The Role of m6A Modification and m6A Regulators in Esophageal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. m6A Modification and m6A Regulators
2.1. Methyltransferases
2.2. Demethylases
2.3. m6A RNA Binding Proteins
3. m6A Modification and Its Effect on Various RNAs in ESCA
3.1. m6A Modification in ESCA
3.2. The Effect of m6A Modification on mRNA in ESCA
3.3. The Effect of m6A Modification on Non-Coding RNAs in ESCA
4. The Role of m6A Regulators in Development, Progression, Prognosis and Treatment of ESCA
4.1. m6A Regulators and EAC
4.2. m6A Regulators and ESCA (Not including EAC)
4.2.1. The Expression of m6A Regulators and their Association with Clinicopathological Characteristic in ESCA
- Writers
- Erasers
- Readers
4.2.2. The Effect and Mechanism of m6A Regulators in Progression of ESCA
m6A Regulators Regulated Biological Behavior of ESCC Cells in an m6A-Dependent Way
- Writers

- Erasers

- Readers
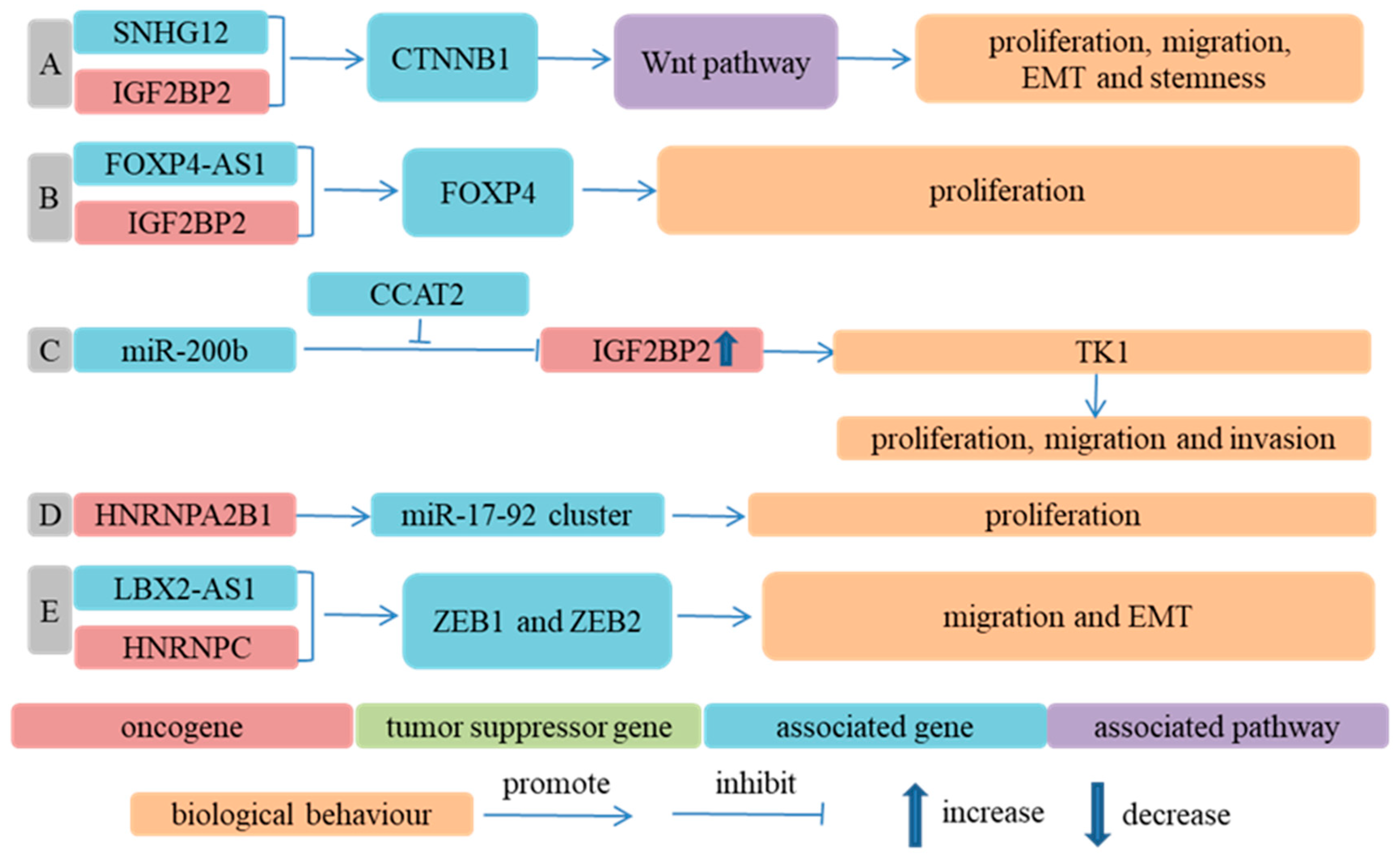
m6A Regulators Regulated Biological Behavior of ESCC Cells in an m6A-Independent Way
- Writers
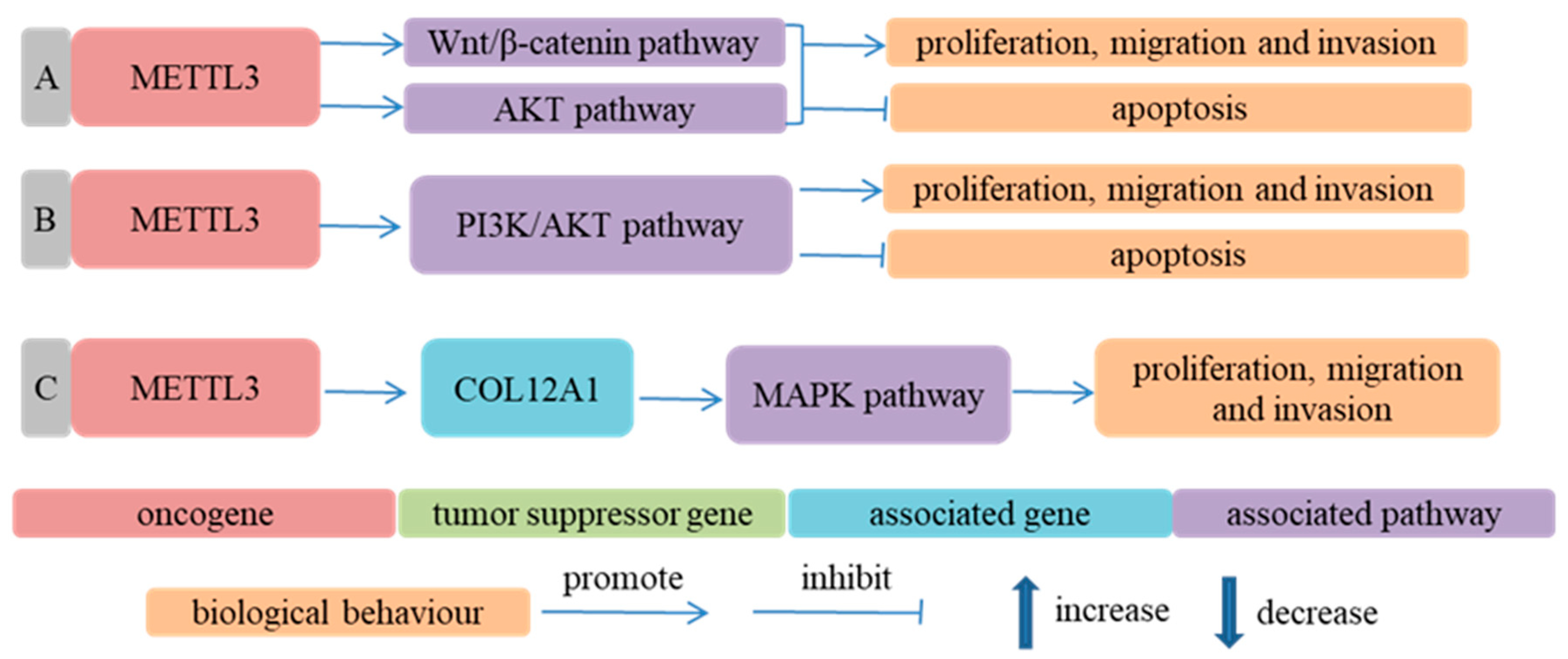
- Erasers
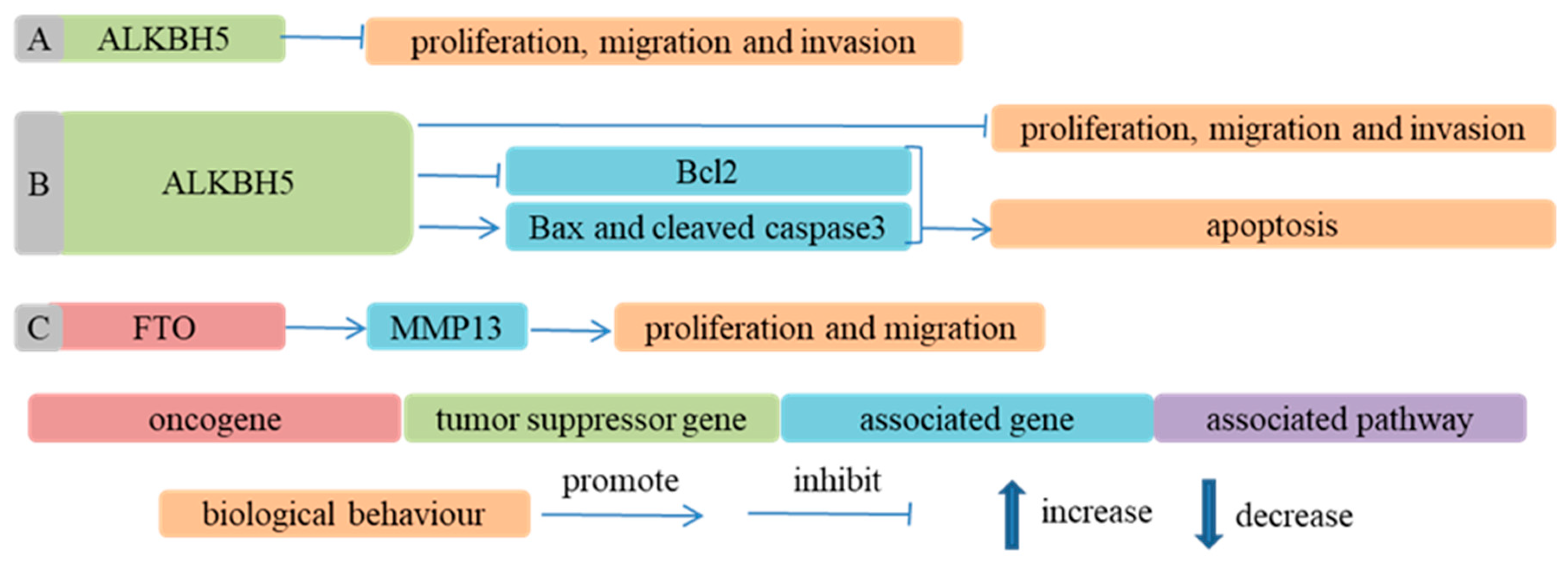
- Readers
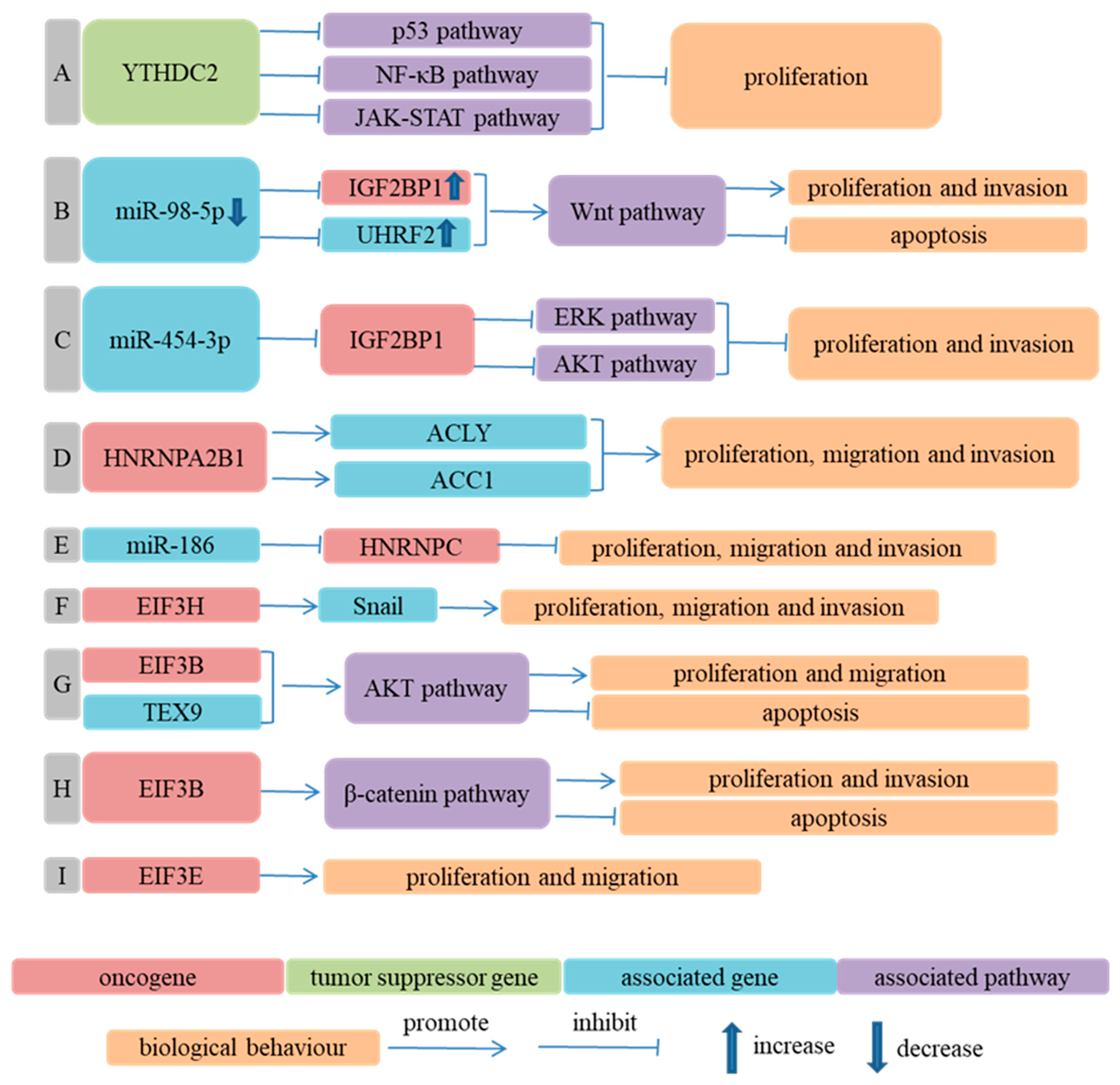
- Combined Effects of m6A Regulators in Progression of ESCA

4.2.3. The Role of m6A Regulators in Treatment of ESCA
4.2.4. The Association between Expression of m6A Regulators and Prognosis of ESCA Patients
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Krug, R.M.; Morgan, M.A.; Shatkin, A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976, 20, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beemon, K.; Keith, J. Localization of N6-methyladenosine in the Rous sarcoma virus genome. J. Mol. Biol. 1977, 113, 165–179. [Google Scholar] [CrossRef]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar] [PubMed]
- Wei, C.M.; Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977, 16, 1672–1676. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Chen, T.; Hao, Y.J.; Zhang, Y.; Li, M.M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16, 289–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Y.; Laurent, B.; Hsu, C.H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.S.; Wang, X.; Beadell, A.V.; Lu, Z.; Shi, H.; Kuuspalu, A.; Ho, R.K.; He, C. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017, 542, 475–478. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wang, L.D.; Zhou, F.Y.; Li, X.M.; Sun, L.D.; Song, X.; Jin, Y.; Li, J.M.; Kong, G.Q.; Qi, H.; Cui, J.; et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet. 2010, 42, 759–763. [Google Scholar] [CrossRef]
- Wu, C.; Hu, Z.; He, Z.; Jia, W.; Wang, F.; Zhou, Y.; Liu, Z.; Zhan, Q.; Liu, Y.; Yu, D.; et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat. Genet. 2011, 43, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Kraft, P.; Zhai, K.; Chang, J.; Wang, Z.; Li, Y.; Hu, Z.; He, Z.; Jia, W.; Abnet, C.C.; et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat. Genet. 2012, 44, 1090–1097. [Google Scholar] [CrossRef]
- Zhou, R.M.; Li, Y.; Wang, N.; Huang, X.; Cao, S.R.; Shan, B.E. Association of programmed death-1 polymorphisms with the risk and prognosis of esophageal squamous cell carcinoma. Cancer Genet. 2016, 209, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Li, Y.; Wang, N.; Niu, C.; Huang, X.; Cao, S.; Huo, X. PARP1 rs1136410 C/C genotype associated with an increased risk of esophageal cancer in smokers. Mol. Biol. Rep. 2021, 48, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Grady, W.M.; Yu, M.; Markowitz, S.D. Epigenetic Alterations in the Gastrointestinal Tract: Current and Emerging Use for Biomarkers of Cancer. Gastroenterology 2021, 160, 690–709. [Google Scholar] [CrossRef]
- Cao, W.; Lee, H.; Wu, W.; Zaman, A.; McCorkle, S.; Yan, M.; Chen, J.; Xing, Q.; Sinnott-Armstrong, N.; Xu, H.; et al. Multi-faceted epigenetic dysregulation of gene expression promotes esophageal squamous cell carcinoma. Nat. Commun. 2020, 11, 3675. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schöller, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 2018, 24, 499–512. [Google Scholar] [CrossRef] [Green Version]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Tang, H.W.; Li, J.; Perrimon, N.; Yan, D. Xio is a component of the Drosophila sex determination pathway and RNA N(6)-methyladenosine methyltransferase complex. Proc. Natl. Acad. Sci. USA 2018, 115, 3674–3679. [Google Scholar] [CrossRef] [Green Version]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e1026. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Kan, L.; Grozhik, A.V.; Vedanayagam, J.; Patil, D.P.; Pang, N.; Lim, K.S.; Huang, Y.C.; Joseph, B.; Lin, C.J.; Despic, V.; et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat. Commun. 2017, 8, 15737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef]
- Shima, H.; Matsumoto, M.; Ishigami, Y.; Ebina, M.; Muto, A.; Sato, Y.; Kumagai, S.; Ochiai, K.; Suzuki, T.; Igarashi, K. S-Adenosylmethionine Synthesis Is Regulated by Selective N(6)-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 2017, 21, 3354–3363. [Google Scholar] [CrossRef] [Green Version]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 2019, 363, eaav0080. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, M.; Li, K.; Bai, D.; Yi, C. Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 2019, 29, 80–82. [Google Scholar] [CrossRef] [Green Version]
- van Tran, N.; Ernst, F.G.M.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef] [Green Version]
- Pinto, R.; Vågbø, C.B.; Jakobsson, M.E.; Kim, Y.; Baltissen, M.P.; O’Donohue, M.F.; Guzmán, U.H.; Małecki, J.M.; Wu, J.; Kirpekar, F.; et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020, 48, 830–846. [Google Scholar] [CrossRef]
- Ma, H.; Wang, X.; Cai, J.; Dai, Q.; Natchiar, S.K.; Lv, R.; Chen, K.; Lu, Z.; Chen, H.; Shi, Y.G.; et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019, 15, 88–94. [Google Scholar] [CrossRef]
- Fedeles, B.I.; Singh, V.; Delaney, J.C.; Li, D.; Essigmann, J.M. The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. J. Biol. Chem. 2015, 290, 20734–20742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, G.; Yang, C.G.; Yang, S.; Jian, X.; Yi, C.; Zhou, Z.; He, C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008, 582, 3313–3319. [Google Scholar] [CrossRef] [Green Version]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m(6)A(m) in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Wang, Z.; Zhou, N.; Li, G.; Kou, Y.; Luo, Y.; Wang, Y.; Yang, J.; Tian, F. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 2019, 18, 168. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Gu, J.; Jin, Y.; Yuan, Q.; Ma, G.; Du, M.; Ge, Y.; Qin, C.; Lv, Q.; Fu, G.; et al. Genetic variants in N6-methyladenosine are associated with bladder cancer risk in the Chinese population. Arch. Toxicol. 2021, 95, 299–309. [Google Scholar] [CrossRef]
- Tian, J.; Ying, P.; Ke, J.; Zhu, Y.; Yang, Y.; Gong, Y.; Zou, D.; Peng, X.; Yang, N.; Wang, X.; et al. ANKLE1 N(6) -Methyladenosine-related variant is associated with colorectal cancer risk by maintaining the genomic stability. Int. J. Cancer 2020, 146, 3281–3293. [Google Scholar] [CrossRef]
- Chen, X.; Huang, L.; Yang, T.; Xu, J.; Zhang, C.; Deng, Z.; Yang, X.; Liu, N.; Chen, S.; Lin, S. METTL3 Promotes Esophageal Squamous Cell Carcinoma Metastasis through Enhancing GLS2 Expression. Front. Oncol. 2021, 11, 667451. [Google Scholar] [CrossRef]
- Han, H.; Yang, C.; Zhang, S.; Cheng, M.; Guo, S.; Zhu, Y.; Ma, J.; Liang, Y.; Wang, L.; Zheng, S.; et al. METTL3-mediated m(6)A mRNA modification promotes esophageal cancer initiation and progression via Notch signaling pathway. Mol. Ther.-Nucleic Acids 2021, 26, 333–346. [Google Scholar] [CrossRef]
- Li, R.; Zeng, L.; Zhao, H.; Deng, J.; Pan, L.; Zhang, S.; Wu, G.; Ye, Y.; Zhang, J.; Su, J.; et al. ATXN2-mediated translation of TNFR1 promotes esophageal squamous cell carcinoma via m(6)A-dependent manner. Mol. Ther. J. Am. Soc. Gene Ther. 2022, 30, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Nagaki, Y.; Motoyama, S.; Yamaguchi, T.; Hoshizaki, M.; Sato, Y.; Sato, T.; Koizumi, Y.; Wakita, A.; Kawakita, Y.; Imai, K.; et al. m(6) A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells Devoted Mol. Cell. Mech. 2020, 25, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Dong, M.; Wang, Z.; Wan, J.; Xie, Y.; Jiao, Y.; Yan, D. Long non-coding RNA CASC15 facilitates esophageal squamous cell carcinoma tumorigenesis via decreasing SIM2 stability via FTO-mediated demethylation. Oncol. Rep. 2021, 45, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; He, X.; Wang, W.; Hu, X.; Wang, K.; Wang, M. Long noncoding RNA SNHG12 induces proliferation, migration, epithelial-mesenchymal transition, and stemness of esophageal squamous cell carcinoma cells via post-transcriptional regulation of BMI1 and CTNNB1. Mol. Oncol. 2020, 14, 2332–2351. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, T.; Yang, Y.; Kang, W.; Dong, S.; Cheng, S. YY1-induced upregulation of FOXP4-AS1 and FOXP4 promote the proliferation of esophageal squamous cell carcinoma cells. Cell Biol. Int. 2020, 44, 1447–1457. [Google Scholar] [CrossRef]
- Wu, X.; Fan, Y.; Liu, Y.; Shen, B.; Lu, H.; Ma, H. Long Non-Coding RNA CCAT2 Promotes the Development of Esophageal Squamous Cell Carcinoma by Inhibiting miR-200b to Upregulate the IGF2BP2/TK1 Axis. Front. Oncol. 2021, 11, 680642. [Google Scholar] [CrossRef]
- Qian, L.X.; Cao, X.; Du, M.Y.; Ma, C.X.; Zhu, H.M.; Peng, Y.; Hu, X.Y.; He, X.; Yin, L. KIF18A knockdown reduces proliferation, migration, invasion and enhances radiosensitivity of esophageal cancer. Biochem. Biophys. Res. Commun. 2021, 557, 192–198. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Pan, T.; Wang, H.; Zhang, Y.; Li, C. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem. Biophys. Res. Commun. 2019, 511, 566–572. [Google Scholar] [CrossRef]
- Wang, W.; Shao, F.; Yang, X.; Wang, J.; Zhu, R.; Yang, Y.; Zhao, G.; Guo, D.; Sun, Y.; Wang, J.; et al. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N(6)-methyladenosine-dependent YTHDF binding. Nat. Commun. 2021, 12, 3803. [Google Scholar] [CrossRef]
- Huang, G.W.; Chen, Q.Q.; Ma, C.C.; Xie, L.H.; Gu, J. linc01305 promotes metastasis and proliferation of esophageal squamous cell carcinoma through interacting with IGF2BP2 and IGF2BP3 to stabilize HTR3A mRNA. Int. J. Biochem. Cell Biol. 2021, 136, 106015. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Z.; Shi, W.; Shen, J.; Guan, Y.; Ni, F. HLA complex P5 upregulation is correlated with poor prognosis and tumor progression in esophageal squamous cell carcinoma. Bioengineered 2022, 13, 9301–9311. [Google Scholar] [CrossRef]
- Liao, L.; He, Y.; Li, S.J.; Zhang, G.G.; Yu, W.; Yang, J.; Huang, Z.J.; Zheng, C.C.; He, Q.Y.; Li, Y.; et al. Anti-HIV drug elvitegravir suppresses cancer metastasis via increased proteasomal degradation of m6A methyltransferase METTL3. Cancer Res. 2022, 82, 2444–2457. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Ge, F.; Xie, M.; Li, Z.; Zang, C.; Kong, L.; Pu, Y.; Zheng, X.; Tan, Y. FTO mediated ERBB2 demethylation promotes tumor progression in esophageal squamous cell carcinoma cells. Clin. Exp. Metastasis 2022, 39, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Yang, L.; Wang, L.; Liu, Q.; Zhang, K.; Liu, S.; Liu, C.; Gao, Q.; Li, L.; Qin, G.; et al. m6A demethylase FTO promotes tumor progression via regulation of lipid metabolism in esophageal cancer. Cell Biosci. 2022, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, Z.; Wang, L.; Zhang, S.; Ren, L.; Li, S.; Xu, J.; Lv, S. Mechanism of methyltransferase like 3 in epithelial-mesenchymal transition process, invasion, and metastasis in esophageal cancer. Bioengineered 2021, 12, 10023–10036. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, P.; Li, J.; Qi, Q.; Sun, Z.; Shi, S.; Xie, Y.; Liu, S.; Wang, Y.; Du, L.; et al. Exosomal and intracellular miR-320b promotes lymphatic metastasis in esophageal squamous cell carcinoma. Mol. Ther. Oncolytics 2021, 23, 163–180. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, K.; Gu, S.; Wang, W.; Xie, S.; Lu, T.; Li, L.; Dong, C.; Wang, X.; Zhou, Y. A methyltransferase-like 14/miR-99a-5p/tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. Clin. Transl. Med. 2021, 11, e545. [Google Scholar] [CrossRef]
- Chen, P.; Li, S.; Zhang, K.; Zhao, R.; Cui, J.; Zhou, W.; Liu, Y.; Zhang, L.; Cheng, Y. N(6)-methyladenosine demethylase ALKBH5 suppresses malignancy of esophageal cancer by regulating microRNA biogenesis and RAI1 expression. Oncogene 2021, 40, 5600–5612. [Google Scholar] [CrossRef]
- Li, K.; Chen, J.; Lou, X.; Li, Y.; Qian, B.; Xu, D.; Wu, Y.; Ma, S.; Zhang, D.; Cui, W. HNRNPA2B1 Affects the Prognosis of Esophageal Cancer by Regulating the miR-17-92 Cluster. Front. Cell Dev. Biol. 2021, 9, 658642. [Google Scholar] [CrossRef]
- Xue, J.; Xiao, P.; Yu, X.; Zhang, X. A positive feedback loop between AlkB homolog 5 and miR-193a-3p promotes growth and metastasis in esophageal squamous cell carcinoma. Hum. Cell 2021, 34, 502–514. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, C.; Zhang, S.; Yan, H.; Zhang, L.; Jiang, A.; Liu, Y.; Feng, Y.; Li, D.; Guo, Y.; et al. N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell 2021, 56, 702–715.e708. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, C.; Ma, S.; Li, Z.; Wang, W.; Li, Y.; Ma, Y.; Fang, J.; Wang, Y.; Cao, W.; et al. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. CR 2021, 40, 294. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wang, B.; Xu, K.; Nie, L.; Fu, Y.; Wang, Z.; Wang, Q.; Wang, S.; Zou, X. m(6)A Reader HNRNPA2B1 Promotes Esophageal Cancer Progression via Up-Regulation of ACLY and ACC1. Front. Oncol. 2020, 10, 553045. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhong, X.; Zhou, X.; Xie, Q.; Zhao, Z.; Guo, X.; Duan, Y. The M6A methyltransferase METTL3 regulates proliferation in esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2021, 580, 48–55. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Dong, S.; Zhang, Y.; Li, X.; Wang, J. ALKBH5 Is Lowly Expressed in Esophageal Squamous Cell Carcinoma and Inhibits the Malignant Proliferation and Invasion of Tumor Cells. Comput. Math. Methods Med. 2021, 2021, 1001446. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 19, 53. [Google Scholar] [CrossRef] [Green Version]
- Ge, L.; Zhang, N.; Chen, Z.; Song, J.; Wu, Y.; Li, Z.; Chen, F.; Wu, J.; Li, D.; Li, J.; et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin. Chem. 2020, 66, 342–351. [Google Scholar] [CrossRef]
- Huang, W.; Qi, C.B.; Lv, S.W.; Xie, M.; Feng, Y.Q.; Huang, W.H.; Yuan, B.F. Determination of DNA and RNA Methylation in Circulating Tumor Cells by Mass Spectrometry. Anal. Chem. 2016, 88, 1378–1384. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef]
- Zhu, T.; Roundtree, I.A.; Wang, P.; Wang, X.; Wang, L.; Sun, C.; Tian, Y.; Li, J.; He, C.; Xu, Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014, 24, 1493–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.S.; Ping, X.L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Lesbirel, S.; Viphakone, N.; Parker, M.; Parker, J.; Heath, C.; Sudbery, I.; Wilson, S.A. The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 2018, 8, 13827. [Google Scholar] [CrossRef] [Green Version]
- Wojtas, M.N.; Pandey, R.R.; Mendel, M.; Homolka, D.; Sachidanandam, R.; Pillai, R.S. Regulation of m(6)A Transcripts by the 3′→5′ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol. Cell 2017, 68, 374–387.e312. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pestova, T.V.; Kolupaeva, V.G.; Lomakin, I.B.; Pilipenko, E.V.; Shatsky, I.N.; Agol, V.I.; Hellen, C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 2001, 98, 7029–7036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokabe, M.; Fraser, C.S. Human eukaryotic initiation factor 2 (eIF2)-GTP-Met-tRNAi ternary complex and eIF3 stabilize the 43 S preinitiation complex. J. Biol. Chem. 2014, 289, 31827–31836. [Google Scholar] [CrossRef] [Green Version]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef] [Green Version]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Han, B.; Zhang, Z.; Wang, S.; Bai, Y.; Zhang, Y.; Tang, Y.; Du, L.; Xu, L.; Wu, F.; et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation 2020, 142, 556–574. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, Y.; Liu, N.; Lu, Y.; Ma, W.; Yang, Z.; Gan, W.; Li, D. CircMET promotes tumor proliferation by enhancing CDKN2A mRNA decay and upregulating SMAD3. Mol. Cancer 2022, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.X.; Chen, X.; Xia, L.P.; Zhang, J.X.; Pan, Z.Z.; Ma, X.D.; Han, K.; Chen, J.W.; Judde, J.G.; Deas, O.; et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.N.; Chen, Z.Y.; Chen, X.Y.; Chen, M.; Yi, Y.C.; Zhu, J.S.; Zhang, J. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer 2022, 21, 51. [Google Scholar] [CrossRef]
- Liang, L.; Zhu, Y.; Li, J.; Zeng, J.; Wu, L. ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J. Exp. Clin. Cancer Res. 2022, 41, 261. [Google Scholar] [CrossRef]
- Duan, J.L.; Chen, W.; Xie, J.J.; Zhang, M.L.; Nie, R.C.; Liang, H.; Mei, J.; Han, K.; Xiang, Z.C.; Wang, F.W.; et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer 2022, 21, 93. [Google Scholar] [CrossRef]
- Lin, C.; Ma, M.; Zhang, Y.; Li, L.; Long, F.; Xie, C.; Xiao, H.; Liu, T.; Tian, B.; Yang, K.; et al. The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer 2022, 21, 80. [Google Scholar] [CrossRef]
- Plum, P.S.; Ulase, D.; Bollschweiler, E.; Chon, S.H.; Berlth, F.; Zander, T.; Alakus, H.; Hölscher, A.H.; Bruns, C.J.; Schallenberg, S.; et al. Upregulation of insulin-like growth factor II mRNA-binding protein 3 (IMP3) has negative prognostic impact on early invasive (pT1) adenocarcinoma of the esophagus. J. Cancer Res. Clin. Oncol. 2018, 144, 1731–1739. [Google Scholar] [CrossRef]
- Burdelski, C.; Jakani-Karimi, N.; Jacobsen, F.; Möller-Koop, C.; Minner, S.; Simon, R.; Sauter, G.; Steurer, S.; Clauditz, T.S.; Wilczak, W. IMP3 overexpression occurs in various important cancer types and is linked to aggressive tumor features: A tissue microarray study on 8,877 human cancers and normal tissues. Oncol. Rep. 2018, 39, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Bai, M.; Wang, L.; Lu, N.; Wang, J.; Yan, R.; Cui, M.; Yan, H.; Zhang, L. Targeting SNHG3/miR-186-5p reverses the increased m6A level caused by platinum treatment through regulating METTL3 in esophageal cancer. Cancer Cell Int. 2021, 21, 114. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.L.; Yan, S.M.; Yuan, L.; Zeng, M.S. Upregulation of METTL3 Expression Predicts Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 5729–5737. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Zhao, H.; Yu, X.; Cong, P.; Zhou, Y.; Jiang, Y.; Cheng, Y. METTL3 promotes the proliferation and invasion of esophageal cancer cells partly through AKT signaling pathway. Pathol. Res. Pract. 2020, 216, 153087. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Yuan, L.L.; Gao, Y.; Zhou, L.M.; Yang, J.W.; Pei, Z.J. Overexpression of METTL3 associated with the metabolic status on (18)F-FDG PET/CT in patients with Esophageal Carcinoma. J. Cancer 2020, 11, 4851–4860. [Google Scholar] [CrossRef]
- Hu, W.; Liu, W.; Liang, H.; Zhang, C.; Zou, M.; Zou, B. Silencing of methyltransferase-like 3 inhibits oesophageal squamous cell carcinoma. Exp. Ther. Med. 2020, 20, 138. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Z.; Xu, Y.; Huang, C.; Shan, B. METTL3 Facilitates Tumor Progression by COL12A1/MAPK Signaling Pathway in Esophageal Squamous Cell Carcinoma. J. Cancer 2022, 13, 1972–1984. [Google Scholar] [CrossRef]
- Zhou, Y.; Guo, S.; Li, Y.; Chen, F.; Wu, Y.; Xiao, Y.; An, J. METTL3 Is Associated with the Malignancy of Esophageal Squamous Cell Carcinoma and Serves as a Potential Immunotherapy Biomarker. Front. Oncol. 2022, 12, 824190. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.J.; Pang, Y.; Jin, G.; Zhang, H.Y.; Wang, W.H.; Liu, J.W.; Tuo, G.X.; Wu, P.; Yang, Y.; Wang, Z.Q.; et al. Hypoxia induces chemoresistance of esophageal cancer cells to cisplatin through regulating the lncRNA-EMS/miR-758-3p/WTAP axis. Aging 2021, 13, 17155–17176. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.C.; Pan, J.X.; Pan, H.D. Construction and Validation of an m6A RNA Methylation Regulators-Based Prognostic Signature for Esophageal Cancer. Cancer Manag. Res. 2020, 12, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Fang, T.X.; Lei, Y.; Xiao, S.J.; Xia, J.W.; Lin, T.Y.; Li, Y.L.; Zhai, J.X.; Li, X.Y.; Huang, S.H.; et al. m(6)A demethylase ALKBH5 suppression contributes to esophageal squamous cell carcinoma progression. Aging 2021, 13, 21497–21512. [Google Scholar] [CrossRef]
- Liu, S.; Huang, M.; Chen, Z.; Chen, J.; Chao, Q.; Yin, X.; Quan, M. FTO promotes cell proliferation and migration in esophageal squamous cell carcinoma through up-regulation of MMP13. Exp. Cell Res. 2020, 389, 111894. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Ying, P.; Tian, J.; Wang, X.; Mei, S.; Zou, D.; Peng, X.; Gong, Y.; Yang, Y.; Zhu, Y.; et al. Genetic variants in m6A modification genes are associated with esophageal squamous-cell carcinoma in the Chinese population. Carcinogenesis 2020, 41, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.Y.; Sun, J.J.; Chen, S.Y.; Wu, K.J.; Yu, Y.; Zhang, C.; Duan, C.Z. IGF2BP1/UHRF2 Axis Mediated by miR-98-5p to Promote the Proliferation of and Inhibit the Apoptosis of Esophageal Squamous Cell Carcinoma. Ann. Clin. Lab. Sci. 2021, 51, 329–338. [Google Scholar]
- Yan, A.; Wang, C.; Zheng, L.; Zhou, J.; Zhang, Y. MicroRNA-454-3p inhibits cell proliferation and invasion in esophageal cancer by targeting insulin-like growth factor 2 mRNA-binding protein 1. Oncol. Lett. 2020, 20, 359. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, R.; Wei, Q. Network analysis of miRNA targeting m6A-related genes in patients with esophageal cancer. PeerJ 2021, 9, e11893. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Xu, C.Z.; Gu, J.; Liu, X.; Liu, R.; Huang, E.; Yuan, Y.; Zhao, G.; Jiang, J.; Xu, C.; et al. Eukaryotic translation initiation factor 3B accelerates the progress63ion of esophageal squamous cell carcinoma by activating β-catenin signaling pathway. Oncotarget 2016, 7, 43401–43411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Gu, J.; Wang, L.; Liu, R.; Yuan, Y.; Wang, H.; Jiang, J.; Mao, W.; Lu, C.; Ge, D. Up-regulation Of EIF3e Is Associated with The Progression of Esophageal Squamous Cell Carcinoma and Poor Prognosis in Patients. J. Cancer 2018, 9, 1135–1144. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhu, R.; Luo, A.; Zhou, H.; Ding, F.; Yang, H.; Liu, Z. EIF3H promotes aggressiveness of esophageal squamous cell carcinoma by modulating Snail stability. J. Exp. Clin. Cancer Res. 2020, 39, 175. [Google Scholar] [CrossRef]
- Takata, A.; Takiguchi, S.; Okada, K.; Takahashi, T.; Kurokawa, Y.; Yamasaki, M.; Miyata, H.; Nakajima, K.; Mori, M.; Doki, Y. Expression of insulin-like growth factor-II mRNA-binding protein-3 as a marker for predicting clinical outcome in patients with esophageal squamous cell carcinoma. Oncol. Lett. 2014, 8, 2027–2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Zhang, S.; Liu, Z.; Gu, J.; Li, Y.; Wang, L.; Mao, W.; Zhu, Q.; Shou, H.; Ge, D.; et al. TEX9 and eIF3b functionally synergize to promote the progression of esophageal squamous cell carcinoma. BMC Cancer 2019, 19, 875. [Google Scholar] [CrossRef]
- Wang, H.; Shi, X.; Wu, S. miR-550a-3/NFIC plays a driving role in esophageal squamous cell cancer cells proliferation and metastasis partly through EMT process. Mol. Cell. Biochem. 2020, 472, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, R.; Ueda, N.; Uranishi, K.; Hirasaki, M.; Kurokawa, R. Long noncoding RNA pncRNA-D reduces cyclin D1 gene expression and arrests cell cycle through RNA m(6)A modification. J. Biol. Chem. 2020, 295, 5626–5639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Wu, Z.; Wang, X.; Wang, Y.; Hu, X.; Qin, W.; Lu, S.; Xu, D.; Wu, Y.; Chen, Q.; et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene 2020, 39, 5358–5372. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Hsia, J.; Yang, G. Prevalence of smoking in China in 2010. N. Engl. J. Med. 2011, 364, 2469–2470. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukocheva, O.A.; Li, B.; Due, S.L.; Hussey, D.J.; Watson, D.I. Androgens and esophageal cancer: What do we know? World J. Gastroenterol. 2015, 21, 6146–6156. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Hirai, T.; Mukaida, H.; Kawano, K.; Toge, T.; Niimoto, M.; Hattori, T. Detection of androgen receptors in human esophageal cancer. Jpn. J. Surg. 1989, 19, 195–202. [Google Scholar] [CrossRef]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Makhafola, T.J.; Mbele, M.; Yacqub-Usman, K.; Hendren, A.; Haigh, D.B.; Blackley, Z.; Meyer, M.; Mongan, N.P.; Bates, D.O.; Dlamini, Z. Apoptosis in Cancer Cells Is Induced by Alternative Splicing of hnRNPA2/B1 through Splicing of Bcl-x, a Mechanism that Can Be Stimulated by an Extract of the South African Medicinal Plant, Cotyledon orbiculata. Front. Oncol. 2020, 10, 547392. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Z.; Zhu, H.C.; Dai, W.S.; Zeng, X.N.; Luo, J.H.; Sun, X.C. The mechanisms of radioresistance in esophageal squamous cell carcinoma and current strategies in radiosensitivity. J. Thorac. Dis. 2017, 9, 849–859. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, K.; Motoyama, S.; Koyota, S.; Shibuya, K.; Sato, Y.; Sasaki, T.; Wakita, A.; Saito, H.; Minamiya, Y.; Sugiyama, T.; et al. Identification of insulin-like growth factor 2 mRNA-binding protein 3 as a radioresistance factor in squamous esophageal cancer cells. Dis. Esophagus Off. J. Int. Soc. Dis. Esophagus 2014, 27, 479–484. [Google Scholar] [CrossRef]
- Wakita, A.; Motoyama, S.; Sato, Y.; Nagaki, Y.; Fujita, H.; Terata, K.; Imai, K.; Maeda, E.; Minamiya, Y. IGF2BP3 Expression Correlates with Poor Prognosis in Esophageal Squamous Cell Carcinoma. J. Surg. Res. 2021, 259, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Tan, F.; Huai, Q.; Wang, Z.; Shao, F.; Zhang, G.; Yang, Z.; Li, R.; Xue, Q.; Gao, S.; et al. Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and m6A RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma. Front. Immunol. 2021, 12, 669750. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Lu, X.; Yang, X.; Yang, Y.; Wang, D.; Li, M.; Guan, W.; Xu, M. Estimating the prognosis of esophageal squamous cell carcinoma based on The Cancer Genome Atlas (TCGA) of m6A methylation-associated genes. J. Gastrointest. Oncol. 2022, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Luo, J.; Xu, T.; Sun, K. N6-Methyladenosine RNA Modification in Inflammation: Roles, Mechanisms, and Applications. Front. Cell Dev. Biol. 2021, 9, 670711. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, N.; Crespi, M.; Grassi, A.; Qing, W.G.; Qiong, S.; Cai, L.Z. Precursor lesions of oesophageal cancer in high-risk populations in Iran and China. Lancet 1982, 1, 876–879. [Google Scholar] [CrossRef]
- Mandard, A.M.; Hainaut, P.; Hollstein, M. Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat. Res. 2000, 462, 335–342. [Google Scholar] [CrossRef]
- Wang, L.D.; Bi, X.; Song, X.; Pohl, N.M.; Cheng, Y.; Zhou, Y.; Shears, S.; Ansong, E.; Xing, M.; Wang, S.; et al. A sequence variant in the phospholipase C epsilon C2 domain is associated with esophageal carcinoma and esophagitis. Mol. Carcinog. 2013, 52 (Suppl. S1), E80–E86. [Google Scholar] [CrossRef]
- Rong, Z.X.; Li, Z.; He, J.J.; Liu, L.Y.; Ren, X.X.; Gao, J.; Mu, Y.; Guan, Y.D.; Duan, Y.M.; Zhang, X.P.; et al. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front. Oncol. 2019, 9, 369. [Google Scholar] [CrossRef] [Green Version]
- An, Y.; Duan, H. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 2022, 21, 14. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.S.; Gao, Y.; Wu, L.B.; Wan, H.B.; Yan, P.; Jin, Y.; Guo, S.B.; Wang, Y.L.; Chen, X.Q.; Zhou, L.M.; et al. Comprehensive Analysis of GLUT1 Immune Infiltrates and ceRNA Network in Human Esophageal Carcinoma. Front. Oncol. 2021, 11, 665388. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Campos-Sandoval, J.A.; de Los Santos-Jiménez, J.; Segura, J.A.; Alonso, F.J.; Márquez, J. Metabolic Reprogramming of Cancer by Chemicals that Target Glutaminase Isoenzymes. Curr. Med. Chem. 2020, 27, 5317–5339. [Google Scholar] [CrossRef]
- Baba, Y.; Nomoto, D.; Okadome, K.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Baba, H. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci. 2020, 111, 3132–3141. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Ao, X.; Chen, Q.; Yu, Y.; Ao, L.; Xing, W.; Guo, W.; Wu, X.; Pu, C.; Hu, X.; et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer 2022, 21, 60. [Google Scholar] [CrossRef]
- Ni, Z.; Sun, P.; Zheng, J.; Wu, M.; Yang, C.; Cheng, M.; Yin, M.; Cui, C.; Wang, G.; Yuan, L.; et al. JNK Signaling Promotes Bladder Cancer Immune Escape by Regulating METTL3-Mediated m6A Modification of PD-L1 mRNA. Cancer Res. 2022, 82, 1789–1802. [Google Scholar] [CrossRef]
- Peng, L.; Pan, B.; Zhang, X.; Wang, Z.; Qiu, J.; Wang, X.; Tang, N. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol. Toxicol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Li, H.B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Wang, X.; Liu, Y.; Ren, X.; Wang, A.; Chen, Z.; Yao, J.; Mao, K.; Liu, T.; Meng, F.L.; et al. Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci. Adv. 2021, 7, eabd4742. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Wen, D.; Zeng, L.; Lu, J.; Xiao, X.; Chen, Y.; Song, H.; Liu, Z. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int. J. Biol. Sci. 2022, 18, 5001–5018. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wong, C.C.; Pan, Y.; Chen, H.; Liu, W.; Zhai, J.; Kang, W.; Shi, Y.; Yamamoto, M.; Tsukamoto, T.; et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J. Immunother. Cancer 2022, 10, e003663. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, G.; Xu, H.; Dong, W.; Dong, Z.; Qiu, Z.; Zhang, Z.; Li, F.; Huang, Y.; Li, Y.; et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021, 33, 1221–1233.e1211. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, Y.; Xie, Y.; Zhang, L.; Gao, M.; Li, S.; Wang, F. m6A Regulators Is Differently Immune Response of Esophageal Cancer. Front. Cell Dev. Biol. 2021, 9, 650023. [Google Scholar] [CrossRef] [PubMed]
| m6A Regulator | Target | Type of Target RNA | Function | Molecular Mechanism | Reference |
|---|---|---|---|---|---|
| METTL3 | GLS2 | mRNA | Writer | Increase m6A level of GLS2 mRNA, upregulate GLS2 expression | [59] |
| METTL3 | NOTCH1 | mRNA | Writer | Increase m6A level of NOTCH1 mRNA, upregulate NOTCH1 expression | [60] |
| METTL3 | TNFR1 | mRNA | Writer | Increase m6A level of TNFR1 mRNA, upregulate TNFR1 expression | [61] |
| ALKBH5 | CDKN1A | mRNA | Eraser | Decrease m6A level of CDKN1A mRNA, decrease stability of CDKN1A mRNA, downregulate p21 expression | [62] |
| FTO | SIM2 | mRNA | Eraser | Decrease m6A level of SIM2 mRNA, decrease stability of SIM2 mRNA, downregulate SIM2 expression | [63] |
| IGF2BP2 | CTNNB1 | mRNA | Reader | Increase stability of CTNNB1 mRNA, upregulate CTNNB1 expression | [64] |
| IGF2BP2 | FOXP4 | mRNA | Reader | Increase stability of FOXP4 mRNA, upregulate FOXP4 expression | [65] |
| IGF2BP2 | TK1 | mRNA | Reader | Recognize m6A of TK1 mRNA, upregulate TK1 expression | [66] |
| IGF2BP2 | KIF18A | mRNA | Reader | Increase stability of KIF18A mRNA, upregulate expression of KIF18A | [67] |
| HNRNPC | ZEB1 and ZEB2 | mRNA | Reader | Increase stability of ZEB1 and ZEB2 mRNA, upregulate expression of ZEB1 and ZEB2 | [68] |
| METTL3, METTL14, and YTHDF2 | APC | mRNA | METTL3 and METTL14 as writer; YTHDF2 as reader | METTL3 increases m6A level of APC mRNA in an METTL14-dependent way; YTHDF2 promotes degradation of APC mRNA | [69] |
| IGF2BP2 and IGF2BP3 | HTR3A | mRNA | Reader | Increase stability of HTR3A mRNA, upregulate HTR3A expression | [70] |
| METTL3 and YTHDF1 | HK2 | mRNA | METTL3 as writer; YTHDF1 as reader | Increase stability of HK2 mRNA, upregulate HK2 expression | [71] |
| METTL3 and YTHDF3 | EGR1 | mRNA | METTL3 as writer; YTHDF3 as reader | Increase stability of EGR1 mRNA, upregulate EGR1 expression | [72] |
| FTO and YTHDF1 | ERBB2 | mRNA | FTO as eraser; YTHDF1 as reader | Increase stability of ERBB2 mRNA, upregulate ERBB2 expression | [73] |
| FTO and YTHDF1 | HSD17B11 | mRNA | FTO as eraser; YTHDF1 as reader | Decrease the translation efficiency of HSD17B11 mRNA, downregulate HSD17B11 expression | [74] |
| METTL3 | pri-miR-200-5p | miRNA | Writer | Increase m6A level of pri-miR-200-5p, upregulate miR-200-5p expression | [75] |
| METTL3 | pri-miR-320b | miRNA | Writer | Increase m6A level of pri-miR-320b, upregulate miR-320b expression | [76] |
| METTL14 | pri-miR-99a | miRNA | Writer | Increase m6A level of pri-miR-99a, upregulate miR-99a-5p expression | [77] |
| ALKBH5 | pri-miR-194-2 | miRNA | Eraser | Decrease m6A level of pri-miR-194-2, downregulate miR-194-2 expression | [78] |
| HNRNPA2B1 | miR-17-92 cluster | miRNA | Reader | Bind to m6A of miR-17-92 cluster, upregulate expression of miR-17-92 cluster | [79] |
| METTL3 and ALKBH5 | pri-miR-193a-3p | miRNA | METTL3 as writer; ALKBH5 as eraser | METTL3 increases m6A level of pri-miR-193a-3p, upregulates miR-193a-3p expression; ALKBH5 decreases m6A level of pri-miR-193a-3p, downregulates miR-193a-3p expression | [80] |
| METTL3, METTL14, WTAP, ALKBH5, and YTHDF1 | LINC00278 | lncRNA | METTL3, METTL14, and WTAP as writers; ALKBH5 as eraser; YTHDF1 as reader | METTL3, METTL14, and WTAP increase m6A level of LINC00278; ALKBH5 decreases m6A level of LINC00278; YTHDF1 promotes translation of LINC00278 | [81] |
| METTL3, RBM15, WTAP, and YTHDC2 | MALAT1 | lncRNA | METTL3, RBM15, and WTAP as writer; YTHDC2 as reader | RBM15 interacts with METTL3 in a WTAP-dependent way to deposit m6A onto MALAT1; YTHDC1 binds to m6A of MALAT1 and maintains composition of nuclear speckle | [82] |
| FTO and YTHDF2 | LINC00022 | lncRNA | FTO as eraser; YTHDF2 as reader | FTO reduces the enrichment of m6A at site 2 of LINC00022 transcript; YTHDF2 promotes degradation of LINC00022 | [83] |
| m6A Regulator | Upstream Gene | Type of Upstream Gene | Molecular Mechanism | Reference |
|---|---|---|---|---|
| METTL3 | SNHG3 and miR-186-5p | lncRNA and miRNA | SNHG3 sponges miR-186-5p and alleviates inhibition of METTL3 by miR-186-5p | [122] |
| WTAP | EMS and miR-758-3p | lncRNA and miRNA | EMS sponges miR-758-3p and alleviates inhibition of METTL3 by miR-758-3p | [129] |
| IGF2BP2 | CCAT2 and miR-200b | lncRNA and miRNA | CCAT2 sponges miR-200b and alleviates inhibition of IGF2BP2 by miR-200b | [66] |
| IGF2BP1 | miR-454-3p | miRNA | miR-454-3p inhibits expression of IGF2BP1 | [135] |
| HNRNPC | miR-186 | miRNA | miR-186 inhibits expression of HNRNPC | [136] |
| ALKBH5 | miR-193a-3p | miRNA | miR-193a-3p inhibits expression of ALKBH5 | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Niu, C.; Wang, N.; Huang, X.; Cao, S.; Cui, S.; Chen, T.; Huo, X.; Zhou, R. The Role of m6A Modification and m6A Regulators in Esophageal Cancer. Cancers 2022, 14, 5139. https://doi.org/10.3390/cancers14205139
Li Y, Niu C, Wang N, Huang X, Cao S, Cui S, Chen T, Huo X, Zhou R. The Role of m6A Modification and m6A Regulators in Esophageal Cancer. Cancers. 2022; 14(20):5139. https://doi.org/10.3390/cancers14205139
Chicago/Turabian StyleLi, Yuekao, Chaoxu Niu, Na Wang, Xi Huang, Shiru Cao, Saijin Cui, Tianyu Chen, Xiangran Huo, and Rongmiao Zhou. 2022. "The Role of m6A Modification and m6A Regulators in Esophageal Cancer" Cancers 14, no. 20: 5139. https://doi.org/10.3390/cancers14205139





