Casimiro, M.C.; Casimiro, M.C.; Crosariol, M.; Loro, E.; Li, Z.; Pestell, R.G. Cyclins and cell cycle control in cancer and disease. Genes Cancer 2012, 3, 649–657. [Google Scholar] [CrossRef]
- Saeki, T.; Ouchi, M.; Ouchi, T. Physiological and oncogenic Aurora-A pathway. Int. J. Biol. Sci. 2009, 5, 758–762. [Google Scholar] [CrossRef] [Green Version]
- Schmit, T.L.; Zhong, W.; Nihal, M.; Ahmad, N. Polo-like kinase 1 (Plk1) in non-melanoma skin cancers. Cell Cycle 2009, 8, 2697–2702. [Google Scholar] [CrossRef]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef]
- Wang, S.; Wang, W.-L.; Chang, Y.-L.; Wu, C.-T.; Chao, Y.-C.; Kao, S.-H.; Yuan, A.; Lin, C.-W.; Yang, S.-C.; Chan, W.-K.; et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol. 2009, 11, 694–704. [Google Scholar] [CrossRef]
- Wu, W.S.; Heinrichs, S.; Xu, D.; Garrison, S.P.; Zambetti, G.P.; Adams, J.M.; Look, A.T. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 2005, 123, 641–653. [Google Scholar] [CrossRef] [Green Version]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Damodaran, S.; Damaschke, N.; Gawdzik, J.; Yang, B.; Shi, C.; Allen, G.O.; Huang, W.; Denu, J.; Jarrard, D. Dysregulation of Sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer 2017, 17, 874. [Google Scholar] [CrossRef] [PubMed]
- Weir, H.J.; Lane, J.D.; Balthasar, N. SIRT3: A Central Regulator of Mitochondrial Adaptation in Health and Disease. Genes Cancer 2013, 4, 118–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcus, J.M.; Andrabi, S.A. SIRT3 Regulation Under Cellular Stress: Making Sense of the Ups and Downs. Front. Neurosci. 2018, 12, 799. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.; Xie, S.; Qiu, X.; Mohrin, M.; Shin, J.; Liu, Y.; Zhang, D.; Scadden, D.T.; Chen, D. SIRT3 reverses aging-associated degeneration. Cell Rep. 2013, 3, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Sidorova-Darmos, E.; Sommer, R.; Eubanks, J.H. The Role of SIRT3 in the Brain Under Physiological and Pathological Conditions. Front. Cell Neurosci. 2018, 12, 196. [Google Scholar] [CrossRef] [Green Version]
- Torrens-Mas, M.; Oliver, J.; Roca, P.; Sastre-Serra, J. SIRT3: Oncogene and Tumor Suppressor in Cancer. Cancers 2017, 9, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Patel, K.; Muldoon-Jacobs, K.; Bisht, K.S.; Aykin-Burns, N.; Pennington, J.D.; van der Meer, R.; Nguyen, P.; Savage, J.; Owens, K.M.; et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010, 17, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Finley, L.W.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.; Cardoso, S.M.; Clish, C.; et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef] [Green Version]
- Mehner, C.; Hockla, A.; Miller, E.; Ran, S.; Radisky, D.C.; Radisky, E.S. Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget 2014, 5, 2736–2749. [Google Scholar] [CrossRef] [Green Version]
- Nasrin, N.; Wu, X.; Fortier, E.; Feng, Y.; Bare, O.C.; Chen, S.; Ren, X.; Wu, Z.; Streeper, R.S.; Bordone, L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 2010, 285, 31995–32002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Zhu, G. Sirtuin-4 (SIRT4), a therapeutic target with oncogenic and tumor-suppressive activity in cancer. Onco Targets Ther. 2018, 11, 3395–3400. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.M.; Xiao, C.; Finley, L.W.; Lahusen, T.; Souza, A.L.; Pierce, K.; Li, Y.-H.; Wang, X.; Laurent, G.; German, N.J.; et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 2013, 23, 450–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.M.; Lee, A.; Lee, J.; Haigis, M.C. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J. Biol. Chem. 2014, 289, 4135–4144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyo, M.; Yamamoto, H.; Konno, M.; Colvin, H.; Nishida, N.; Koseki, J.; Kawamoto, K.; Ogawa, H.; Hamabe, A.; Uemura, M.; et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br. J. Cancer 2015, 113, 492–499. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Lin, J.; Feng, S.; Chen, X.; Huang, H.; Wang, C.; Yu, Y.; He, Y.; Han, S.; Zheng, L. SIRT4 inhibits the proliferation, migration, and invasion abilities of thyroid cancer cells by inhibiting glutamine metabolism. Onco Targets Ther. 2019, 12, 2397–2408. [Google Scholar] [CrossRef] [Green Version]
- Shi, Q.; Liu, T.; Zhang, X.; Geng, J.; He, X.; Nu, M.; Pang, D. Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer. Oncol. Lett. 2016, 12, 2606–2612. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Lin, Y.; Zhu, G. SIRT4 is upregulated in breast cancer and promotes the proliferation, migration and invasion of breast cancer cells. Int. J. Clin. Exp. Pathol. 2017, 10, 11849–11856. [Google Scholar]
- Igci, M.; Kalender, M.E.; Borazan, E.; Bozgeyik, I.; Bayraktar, R.; Bozgeyik, E.; Camci, C.; Arslan, A. High-throughput screening of Sirtuin family of genes in breast cancer. Gene 2016, 586, 123–128. [Google Scholar] [CrossRef]
- Prasad, V.V.; Gopalan, R.O. Continued use of MDA-MB-435, a melanoma cell line, as a model for human breast cancer, even in year, 2014. NPJ Breast Cancer 2015, 1, 15002. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014, 19, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Ma, X.; He, Y.; Yuan, C.; Chen, Q.; Li, G.; Chen, X. Sirtuin 5: A review of structure, known inhibitors and clues for developing new inhibitors. Sci. China Life Sci. 2017, 60, 249–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.C.; Lin, P.-M.; Lin, S.-F.; Hsu, C.-H.; Lin, H.-C.; Hu, M.-L.; Yang, M.-Y. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour. Biol. 2013, 34, 1847–1854. [Google Scholar] [CrossRef] [PubMed]
- Bartosch, C.; Monteiro-Reis, S.; Almeida-Rios, D.; Vieira, R.; Castro, A.; Moutinho, M.; Rodrigues, M.; Graça, I.; Lopes, J.M.; Jerónimo, C. Assessing sirtuin expression in endometrial carcinoma and non-neoplastic endometrium. Oncotarget 2016, 7, 1144–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temel, M.; Koç, M.N.; Ulutaş, S.; Göğebakan, B. The expression levels of the sirtuins in patients with BCC. Tumour Biol. 2016, 37, 6429–6435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, C.; Tian, Y.; Yao, Y.; Mao, J.; Wang, H.; Li, Z.; Xu, Y.; Ye, M.; Wang, L. SIRT5 Promotes Hepatocellular Carcinoma Progression by Regulating Mitochondrial Apoptosis. J. Cancer 2019, 10, 3871–3882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abril, Y.L.N.; Fernandez, I.R.; Hong, J.Y.; Chiang, Y.-L.; Kutateladze, D.A.; Zhao, Q.; Yang, M.; Hu, J.; Sadhukhan, S.; Li, B.; et al. Pharmacological and genetic perturbation establish SIRT5 as a promising target in breast cancer. Oncogene 2021, 40, 1644–1658. [Google Scholar] [CrossRef]
- Garte, S.J. The c-myc oncogene in tumor progression. Crit. Rev. Oncog. 1993, 4, 435–449. [Google Scholar]
- Chou, C.C.; Lee, K.-H.; Lai, I.-L.; Wang, D.; Mo, X.; Kulp, S.K.; Shapiro, C.L.; Chen, C.-S. AMPK reverses the mesenchymal phenotype of cancer cells by targeting the Akt-MDM2-Foxo3a signaling axis. Cancer Res. 2014, 74, 4783–4795. [Google Scholar] [CrossRef] [Green Version]
- Dehner, M.; Hadjihannas, M.; Weiske, J.; Huber, O.; Behrens, J. Wnt signaling inhibits Forkhead box O3a-induced transcription and apoptosis through up-regulation of serum- and glucocorticoid-inducible kinase 1. J. Biol. Chem. 2008, 283, 19201–19210. [Google Scholar] [CrossRef] [Green Version]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevrollier, A.; Loiseau, D.; Chabi, B.; Renier, G.; Douay, O.; Malthièry, Y.; Stepien, G. ANT2 Isoform Required for Cancer Cell Glycolysis. J. Bioenerg. Biomembr. 2005, 37, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.; Schulze, A. Cholesteryl Esters: Fueling the Fury of Prostate Cancer. Cell Metabolism 2014, 19, 350–352. [Google Scholar] [CrossRef] [Green Version]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lee, R.; Feinbaum, R.; Ambros, V. A short history of a short RNA. Cell 2004, 116, S89–S92. [Google Scholar] [CrossRef] [Green Version]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. Calin, MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res. 2011, 717, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome. Biol. 2014, 15, R57. [Google Scholar] [CrossRef] [Green Version]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Ipsaro, J.J.; Joshua-Tor, L. Joshua-Tor, From guide to target: Molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 2015, 22, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Lucas, A.S.; Wang, Z.; Liu, Y. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014, 15, S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhou, W.; Liu, Y.; Liu, T.; Li, C.; Wang, L. Oncogenic role of microRNA-532-5p in human colorectal cancer via targeting of the 5’UTR of RUNX3. Oncol. Lett. 2018, 15, 7215–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS ONE 2013, 8, e79467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, G.; Brkić, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [PubMed]
- Rasko, J.E.; Wong, J.J. Nuclear microRNAs in normal hemopoiesis and cancer. J. Hematol. Oncol. 2017, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, S.K.; Smith, M.A.; Wehrkamp, C.J.; Mohr, A.M.; Mott, J.L. MicroRNA Function in Human Diseases. Med. Epigenetics 2013, 1, 106–115. [Google Scholar] [CrossRef]
- Sumathipala, M.; Weiss, S.T. Predicting miRNA-based disease-disease relationships through network diffusion on multi-omics biological data. Sci. Rep. 2020, 10, 8705. [Google Scholar] [CrossRef]
- Angelucci, F.; Cechova, K.; Valis, M.; Kuca, K.; Zhang, B.; Hort, J. MicroRNAs in Alzheimer’s Disease: Diagnostic Markers or Therapeutic Agents? Front. Pharm. 2019, 10, 665. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, S.; Purohit, P.; Sharma, P. MicroRNAs and Diseases: Promising Biomarkers for Diagnosis and Therapeutics. Indian J. Clin. Biochem. 2019, 34, 243–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Chen, F.; Hu, T.; Peng, W.; Gu, Q.; Sun, Y. The Diverse Oncogenic and Tumor Suppressor Roles of microRNA-105 in Cancer. Front. Oncol. 2019, 9, 518. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, Y.; Sun, D.; Wang, S.; Ding, K.; Liu, M.; Zhang, Y.; Miao, Y.; Liu, H.; Zhou, F. MicroRNA-181a Functions as an Oncogene in Gastric Cancer by Targeting Caprin-1. Front. Pharm. 2018, 9, 1565. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Z. Effect of miRNAs in lung cancer suppression and oncogenesis. Open Life Sci. 2016, 11, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Hannafon, B.N.; Cai, A.; Calloway, C.L.; Xu, Y.-F.; Zhang, R.; Fung, K.-M.; Ding, W.-Q. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: Evidence from a CRISPR/Cas9 deletion study. BMC Cancer 2019, 19, 642. [Google Scholar]
- Kanchan, R.K.; Siddiqui, J.A.; Mahapatra, S.; Batra, S.K.; Nasser, M.W. microRNAs Orchestrate Pathophysiology of Breast Cancer Brain Metastasis: Advances in Therapy. Mol. Cancer 2020, 19, 29. [Google Scholar] [CrossRef] [Green Version]
- Loh, H.Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.M.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef] [Green Version]
- Maskey, N.; Li, D.; Xu, H.; Song, H.; Wu, C.; Hua, K.; Song, J.; Fang, L. MicroRNA-340 inhibits invasion and metastasis by downregulating ROCK1 in breast cancer cells. Oncol. Lett. 2017, 14, 2261–2267. [Google Scholar] [CrossRef] [Green Version]
- Hoey, C.; Ahmed, M.; Ghiam, A.F.; Vesprini, D.; Huang, X.; Commisso, K.; Commisso, A.; Ray, J.; Fokas, E.; Loblaw, D.A.; et al. Circulating miRNAs as non-invasive biomarkers to predict aggressive prostate cancer after radical prostatectomy. J. Transl. Med. 2019, 17, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidarra, D.; Constâncio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Maurício, J.; Oliveira, J.; Henrique, R.; Jerónimo, C.; et al. Circulating MicroRNAs as Biomarkers for Prostate Cancer Detection and Metastasis Development Prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, U.-G.; Yang, D.; Hsieh, J.-T. The role of microRNAs in prostate cancer progression. Transl. Androl. Urol. 2013, 2, 228–241. [Google Scholar] [PubMed]
- Richardsen, E.; Andersen, S.; Melbø-Jørgensen, C.; Rakaee, M.; Ness, N.; Al-Saad, S.; Nordby, Y.; Pedersen, M.I.; Dønnem, T.; Bremnes, R.M.; et al. MicroRNA 141 is associated to outcome and aggressive tumor characteristics in prostate cancer. Sci. Rep. 2019, 9, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009, 28, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Yarahmadi, S.; Abdolvahabi, Z.; Hesari, Z.; Tavakoli-Yaraki, M.; Yousefi, Z.; Seiri, P.; Hosseinkhani, S.; Nourbakhsh, M. Inhibition of sirtuin 1 deacetylase by miR-211-5p provides a mechanism for the induction of cell death in breast cancer cells. Gene 2019, 711, 143939. [Google Scholar] [CrossRef]
- Abdolvahabi, Z.; Nourbakhsh, M.; Hosseinkhani, S.; Hesari, Z.; Alipour, M.; Jafarzadeh, M.; Ghorbanhosseini, S.S.; Seiri, P.; Yousefi, Z.; Yarahmadi, S.; et al. MicroRNA-590-3P suppresses cell survival and triggers breast cancer cell apoptosis via targeting sirtuin-1 and deacetylation of p53. J. Cell Biochem. 2019, 120, 9356–9368. [Google Scholar] [CrossRef]
- Liang, Y.; Song, X.; Li, Y.; Sang, Y.; Zhang, N.; Zhang, H.; Liu, Y.; Duan, Y.; Chen, B.; Guo, R.; et al. A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 2018, 9, 563. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, Y.; Wang, D.; Wei, X. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol. Res. 2017, 50, 27. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yuan, L.; Luo, J.; Gao, J.; Guo, J.; Xie, X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 2013, 13, 109–117. [Google Scholar] [CrossRef]
- Eades, G.; Yao, Y.; Yang, M.; Zhang, Y.; Chumsri, S.; Zhou, Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J. Biol. Chem. 2011, 286, 25992–26002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Li, L. MicroRNA-3666 inhibits breast cancer cell proliferation by targeting sirtuin 7. Mol. Med. Rep. 2017, 16, 8493–8500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Kojima, K.; Hamada, N.; Ohhashi, R.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem. Biophys. Res. Commun. 2008, 377, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, B.; Ma, B. The UCA1/miR-204/Sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Cancer Chemother. Pharm. 2016, 78, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Ma, B.; Jiang, X.; Li, X.; Jia, Y. Astragalus Polysaccharides Inhibits Tumorigenesis and Lipid Metabolism Through miR-138-5p/SIRT1/SREBP1 Pathway in Prostate Cancer. Front. Pharmacol. 2020, 11, 598. [Google Scholar] [CrossRef]
- Shu, Y.; Ren, L.; Xie, B.; Liang, Z.; Chen, J. MiR-204 enhances mitochondrial apoptosis in doxorubicin-treated prostate cancer cells by targeting SIRT1/p53 pathway. Oncotarget 2017, 8, 97313–97322. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Sharad, S.; Petrovics, G.; Mohamed, A.; Dobi, A.; Sreenath, T.L.; Srivastava, S.; Biswas, R. Loss of miR-449a in ERG-associated prostate cancer promotes the invasive phenotype by inducing SIRT1. Oncotarget 2016, 7, 22791–22806. [Google Scholar] [CrossRef] [Green Version]
- Ramalinga, M.; Roy, A.; Srivastava, A.; Bhattarai, A.; Harish, V.; Suy, S.; Collins, S.; Kumar, D. MicroRNA-212 negatively regulates starvation induced autophagy in prostate cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis and cellular senescence. Oncotarget 2015, 6, 34446–34457. [Google Scholar] [CrossRef] [Green Version]
- Duan, K.; Ge, Y.-C.; Zhang, X.-P.; Wu, S.-Y.; Feng, J.-S.; Chen, S.-L.; Zhang, L.; Yuan, Z.-H.; Fu, C.-H. miR-34a inhibits cell proliferation in prostate cancer by downregulation of SIRT1 expression. Oncol. Lett. 2015, 10, 3223–3227. [Google Scholar] [CrossRef] [Green Version]
- Slabáková, E.; Culig, Z.; Remšík, J.; Souček, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100, Erratum in Cell Death Dis. 2018, 9, 783. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Cao, Y.; Wu, B.; Cao, Y. MicroRNA-3666 Suppresses Cell Growth in Head and Neck Squamous Cell Carcinoma Through Inhibition of PFKFB3-Mediated Warburg Effect. Onco. Targets Ther. 2020, 13, 9029–9041. [Google Scholar] [CrossRef] [PubMed]
- Shou, T.; Yang, H.; Lv, J.; Liu, D.; Sun, X. MicroRNA-3666 suppresses the growth and migration of glioblastoma cells by targeting KDM2A. Mol. Med. Rep. 2019, 19, 1049–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, P.; Liu, K.; Lin, Q.; Yang, W.; Liu, D.; Lin, Z.; Shao, W.; Ji, T. Sirtuin 7 promotes glioma proliferation and invasion through activation of the ERK/STAT3 signaling pathway. Oncol. Lett. 2019, 17, 1445–1452. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, Y.-S. miR-490-3p modulates the progression of prostate cancer through regulating histone deacetylase 2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 539–546. [Google Scholar]
- Luu, H.N.; Lin, H.-Y.; Sørensen, K.D.; Ogunwobi, O.; Kumar, N.; Chornokur, G.; Phelan, C.; Jones, D.; Kidd, L.; Batra, J.; et al. miRNAs associated with prostate cancer risk and progression. BMC Urol. 2017, 17, 18. [Google Scholar]
- Nayak, B.; Khan, N.; Garg, H.; Rustagi, Y.; Singh, P.; Seth, A.; Dinda, A.K.; Kaushal, S. Role of miRNA-182 and miRNA-187 as potential biomarkers in prostate cancer and its correlation with the staging of prostate cancer. Int. Braz. J. Urol. 2020, 46, 614–623. [Google Scholar] [CrossRef]
- Wagner, S.; Ngezahayo, A.; Escobar, H.M.; Nolte, I. Role of miRNA let-7 and its major targets in prostate cancer. Biomed. Res. Int. 2014, 2014, 376326. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yang, Y.; Gan, R.; Zhao, L.; Li, W.; Zhou, H.; Wang, X.; Lü, J.; Meng, Q.H. Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS ONE 2014, 9, e98833. [Google Scholar] [CrossRef]



| Sirtuins | Mechanism/Target | Function | References |
|---|---|---|---|
| SIRT1 | Deacetylates and stabilizes PRRX1 | Inhibits breast cancer stemness and metastasis | [46] |
| SIRT1 | Upregulates V-ATPase expression and activity | Inhibits secretion of exosomes, which cause aggressiveness of breast cancer cells | [47] |
| SIRT1 | Upregulates constitutive high FOXO3 expression | Promotes drug (lapatinib) resistance of breast cancer | [48] |
| SIRT1 | Upregulates DNA POLD1 | Promotes proliferation, migration, and invasion of breast cancer | [49] |
| SIRT1 | Upregulates Akt activity | Promotes formation of breast cancer | [50] |
| SIRT1 | Downregulates Bcl-2 protein | Suppresses growth of breast cancer | [51] * |
| SIRT1 | Upregulates CYP19A1 expression | Promotes growth of estrogen-dependent breast cancer | [52] |
| SIRT1 | Prevents p53 activation and increased p21 expression | Promotes growth of breast cancer | [53] |
| SIRT1 | Upregulates expression of Frizzled 7 | Promotes proliferation of breast cancer | [54] |
| SIRT1 | Represses ERα activation via Akt deactivation | Suppresses growth of estrogen-dependent breast cancer | [55] |
| SIRT1 | Upregulates EMT-related proteins | Promotes invasion and metastasis of breast cancer | [56] |
| SIRT1 | Prevents c-Myc-mediated upregulation of MTDH | Inhibits proliferation of breast cancer | [57] |
| SIRT1 | Upregulates expression of CD36 | Inhibits proliferation of breast cancer | [58] |
| SIRT1 | Inactivates cyclin G2 and p53 | Promotes growth of estrogen-dependent breast cancer | [59] |
| SIRT2 | Deacetylates and inhibits the peroxidase activity of peroxiredoxin-1 | Suppresses growth of breast cancer | [60] |
| SIRT2 | Deacetylates K116 in the SLUG domain to stabilize slug | Promotes growth and aggressiveness of basal-like breast cancer | [61] |
| SIRT2 | Deacetylates APCCDH1 and APCCDC2 | Suppresses growth of breast cancer | [62] |
| SIRT2 | Deacetylates and activates pyruvate kinase (PKM2) | Promotes growth of breast cancer | [63] |
| SIRT2 | Deacetylates K116 in the SLUG domain to stabilize slug | Promotes growth and aggressiveness of basal-like breast cancer | [61] |
| SIRT2 | Deacetylates APCCDH1 and APCCDC2 | Suppresses growth of breast cancer | [62] |
| SIRT2 | Promotes BRCA1-BARD1 heterodimerization via deacetylation | Suppresses growth of breast cancer | [64] |
| SIRT2 | Regulates CD8+ effector memory T-cells differentiation | Induces immune response against breast cancer | [65] |
| SIRT3 | Downregulates ROS production and upregulates MnSOD, IDH2, PGC1-α, and TFAM | Increases cell viability and inhibits autophagy and apoptosis of breast cancer | [66] |
| SIRT3 | Deacetylates and increases IDH2 activities | Suppresses the growth of breast cancer | [67] |
| SIRT3 | Downregulates expression of the angiogenic gene vegfr1, EMT markers (vimentin and slug), LDHA, antioxidant genes (sod2 and cat), SIRT1 and PGC1α | Inhibits survival, proliferation, and mitochondrial function in breast cancer | [68] |
| SIRT3 | Upregulates expression of SIRT1, LDHA, and PGC-1α; increases mitochondrial ROS, induces DNA damage, timp-1 expression, formation of multinucleated cells, and apoptosis. | Improves mitochondrial function but inhibits proliferation of breast cancer | [69] |
| SIRT3 | Upregulates p53 and attenuates the response to estrogen | Suppresses growth of estrogen-dependent breast cancer | [70] |
| SIRT3 | Participates in PGC-1α suppression of glycolytic metabolism | Inhibits proliferation of breast cancer | [71] |
| SIRT4 | Downregulates IL-6, STAT3 Y705 phosphorylation as well as transcription and translation of STAT3 target genes (MYC and CNDD1) | Enhances the sensitivity of breast cancer cells to tamoxifen | [72] |
| SIRT4 | Deacetylates MTHFD2 at K50 | Inhibits growth of breast cancer | [73] |
| SIRT4 | Downregulates SIRT1 expression and stem cell markers (Oct4, Sox2, and Nanog) | Suppresses the self-renewal of breast cancer stem cells | [74] |
| SIRT5 | Regulates glutamine metabolism, suppresses LC3-II and GABARAPL2 accumulation as well as sequestosome 1 degradation | Induces mitophagy and autophagy of breast cancer | [75] |
| SIRT5 | Desuccinylates and stabilizes mitochondrial enzyme GLS | Promotes breast cancer tumorigenesis | [76] |
| SIRT5 | Reduces ROS generation and increases NADPH and GSH levels | Promotes tumor progression and metastasis of breast cancer | [77] |
| SIRT6 | Deacetylates and activates NAMPT and glucose-6-phosphate dehydrogenase activities and increases NADH levels | Promotes breast cancer survival and resistance to oxidative stress | [78] |
| SIRT6 | Downregulates PI3K signaling at the transcriptional level independent of its deacetylase activity | Suppresses progression and stemness of breast cancer | [79] |
| SIRT6 | Enhances pyruvate dehydrogenase expression and activity, oxidative phosphorylation, and ATP/AMP ratio | Promotes growth of breast cancer | [80] |
| SIRT7 | Deacetylates and promotes SMAD4 degradation mediated by β-TrCP1; downregulates TGFβ and prevents epithelial-to-mesenchymal transition | Inhibits metastasis of breast cancer | [81] |
| SIRT7 | Deacetylates the TEK promoter at H3K18 | Promotes Adriamycin-induced metastasis of breast cancer | [82] |
| Sirtuins | Mechanism/Target | Function | References |
|---|---|---|---|
| SIRT1 | Deacetylates and upregulates Matrix Metalloproteinase-2 (MMP2) expression | Promotes prostate cancer cell invasion | [83] |
| SIRT1 | Downregulates EMT-related protein (E-cadherin) and upregulates mesenchymal markers (vimentin and N-cadherin) | Promotes movement, migration, and invasion of prostate cancer cells | [84] |
| SIRT1 | Downregulates expression of epithelial marker (E-cadherin) and upregulates expression of mesenchymal markers (N-cadherin and fibronectin) and EMT-inducing transcription factor (ZEB1) | Promotes migration and metastasis of prostate cancer | [85] |
| SIRT1 | Deacetylates and deactivates p53 and FOXO1 | Promotes development of prostate cancer | [86] |
| SIRT1 | Upregulates phosphorylation of S6K and 4EBP1 | Suppresses cell proliferation and induces autophagy of prostate cancer | [87] |
| SIRT1 | Antagonizes PCAF-catalyzed MPP8-K439 acetylation | Promotes migration, invasion, and EMT of prostate cancer | [88] |
| SIRT1 | Deacetylates AR and histone H3, and suppresses AR-mediated gene transcription | Inhibits proliferation of prostate cancer | [89] |
| SIRT1 | Deacetylates AR and inhibits coactivator-induced interactions between AR amino and carboxyl termini | Inhibits proliferation of prostate cancer | [90] |
| SIRT1 | Upregulates AR signaling | Promotes progression of prostate cancer | [91] |
| SIRT2 | Not known | Not Known | Not Known |
| SIRT3 | Inhibits phosphorylation of Akt, leading to ubiquitination and degradation of oncoprotein c-Myc | Inhibits proliferation of prostate cancer | [92] |
| SIRT3 | Promotes FOXO3A expression by suppressing Wnt/β-catenin pathway; Downregulates EMT | Inhibits migration and metastasis of prostate cancer | [93] |
| SIRT2 | Not known | Not Known | Not Known |
| SIRT3 | Inhibits phosphorylation of Akt, leading to ubiquitination and degradation of oncoprotein c-Myc | Inhibits proliferation of prostate cancer | [92] |
| SIRT3 | Promotes FOXO3A expression by suppressing Wnt/β-catenin pathway; Downregulates EMT | Inhibits migration and metastasis of prostate cancer | [93] |
| SIRT3 | Inhibits cleavage of poly (ADP-ribose) polymerase (PARP) and upregulates expression of proliferating cell nuclear antigen | Promotes proliferation and suppresses apoptosis of prostate cancer | [94] |
| SIRT3 | Reduces the level of acetylated mitochondrial ACO2 | Reduces growth and survival of prostate cancer | [95] |
| SIRT3 | Inhibits RIPK3-mediated necroptosis and innate immune response | Promotes progression of prostate cancer | [96] |
| SIRT4 | Decreases glutamine uptake and metabolism | Inhibits proliferation of prostate cancer | [97] |
| SIRT4 | Deacetylates ANT2 to promote its ubiquitination and degradation | Suppresses proliferation and promotes mitochondrial-mediated apoptosis of prostate cancer | [98] |
| SIRT5 | Upregulates cyclin D1, MMP9, and MAPK signaling proteins; downregulates ACAT1 protein expression | Promotes proliferation, invasion, and migration of prostate cancer | [99] |
| SIRT5 | Inhibits the PI3K/AKT signaling | Suppresses growth and metastasis of prostate cancer | [100] |
| SIRT5 | Desuccinylates LDHA | Reduces migration and invasion of prostate cancer | [101] |
| SIRT6 | Increases Bcl-2 gene expression and induces cell cycle arrest at the sub-G1 phase | Inhibits apoptosis and promotes survival and proliferation of prostate cancer | [102] |
| SIRT6 | Downregulates E-cadherin level and upregulates N-cadherin level | Promotes migration and invasion of prostate cancer | [103] |
| SIRT6 | Inhibits RIPK3-mediated necroptosis and innate immune response | Promotes progression of prostate cancer | [96] |
| SIRT6 | Upregulates the Wnt/β-catenin signaling | Promotes progression of prostate cancer | [104] |
| SIRT7 | Upregulates expression of EMT marker (fibronectin) | Promotes aggressiveness of prostate cancer | [105] |
| SIRT7 | Upregulates AR signaling, LC3BI to LC3BII conversion, vimentin, slug, MMP2, and MMP9; downregulates SMAD4 protein expression | Promotes cell proliferation, metastasis, and androgen-induced autophagy of prostate cancer | [106] |
| SIRT7 | Downregulates EMT-related protein (E-cadherin) and upregulates mesenchymal markers (vimentin) and slug | Promotes migration and metastasis of prostate cancer | [107] |
| Sirtuins | Regulatory microRNA | Function | References |
|---|---|---|---|
| SIRT1 | miR-211-5p | Reduces cell viability and induces apoptosis of breast cancer | [206] |
| SIRT1 | miR-590-3P | Induces apoptosis and reduces survival of breast cancer | [207] |
| SIRT1 | miR-4766-5p | Suppresses cell proliferation, metastasis, and chemoresistance in breast cancer | [208] |
| SIRT1 | miR-22 | Suppresses tumorigenesis and improves radiosensitivity of breast cancer | [209] |
| SIRT1 | miR-34a | Inhibits proliferation and migration of breast cancer | [210] |
| SIRT1 | miR-200a | Prevents growth and EMT-like transformation in breast cancer | [211] |
| SIRT2 | Not Known | Not Known | Not Known |
| SIRT3 | Not Known | Not Known | Not Known |
| SIRT4 | Not Known | Not Known | Not Known |
| SIRT5 | Not Known | Not Known | Not Known |
| SIRT6 | Not Known | Not Known | Not Known |
| SIRT7 | miR-3666 | Inhibits proliferation and promotes apoptosis of breast cancer | [212] |
| Sirtuins | Regulatory microRNA | Function | References |
|---|---|---|---|
| SIRT1 | miR-34a | Reduces growth and chemoresistance of prostate cancer | [213] |
| SIRT1 | miR-204 | Enhances docetaxel-induced apoptosis of prostate cancer | [214] |
| SIRT1 | miR-138-5p | Inhibits proliferation and lipid metabolism of prostate cancer | [215] |
| SIRT1 | miR-204 | Enhances doxorubicin-induced mitochondrial-mediated apoptosis of prostate cancer | [216] |
| SIRT1 | miR-449a | Suppresses invasiveness of prostate cancer | [217] |
| SIRT1 | miR-212 | Inhibits starvation- and SIRT1-induced autophagy of prostate cancer | [218] |
| SIRT1 | miR-34a | Inhibits proliferation of prostate cancer | [219] |
| SIRT2 | Not known | Not Known | Not Known |
| SIRT3 | Not Known | Not Known | Not Known |
| SIRT4 | Not Known | Not Known | Not Known |
| SIRT5 | Not Known | Not Known | Not Known |
| SIRT6 | Not Known | Not Known | Not Known |
| SIRT7 | Not Known | Not Known | Not Known |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onyiba, C.I.; Scarlett, C.J.; Weidenhofer, J. The Mechanistic Roles of Sirtuins in Breast and Prostate Cancer. Cancers 2022, 14, 5118. https://doi.org/10.3390/cancers14205118
Onyiba CI, Scarlett CJ, Weidenhofer J. The Mechanistic Roles of Sirtuins in Breast and Prostate Cancer. Cancers. 2022; 14(20):5118. https://doi.org/10.3390/cancers14205118
Chicago/Turabian StyleOnyiba, Cosmos Ifeanyi, Christopher J. Scarlett, and Judith Weidenhofer. 2022. "The Mechanistic Roles of Sirtuins in Breast and Prostate Cancer" Cancers 14, no. 20: 5118. https://doi.org/10.3390/cancers14205118
APA StyleOnyiba, C. I., Scarlett, C. J., & Weidenhofer, J. (2022). The Mechanistic Roles of Sirtuins in Breast and Prostate Cancer. Cancers, 14(20), 5118. https://doi.org/10.3390/cancers14205118







