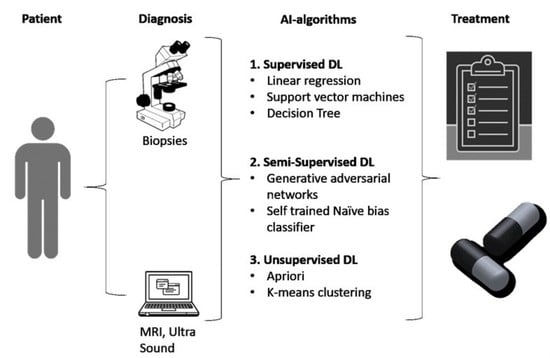
Artificial Intelligence for Clinical Diagnosis and Treatment of Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Methodology
3. Results
3.1. Application of AI in Prostate Cancer Diagnosis
3.1.1. AI in Biopsy-Based Detection of Prostate Cancer
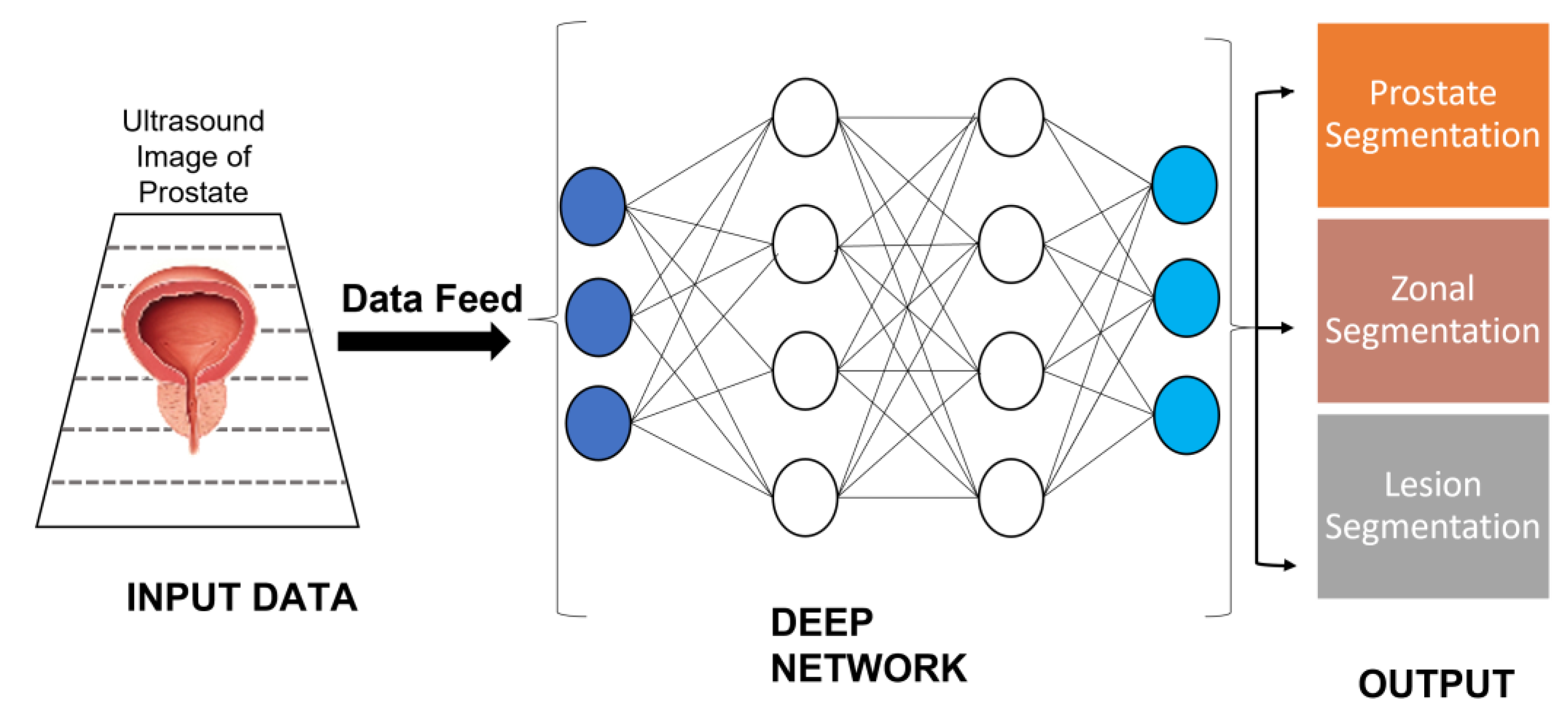
3.1.2. Artificial Intelligence in MRI-Guided PC Detection
3.1.3. Artificial Intelligence in Transrectal Ultrasound-Guided Biopsy-Based PC Detection
3.1.4. Artificial Intelligence in 3D Pathology Based PC Detection
3.1.5. Artificial Intelligence in Genomics-Based and Proteomics-Based PC Detection
3.1.6. Artificial Intelligence in CT Scan-Based PC Detection
3.2. Artificial Intelligence in PC Treatment
3.3. Recent Advancements and Future Aspects
3.4. Available Codes and Programs
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Laversanne, M.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available online: https://gco.iarc.fr/today (accessed on 27 June 2022).
- Cancer Stat Facts: Prostate Cancer, NIH. Available online: https://seer.cancer.gov/statfacts/html/prost.html (accessed on 27 June 2022).
- Barsouk, A.; Padala, S.A.; Vakiti, A.; Mohammed, A.; Saginala, K.; Thandra, K.C.; Rawla, P.; Barsouk, A. Epidemiology, Staging and Management of Prostate Cancer. Med. Sci. 2020, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Dess, R.T.; Hartman, H.E.; Mahal, B.A.; Soni, P.D.; Jackson, W.C.; Cooperberg, M.R.; Amling, C.L.; Aronson, W.J.; Kane, C.J.; Terris, M.K.; et al. Association of Black Race With Prostate Cancer–Specific and Other-Cause Mortality. JAMA Oncol. 2019, 5, 975. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, D.J.; Gaudet, M.M.; Pal, P.; Kirchhoff, T.; Balistreri, L.; Vora, K.; Bhatia, J.; Stadler, Z.; Fine, S.W.; Reuter, V.; et al. Germline BRCA Mutations Denote a Clinicopathologic Subset of Prostate Cancer. Clin. Cancer Res. 2010, 16, 2115–2121. [Google Scholar] [CrossRef] [PubMed]
- Watkins Bruner, D.; Moore, D.; Parlanti, A.; Dorgan, J.; Engstrom, P. Relative Risk of Prostate Cancer for Men with Affected Relatives: Systematic Review and Meta-Analysis. Int. J. Cancer 2003, 107, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Plaskon, L.A.; Penson, D.F.; Vaughan, T.L.; Stanford, J.L. Cigarette Smoking and Risk of Prostate Cancer in Middle-Aged Men. Cancer Epidemiol. Biomark. Prev. 2003, 12, 604–609. [Google Scholar]
- Allott, E.H.; Masko, E.M.; Freedland, S.J. Obesity and Prostate Cancer: Weighing the Evidence. Eur. Urol. 2013, 63, 800–809. [Google Scholar] [CrossRef]
- Ross, A.E.; Feng, Z.; Pierorazio, P.M.; Landis, P.; Walsh, P.C.; Carter, H.B.; Trock, B.J.; Schaeffer, E.M. Effect of Treatment with 5-α Reductase Inhibitors on Progression in Monitored Men with Favourable-Risk Prostate Cancer: 5-ARI USE IN FAVOURABLE RISK PROSTATE CANCER. BJU Int. 2012, 110, 651–657. [Google Scholar] [CrossRef]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Koteluk, O.; Wartecki, A.; Mazurek, S.; Kołodziejczak, I.; Mackiewicz, A. How Do Machines Learn? Artificial Intelligence as a New Era in Medicine. JPM 2021, 11, 32. [Google Scholar] [CrossRef]
- Panch, T.; Szolovits, P.; Atun, R. Artificial Intelligence, Machine Learning and Health Systems. J. Glob. Health 2018, 8, 020303. [Google Scholar] [CrossRef]
- Uddin, S.; Khan, A.; Hossain, M.E.; Moni, M.A. Comparing Different Supervised Machine Learning Algorithms for Disease Prediction. BMC Med. Inform. Decis. Mak. 2019, 19, 281. [Google Scholar] [CrossRef] [PubMed]
- Tătaru, O.S.; Vartolomei, M.D.; Rassweiler, J.J.; Virgil, O.; Lucarelli, G.; Porpiglia, F.; Amparore, D.; Manfredi, M.; Carrieri, G.; Falagario, U.; et al. Artificial Intelligence and Machine Learning in Prostate Cancer Patient Management—Current Trends and Future Perspectives. Diagnostics 2021, 11, 354. [Google Scholar] [CrossRef] [PubMed]
- Kartasalo, K.; Bulten, W.; Delahunt, B.; Chen, P.-H.C.; Pinckaers, H.; Olsson, H.; Ji, X.; Mulliqi, N.; Samaratunga, H.; Tsuzuki, T.; et al. Artificial Intelligence for Diagnosis and Gleason Grading of Prostate Cancer in Biopsies—Current Status and Next Steps. Eur. Urol. Focus 2021, 7, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Biopsy. National Health Service. (Last Date Accessed). Available online: Https://Www.Nhs.Uk/Conditions/Biopsy/ (accessed on 27 June 2022).
- Types of Endoscopy. American Society of Clinical Oncology. Available online: http://www.cancer.net/navigating-cancer-care/diagnosing-cancer/tests-and-procedures/types-endoscopy (accessed on 26 June 2022).
- Alguire, P.C.; Mathes, B.M. Skin biopsy techniques for the internist. J. Gen. Intern. Med. 1998, 13, 46–54. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rindy, L.J. Chambers AR Bone Marrow Aspiration and Biopsy. Available online: https://www.ncbi.nlm.nih.gov/books/NBK559232/ (accessed on 27 April 2022).
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. JEGH 2019, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-H.; Bhattacharjee, S.; Prakash, D.; Kang, S.; Cho, N.-H.; Kim, H.-C.; Choi, H.-K. Artificial Intelligence Techniques for Prostate Cancer Detection through Dual-Channel Tissue Feature Engineering. Cancers 2021, 13, 1524. [Google Scholar] [CrossRef]
- Montironi, R.; Mazzuccheli, R.; Scarpelli, M.; Lopez-Beltran, A.; Fellegara, G.; Algaba, F. Gleason Grading of Prostate Cancer in Needle Biopsies or Radical Prostatectomy Specimens: Contemporary Approach, Current Clinical Significance and Sources of Pathology Discrepancies. BJU Int. 2005, 95, 1146–1152. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H.; et al. Development and Validation of a Deep Learning Algorithm for Improving Gleason Scoring of Prostate Cancer. npj Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef]
- Bulten, W.; Balkenhol, M.; Belinga, J.-J.A.; Brilhante, A.; Çakır, A.; Egevad, L.; Eklund, M.; Farré, X.; Geronatsiou, K.; Molinié, V.; et al. Artificial Intelligence Assistance Significantly Improves Gleason Grading of Prostate Biopsies by Pathologists. Mod. Pathol. 2021, 34, 660–671. [Google Scholar] [CrossRef]
- Doyle, S.; Feldman, M.D.; Shih, N.; Tomaszewski, J.; Madabhushi, A. Cascaded Discrimination of Normal, Abnormal, and Confounder Classes in Histopathology: Gleason Grading of Prostate Cancer. BMC Bioinform. 2012, 13, 282. [Google Scholar] [CrossRef]
- Marginean, F.; Arvidsson, I.; Simoulis, A.; Christian Overgaard, N.; Åström, K.; Heyden, A.; Bjartell, A.; Krzyzanowska, A. An Artificial Intelligence–Based Support Tool for Automation and Standardisation of Gleason Grading in Prostate Biopsies. Eur. Urol. Focus 2021, 7, 995–1001. [Google Scholar] [CrossRef]
- Lucas, M.; Jansen, I.; Savci-Heijink, C.D.; Meijer, S.L.; de Boer, O.J.; van Leeuwen, T.G.; de Bruin, D.M.; Marquering, H.A. Deep Learning for Automatic Gleason Pattern Classification for Grade Group Determination of Prostate Biopsies. Virchows Arch. 2019, 475, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Schnall, M.D.; Imai, Y.; Tomaszewski, J.; Pollack, H.M.; Lenkinski, R.E.; Kressel, H.Y. Prostate Cancer: Local Staging with Endorectal Surface Coil MR Imaging. Radiology 1991, 178, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.D.; Crukley, C.; McKenzie, C.A.; Montreuil, J.; Gibson, E.; Romagnoli, C.; Gomez, J.A.; Moussa, M.; Chin, J.; Bauman, G.; et al. Prostate: Registration of Digital Histopathologic Images to in Vivo MR Images Acquired by Using Endorectal Receive Coil. Radiology 2012, 263, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Toth, R.; van de Ven, W.; Hoeks, C.; Kerkstra, S.; van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J.; et al. Evaluation of Prostate Segmentation Algorithms for MRI: The PROMISE12 Challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Greer, M.D.; Brown, A.M.; Shih, J.H.; Summers, R.M.; Marko, J.; Law, Y.M.; Sankineni, S.; George, A.K.; Merino, M.J.; Pinto, P.A.; et al. Accuracy and Agreement of PIRADSv2 for Prostate Cancer MpMRI: A Multireader Study: PIRADSv2 for Prostate Tumor Detection. J. Magn. Reson. Imaging 2017, 45, 579–585. [Google Scholar] [CrossRef]
- Gaur, S.; Lay, N.; Harmon, S.A.; Doddakashi, S.; Mehralivand, S.; Argun, B.; Barrett, T.; Bednarova, S.; Girometti, R.; Karaarslan, E.; et al. Can Computer-Aided Diagnosis Assist in the Identification of Prostate Cancer on Prostate MRI? A Multi-Center, Multi-Reader Investigation. Oncotarget 2018, 9, 33804–33817. [Google Scholar] [CrossRef]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef]
- Rouvière, O.; Puech, P.; Renard-Penna, R.; Claudon, M.; Roy, C.; Mège-Lechevallier, F.; Decaussin-Petrucci, M.; Dubreuil-Chambardel, M.; Magaud, L.; Remontet, L.; et al. Use of Prostate Systematic and Targeted Biopsy on the Basis of Multiparametric MRI in Biopsy-Naive Patients (MRI-FIRST): A Prospective, Multicentre, Paired Diagnostic Study. Lancet Oncol. 2019, 20, 100–109. [Google Scholar] [CrossRef]
- van der Leest, M.; Cornel, E.; Israël, B.; Hendriks, R.; Padhani, A.R.; Hoogenboom, M.; Zamecnik, P.; Bakker, D.; Setiasti, A.Y.; Veltman, J.; et al. Head-to-Head Comparison of Transrectal Ultrasound-Guided Prostate Biopsy Versus Multiparametric Prostate Resonance Imaging with Subsequent Magnetic Resonance-Guided Biopsy in Biopsy-Naïve Men with Elevated Prostate-Specific Antigen: A Large Prospective Multicenter Clinical Study. Eur. Urol. 2019, 75, 570–578. [Google Scholar] [CrossRef]
- Mottet, N.; Bellmunt, J.; Bolla, M.; Briers, E.; Cumberbatch, M.G.; De Santis, M.; Fossati, N.; Gross, T.; Henry, A.M.; Joniau, S.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017, 71, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Twilt, J.J.; van Leeuwen, K.G.; Huisman, H.J.; Fütterer, J.J.; de Rooij, M. Artificial Intelligence Based Algorithms for Prostate Cancer Classification and Detection on Magnetic Resonance Imaging: A Narrative Review. Diagnostics 2021, 11, 959. [Google Scholar] [CrossRef] [PubMed]
- Mehralivand, S.; Yang, D.; Harmon, S.A.; Xu, D.; Xu, Z.; Roth, H.; Masoudi, S.; Sanford, T.H.; Kesani, D.; Lay, N.S.; et al. A Cascaded Deep Learning–Based Artificial Intelligence Algorithm for Automated Lesion Detection and Classification on Biparametric Prostate Magnetic Resonance Imaging. Acad. Radiol. 2021, 29, S1076633221003779. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, H.; Mofid, B.; Shiri, I.; Razzaghdoust, A.; Saadipoor, A.; Mahdavi, A.; Galandooz, H.M.; Mahdavi, S.R. Machine Learning-Based Radiomic Models to Predict Intensity-Modulated Radiation Therapy Response, Gleason Score and Stage in Prostate Cancer. Radiol. Med. 2019, 124, 555–567. [Google Scholar] [CrossRef]
- Dulhanty, C.; Wang, L.; Cheng, M.; Gunraj, H.; Khalvati, F.; Haider, M.A.; Wong, A. Radiomics Driven Diffusion Weighted Imaging Sensing Strategies for Zone-Level Prostate Cancer Sensing. Sensors 2020, 20, E1539. [Google Scholar] [CrossRef]
- Aldoj, N.; Lukas, S.; Dewey, M.; Penzkofer, T. Semi-Automatic Classification of Prostate Cancer on Multi-Parametric MR Imaging Using a Multi-Channel 3D Convolutional Neural Network. Eur. Radiol. 2020, 30, 1243–1253. [Google Scholar] [CrossRef]
- Kasel-Seibert, M.; Lehmann, T.; Aschenbach, R.; Guettler, F.V.; Abubrig, M.; Grimm, M.-O.; Teichgraeber, U.; Franiel, T. Assessment of PI-RADS v2 for the Detection of Prostate Cancer. Eur. J. Radiol. 2016, 85, 726–731. [Google Scholar] [CrossRef]
- Chen, Y.-T.; Siu, L.K.; Tsai, Y.-K.; Lin, F.-M.; Koh, T.H.; Chen, J.-H. A Common Flanking Region in Promiscuous Plasmids Encoding Bla NDM-1 in Klebsiella Pneumoniae Isolated in Singapore. Microb. Drug Resist. 2016, 22, 109–114. [Google Scholar] [CrossRef]
- Hötker, A.M.; Da Mutten, R.; Tiessen, A.; Konukoglu, E.; Donati, O.F. Improving Workflow in Prostate MRI: AI-Based Decision-Making on Biparametric or Multiparametric MRI. Insights Imaging 2021, 12, 112. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An Overview of Deep Learning in Medical Imaging Focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef]
- Harvey, C.J.; Pilcher, J.; Richenberg, J.; Patel, U.; Frauscher, F. Applications of Transrectal Ultrasound in Prostate Cancer. Br. J. Radiol. 2012, 85, S3–S17. [Google Scholar] [CrossRef]
- Porter, C.R.; Crawford, E.D. Combining Artificial Neural Networks and Transrectal Ultrasound in the Diagnosis of Prostate Cancer. Oncology 2003, 17, 1395–1399; discussion 1399, 1403–1406. [Google Scholar] [PubMed]
- Xie, J.; Jin, C.; Liu, M.; Sun, K.; Jin, Z.; Ding, Z.; Gong, X. MRI/Transrectal Ultrasound Fusion-Guided Targeted Biopsy and Transrectal Ultrasound-Guided Systematic Biopsy for Diagnosis of Prostate Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Benelli, A.; Vaccaro, C.; Guzzo, S.; Nedbal, C.; Varca, V.; Gregori, A. The Role of MRI/TRUS Fusion Biopsy in the Diagnosis of Clinically Significant Prostate Cancer. Ther. Adv. Urol. 2020, 12, 1756287220916613. [Google Scholar] [CrossRef] [PubMed]
- van Sloun, R.J.G.; Wildeboer, R.R.; Mannaerts, C.K.; Postema, A.W.; Gayet, M.; Beerlage, H.P.; Salomon, G.; Wijkstra, H.; Mischi, M. Deep Learning for Real-Time, Automatic, and Scanner-Adapted Prostate (Zone) Segmentation of Transrectal Ultrasound, for Example, Magnetic Resonance Imaging–Transrectal Ultrasound Fusion Prostate Biopsy. Eur. Urol. Focus 2021, 7, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Bourne, R.M.; Wang, S.; Devaraj, A.; Gallan, A.J.; Antic, T.; Karczmar, G.S.; Oto, A. Diagnosis of Prostate Cancer with Noninvasive Estimation of Prostate Tissue Composition by Using Hybrid Multidimensional MR Imaging: A Feasibility Study. Radiology 2018, 287, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Mata, L.A.; Retamero, J.A.; Gupta, R.T.; García Figueras, R.; Luna, A. Artificial Intelligence–Assisted Prostate Cancer Diagnosis: Radiologic-Pathologic Correlation. RadioGraphics 2021, 41, 1676–1697. [Google Scholar] [CrossRef]
- Steiner, D.F.; Nagpal, K.; Sayres, R.; Foote, D.J.; Wedin, B.D.; Pearce, A.; Cai, C.J.; Winter, S.R.; Symonds, M.; Yatziv, L.; et al. Evaluation of the Use of Combined Artificial Intelligence and Pathologist Assessment to Review and Grade Prostate Biopsies. JAMA Netw. Open 2020, 3, e2023267. [Google Scholar] [CrossRef]
- Yao, F.; Bian, S.; Zhu, D.; Yuan, Y.; Pan, K.; Pan, Z.; Feng, X.; Tang, K.; Yang, Y. Machine Learning-Based Radiomics for Multiple Primary Prostate Cancer Biological Characteristics Prediction with 18F-PSMA-1007 PET: Comparison among Different Volume Segmentation Thresholds. Radiol. Med. 2022, 127, 1170–1178. [Google Scholar] [CrossRef]
- Wang, X.; Xie, Y.; Zheng, X.; Liu, B.; Chen, H.; Li, J.; Ma, X.; Xiang, J.; Weng, G.; Zhu, W.; et al. A Prospective Multi-Center Randomized Comparative Trial Evaluating Outcomes of Transrectal Ultrasound (TRUS)-Guided 12-Core Systematic Biopsy, MpMRI-Targeted 12-Core Biopsy, and Artificial Intelligence Ultrasound of Prostate (AIUSP) 6-Core Targeted Biopsy for Prostate Cancer Diagnosis. World J. Urol. 2022. [Google Scholar] [CrossRef]
- Luo, R.; Zeng, Q.; Chen, H. Artificial Intelligence Algorithm-Based MRI for Differentiation Diagnosis of Prostate Cancer. Comput. Math. Methods Med. 2022, 2022, 123643. [Google Scholar] [CrossRef] [PubMed]
- Moroianu, Ş.L.; Bhattacharya, I.; Seetharaman, A.; Shao, W.; Kunder, C.A.; Sharma, A.; Ghanouni, P.; Fan, R.E.; Sonn, G.A.; Rusu, M. Computational Detection of Extraprostatic Extension of Prostate Cancer on Multiparametric MRI Using Deep Learning. Cancers 2022, 14, 2821. [Google Scholar] [CrossRef] [PubMed]
- Dadhania, V.; Gonzalez, D.; Yousif, M.; Cheng, J.; Morgan, T.M.; Spratt, D.E.; Reichert, Z.R.; Mannan, R.; Wang, X.; Chinnaiyan, A.; et al. Leveraging Artificial Intelligence to Predict ERG Gene Fusion Status in Prostate Cancer. BMC Cancer 2022, 22, 494. [Google Scholar] [CrossRef] [PubMed]
- Carroll, P.R.; Parsons, J.K.; Andriole, G.; Bahnson, R.R.; Castle, E.P.; Catalona, W.J.; Dahl, D.M.; Davis, J.W.; Epstein, J.I.; Etzioni, R.B.; et al. NCCN Guidelines Insights: Prostate Cancer Early Detection, Version 2.2016. J. Natl. Compr. Canc. Netw. 2016, 14, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.T.C.; Glaser, A.K.; Bera, K.; True, L.D.; Reder, N.P.; Eliceiri, K.W.; Madabhushi, A. Harnessing Non-Destructive 3D Pathology. Nat. Biomed. Eng. 2021, 5, 203–218. [Google Scholar] [CrossRef]
- Pietzsch, T.; Saalfeld, S.; Preibisch, S.; Tomancak, P. BigDataViewer: Visualization and Processing for Large Image Data Sets. Nat. Methods 2015, 12, 481–483. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next Generation of Scientific Image Data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Reder, N.P.; Glaser, A.K.; McCarty, E.F.; Chen, Y.; True, L.D.; Liu, J.T.C. Open-Top Light-Sheet Microscopy Image Atlas of Prostate Core Needle Biopsies. Arch. Pathol. Lab. Med. 2019, 143, 1069–1075. [Google Scholar] [CrossRef]
- Edelstein, A.; Amodaj, N.; Hoover, K.; Vale, R.; Stuurman, N. Computer Control of Microscopes Using ΜManager. Curr. Protoc. Mol. Biol. 2010, 92, 14–20. [Google Scholar] [CrossRef]
- Kaneko, M.; Fukuda, N.; Nagano, H.; Yamada, K.; Yamada, K.; Konishi, E.; Sato, Y.; Ukimura, O. Artificial Intelligence Trained with Integration of Multiparametric MR-US Imaging Data and Fusion Biopsy Trajectory-proven Pathology Data for 3D Prediction of Prostate Cancer: A Proof-of-concept Study. Prostate 2022, 82, 793–803. [Google Scholar] [CrossRef]
- Epstein, J.I. A New Contemporary Prostate Cancer Grading System. Ann. Pathol. 2015, 35, 474–476. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.B.; Leandro, G.; Romerocaces, G.; Bentley, J.; Yoon, J.; Mendrinos, S.; Tadros, Y.; Tian, W.; Lash, R. Improvement of Diagnostic Agreement among Pathologists in Resolving an “Atypical Glands Suspicious for Cancer” Diagnosis in Prostate Biopsies Using a Novel “Disease-Focused Diagnostic Review” Quality Improvement Process. Hum. Pathol. 2016, 56, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, T.A.; Eruyar, A.T.; Cebeci, O.O.; Memik, O.; Ozcan, L.; Kuskonmaz, I. Interobserver Variability in Gleason Histological Grading of Prostate Cancer. Scand. J. Urol. 2016, 50, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Kane, C.J.; Eggener, S.E.; Shindel, A.W.; Andriole, G.L. Variability in Outcomes for Patients with Intermediate-Risk Prostate Cancer (Gleason Score 7, International Society of Urological Pathology Gleason Group 2–3) and Implications for Risk Stratification: A Systematic Review. Eur. Urol. Focus 2017, 3, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, P.C. Treatment of Localized Prostate Cancer: When Is Active Surveillance Appropriate? Nat. Rev. Clin. Oncol. 2010, 7, 394–400. [Google Scholar] [CrossRef]
- Haffner, M.C.; De Marzo, A.M.; Yegnasubramanian, S.; Epstein, J.I.; Carter, H.B. Diagnostic Challenges of Clonal Heterogeneity in Prostate Cancer. JCO 2015, 33, e38–e40. [Google Scholar] [CrossRef]
- Bill-Axelson, A.; Holmberg, L.; Garmo, H.; Rider, J.R.; Taari, K.; Busch, C.; Nordling, S.; Häggman, M.; Andersson, S.-O.; Spångberg, A.; et al. Radical Prostatectomy or Watchful Waiting in Early Prostate Cancer. N. Engl. J. Med. 2014, 370, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.U.; Sønksen, J.; Fode, M. Neglected Side Effects After Radical Prostatectomy: A Systematic Review. J. Sex. Med. 2014, 11, 374–385. [Google Scholar] [CrossRef]
- Xie, W.; Reder, N.P.; Koyuncu, C.; Leo, P.; Hawley, S.; Huang, H.; Mao, C.; Postupna, N.; Kang, S.; Serafin, R.; et al. Prostate Cancer Risk Stratification via Nondestructive 3D Pathology with Deep Learning–Assisted Gland Analysis. Cancer Res. 2022, 82, 334–345. [Google Scholar] [CrossRef]
- Checcucci, E.; Autorino, R.; Cacciamani, G.E.; Amparore, D.; De Cillis, S.; Piana, A.; Piazzolla, P.; Vezzetti, E.; Fiori, C.; Veneziano, D.; et al. Artificial Intelligence and Neural Networks in Urology: Current Clinical Applications. Minerva Urol. Nefrol. 2020, 72, 49–57. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A Systematic Analysis of Human Disease-Associated Gene Sequences In Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Choyke, P.L.; Loeb, S. Active Surveillance of Prostate Cancer. Oncology 2017, 31, 67–70. [Google Scholar] [PubMed]
- Jin, W.; Fei, X.; Wang, X.; Song, Y.; Chen, F. Detection and Prognosis of Prostate Cancer Using Blood-Based Biomarkers. Mediat. Inflamm. 2020, 2020, 8730608. [Google Scholar] [CrossRef]
- McDonald, A.C.; Vira, M.; Shen, J.; Sanda, M.; Raman, J.D.; Liao, J.; Patil, D.; Taioli, E. Circulating MicroRNAs in Plasma as Potential Biomarkers for the Early Detection of Prostate Cancer. Prostate 2018, 78, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Martignano, F.; Rossi, L.; Maugeri, A.; Gallà, V.; Conteduca, V.; De Giorgi, U.; Casadio, V.; Schepisi, G. Urinary RNA-Based Biomarkers for Prostate Cancer Detection. Clin. Chim. Acta 2017, 473, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, N.A.; Hamdy, F.C.; Bryant, R.J. Novel Biomarkers for the Detection of Prostate Cancer. J. Clin. Urol. 2016, 9, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Reed, A.B.; Parekh, D.J. Biomarkers for Prostate Cancer Detection. Expert Rev. Anticancer Ther. 2010, 10, 103–114. [Google Scholar] [CrossRef]
- Margreiter, M.; Stangelberger, A.; Valimberti, E.; Herwig, R.; Djavan, B. Biomarkers for Early Prostate Cancer Detection. Minerva Urol. Nefrol. 2008, 60, 51–60. [Google Scholar]
- Parekh, D.J.; Ankerst, D.P.; Troyer, D.; Srivastava, S.; Thompson, I.M. Biomarkers for Prostate Cancer Detection. J. Urol. 2007, 178, 2252–2259. [Google Scholar] [CrossRef]
- Shariat, S.F.; Karam, J.A.; Roehrborn, C.G. Blood Biomarkers for Prostate Cancer Detection and Prognosis. Future Oncol. 2007, 3, 449–461. [Google Scholar] [CrossRef]
- Stephan, C.; Cammann, H.; Meyer, H.-A.; Lein, M.; Jung, K. PSA and New Biomarkers within Multivariate Models to Improve Early Detection of Prostate Cancer. Cancer Lett. 2007, 249, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Filella, X.; Foj, L. Prostate Cancer Detection and Prognosis: From Prostate Specific Antigen (PSA) to Exosomal Biomarkers. IJMS 2016, 17, 1784. [Google Scholar] [CrossRef] [PubMed]
- Frugé, A.D.; Smith, K.S.; Bail, J.R.; Rais-Bahrami, S.; Demark-Wahnefried, W. Biomarkers Associated With Tumor Ki67 and Cathepsin L Gene Expression in Prostate Cancer Patients Participating in a Presurgical Weight Loss Trial. Front. Oncol. 2020, 10, 544201. [Google Scholar] [CrossRef]
- Green, W.J.; Ball, G.; Hulman, G.; Johnson, C.; Van Schalwyk, G.; Ratan, H.L.; Soria, D.; Garibaldi, J.M.; Parkinson, R.; Hulman, J.; et al. KI67 and DLX2 Predict Increased Risk of Metastasis Formation in Prostate Cancer–a Targeted Molecular Approach. Br. J. Cancer 2016, 115, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhu, S.; Ma, K.; Liu, W.; Yue, Y.; Hu, G.; Lu, H.; Chen, W. Identification of 12 Cancer Types through Genome Deep Learning. Sci. Rep. 2019, 9, 17256. [Google Scholar] [CrossRef]
- Jurtz, V.I.; Johansen, A.R.; Nielsen, M.; Almagro Armenteros, J.J.; Nielsen, H.; Sønderby, C.K.; Winther, O.; Sønderby, S.K. An Introduction to Deep Learning on Biological Sequence Data: Examples and Solutions. Bioinformatics 2017, 33, 3685–3690. [Google Scholar] [CrossRef]
- Park, Y.; Heider, D.; Hauschild, A.-C. Integrative Analysis of Next-Generation Sequencing for Next-Generation Cancer Research toward Artificial Intelligence. Cancers 2021, 13, 3148. [Google Scholar] [CrossRef]
- Hussain, F.; Saeed, U.; Muhammad, G.; Islam, N.; Sheikh, G.S. Classifying Cancer Patients Based on DNA Sequences Using Machine Learning. J. Med. Imaging Health Inform. 2019, 9, 436–443. [Google Scholar] [CrossRef]
- Kim, Y.; Jeon, J.; Mejia, S.; Yao, C.Q.; Ignatchenko, V.; Nyalwidhe, J.O.; Gramolini, A.O.; Lance, R.S.; Troyer, D.A.; Drake, R.R.; et al. Targeted Proteomics Identifies Liquid-Biopsy Signatures for Extracapsular Prostate Cancer. Nat. Commun. 2016, 7, 11906. [Google Scholar] [CrossRef]
- Kim, Y.; Ignatchenko, V.; Yao, C.Q.; Kalatskaya, I.; Nyalwidhe, J.O.; Lance, R.S.; Gramolini, A.O.; Troyer, D.A.; Stein, L.D.; Boutros, P.C.; et al. Identification of Differentially Expressed Proteins in Direct Expressed Prostatic Secretions of Men with Organ-Confined Versus Extracapsular Prostate Cancer. Mol. Cell. Proteom. 2012, 11, 1870–1884. [Google Scholar] [CrossRef]
- Scher, H.I.; Heller, G.; Molina, A.; Attard, G.; Danila, D.C.; Jia, X.; Peng, W.; Sandhu, S.K.; Olmos, D.; Riisnaes, R.; et al. Circulating Tumor Cell Biomarker Panel As an Individual-Level Surrogate for Survival in Metastatic Castration-Resistant Prostate Cancer. JCO 2015, 33, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Cortese, R.; Kwan, A.; Lalonde, E.; Bryzgunova, O.; Bondar, A.; Wu, Y.; Gordevicius, J.; Park, M.; Oh, G.; Kaminsky, Z.; et al. Epigenetic Markers of Prostate Cancer in Plasma Circulating DNA. Hum. Mol. Genet. 2012, 21, 3619–3631. [Google Scholar] [CrossRef] [PubMed]
- Korevaar, S.; Tennakoon, R.; Page, M.; Brotchie, P.; Thangarajah, J.; Florescu, C.; Sutherland, T.; Kam, N.M.; Bab-Hadiashar, A. Incidental Detection of Prostate Cancer with Computed Tomography Scans. Sci. Rep. 2021, 11, 7956. [Google Scholar] [CrossRef]
- Hamdy, F.C.; Donovan, J.L.; Lane, J.A.; Mason, M.; Metcalfe, C.; Holding, P.; Davis, M.; Peters, T.J.; Turner, E.L.; Martin, R.M.; et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N. Engl. J. Med. 2016, 375, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future — Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Auffenberg, G.B.; Ghani, K.R.; Ramani, S.; Usoro, E.; Denton, B.; Rogers, C.; Stockton, B.; Miller, D.C.; Singh, K. Michigan Urological Surgery Improvement Collaborative AskMUSIC: Leveraging a Clinical Registry to Develop a New Machine Learning Model to Inform Patients of Prostate Cancer Treatments Chosen by Similar Men. Eur. Urol. 2019, 75, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Roest, C.; Kwee, T.C.; Saha, A.; Fütterer, J.J.; Yakar, D.; Huisman, H. AI-Assisted Biparametric MRI Surveillance of Prostate Cancer: Feasibility Study. Eur. Radiol. 2022. [Google Scholar] [CrossRef]
- Gravina, M.; Spirito, L.; Celentano, G.; Capece, M.; Creta, M.; Califano, G.; Collà Ruvolo, C.; Morra, S.; Imbriaco, M.; Di Bello, F.; et al. Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions. Diagnostics 2022, 12, 1565. [Google Scholar] [CrossRef]
- Chen, S.; Jian, T.; Chi, C.; Liang, Y.; Liang, X.; Yu, Y.; Jiang, F.; Lu, J. Machine Learning-Based Models Enhance the Prediction of Prostate Cancer. Front. Oncol. 2022, 12, 941349. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Shu, X.; Qiao, X.-F.; Ai, G.-Y.; Liu, L.; Liao, J.; Qian, S.; He, X.-J. Radiomics-Based Machine Learning Models for Predicting P504s/P63 Immunohistochemical Expression: A Noninvasive Diagnostic Tool for Prostate Cancer. Front. Oncol. 2022, 12, 911426. [Google Scholar] [CrossRef]
- Secasan, C.C.; Onchis, D.; Bardan, R.; Cumpanas, A.; Novacescu, D.; Botoca, C.; Dema, A.; Sporea, I. Artificial Intelligence System for Predicting Prostate Cancer Lesions from Shear Wave Elastography Measurements. Curr. Oncol. 2022, 29, 4212–4223. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhou, Q.; Yin, H.; Ang, X.; Li, Y.; Xie, G.; Li, G. Texture Analysis Based on PI-RADS 4/5-Scored Magnetic Resonance Images Combined with Machine Learning to Distinguish Benign Lesions from Prostate Cancer. Transl. Cancer Res. TCR 2022, 11, 1146–1161. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.B.; Grazal, C.; Wedin, R.; Kuo, C.; Chen, Y.; Christensen, B.R.; Cullen, J.; Forsberg, J.A. Machine Learning Algorithms to Estimate 10-Year Survival in Patients with Bone Metastases Due to Prostate Cancer: Toward a Disease-Specific Survival Estimation Tool. BMC Cancer 2022, 22, 476. [Google Scholar] [CrossRef] [PubMed]

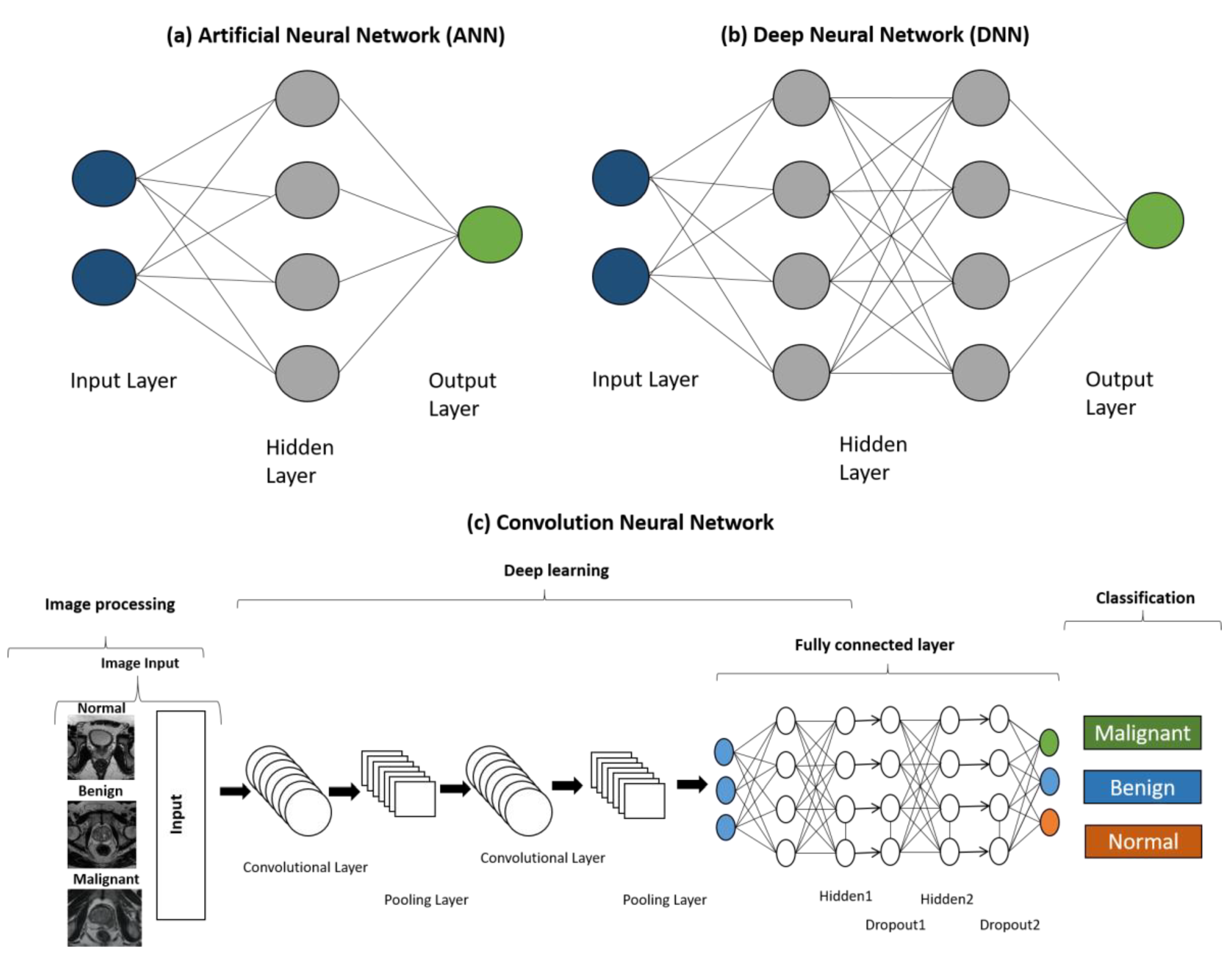



| S. No. | Biopsy Techniques | Summary |
|---|---|---|
| 1 | Needle Biopsy | This method of biopsy inserts a needle into the skin for collecting cells from a suspicious area. This process is also known as a percutaneous tissue biopsy by doctors [16]. |
| 2 | Endoscopic biopsy | Endoscopy is a procedure in which medical staffs use a flexible and thin tube (endoscope) with a light at the terminal to examine structures within the body. Further, special instruments are inserted into the tube to collect a tiny tissue sample to analyze [17]. |
| 3 | Skin biopsy | A skin biopsy collects cells from the surface of the skin. It is mainly used to identify skin diseases, including melanoma. The type of cancer that is detected and the extent of the suspicious cells will determine the sort of skin biopsy that is experienced by the patient [18]. |
| 4 | Bone marrow biopsy | This biopsy method is mainly used after the findings of blood tests or if the doctors propose a malignancy that affects the bone marrow [19]. |
| 5 | Surgical biopsy | A surgical biopsy may be prescribed if other biopsy procedures are ineffective or if the results of the initial tests have been inconclusive. |
| Grade and Gleason Score | Type | Pattern | Size |
|---|---|---|---|
| 1, Score ≤ 6 | Benign | Single glands with sharp boundaries that are well defined and consistent. | Medium |
| 2, Score 3 + 4 = 7 | Benign | Single glands are widely apart and the tumor’s boundaries are not clearly defined; it is less well confined. | Medium |
| 3, Score 4 + 3 = 7 | Malignant | Masses that are single, separated, spherical, irregular, or larger and have a cribriform or papillary pattern. | Small to large |
| 4, Score 4 + 4 or 3 + 5 or 5 + 3 = 8 | Malignant | Fused gland tumour with predominantly pale cells and no architecture. | Small to medium |
| 5, Score 4 + 5 or 5 + 4 = 9 and 5 + 5 = 10 | Malignant | Tumors and cords of comedo cancer, solid sheets and no gland formation. | Small |
| S. No. | Summary | Date Accessed | References |
|---|---|---|---|
| 1. | This study shows that a higher grade PC is related to increased epithelial volume and lower stromal and lumen map volumes measured by hybrid multi-dimensional MRI, thus making this a potential approach in predicting aggressive PC. | 29 August 2022 | [52] |
| 2. | TRUS-Bx remains useful in PC diagnosis when it is paired with mpMRI. This study showed the application of AI algorithms in prostate gland segmentation, lesion identification, and classification using mpMRI and TRUS-Bx, reducing interreader variability and minimising the possible lack of competence of less experienced radiologists. | 29 August 2022 | [53] |
| 3. | This diagnostic study suggested that an AI-based assistive tool can increase the accuracy, speed, and consistency of pathologists’ assessment of prostate biopsy samples. The relatively high number of samples and pathologists involved in this study allowed for a thorough examination of the advantages of an AI-based tool for the contemporaneous assessment of prostate biopsies, as well as insights into potential risks associated with overreliance. | 29 August 2022 | [54] |
| 4. | As per the findings showed in this study, 18F-1007-PSMA PET-based radiomics features with 40–50% standardized uptake value (SUV) max exhibited the most robust predictive ability for evaluating numerous PC biological characteristics. Radiomics properties, when compared to a single PSA model, may give significant benefits in predicting the biological aspects of PC based on the support vector machine. The 50% SUVmax model had the most powerful predictive performance in trained (AUC, 0.82) and tested cohorts for predicting Gleason score (GS) (AUC, 0.80). The 40% SUVmax model has the most significant expected performance for extracapsular extension (ECE) (AUC, 0.77). In terms of vascular invasion (VI), the 50% SUVmax model performed the best (AUC 0.74). | 29 August 2022 | [55] |
| 5. | In this study, artificial intelligence ultrasound of the prostate (AIUSP) detected the PC (49.5%) when it was compared to transrectal ultrasound (TRUS)-guided 12-core systematic biopsy (34.60%) and mpMRI (35.80%). Clinically significant PC (csPC) detection rate in AIUSP group was 32.30%, which was compared to TRUS-SB (26.3%) and mpMRI (23.1%) groups. The overall biopsy core positive rate in the TRUS-SB (11.0%) and mpMRI (12.7%) groups was substantially lower than it was in the AIUSP group (22.7%). | 29 August 2022 | [56] |
| 6. | The weighted low-rank matrix restoration algorithm (RLRE) algorithm was used to de-noise MRI images in this study to identify PC from benign prostatic hyperplasia (BPH) and to evaluate the diagnostic impact of MRI images with varied sequences. The findings showed that the RLRE algorithm might increase MRI images’ presentation effect and resolution. However, RLRE algorithm-based MRI images of the DCE sequence were more useful in the differential diagnosis of PC and BPH, thus facilitating disease therapy. | 29 August 2022 | [57] |
| 7. | The objective of this study was to extend artificial intelligence (AI) models that detect cancer in the prostate that extends to areas outside of it. Herein, by merging different models with image post-processing procedures and clinical judgement criteria, an autonomous strategy was developed to detect cancer spread outside the prostate barrier using prostate MRI images. | 29 August 2022 | [58] |
| 8. | This study observed that a deep learning-based algorithm using only H&E-stained digital slides can correctly predict ERG rearrangement status in most cases of prostatic adenocarcinoma. An artificial intelligence-based model could eliminate the need for extra tumour tissue to be used in ancillary studies to look for ERG gene rearrangement in prostatic adenocarcinoma. All of the models had comparable receiver operating characteristic (ROC) curves with area under the curve (AUC) values ranging from 0.82 to 0.85. These models’ sensitivity and specificity were 0.75 and 0.83, respectively. | 29 August 2022 | [59] |
| S. NO. | Dataset | Method | AUC | References |
|---|---|---|---|---|
| 1. | The bpMRI of 1513 including 73 patients 2 consecutive bpMRI scans with clinical variables (PSA, PSA density, and age) | Deep learning algorithm | 0.86 | [103] |
| 2. | Trans-rectal prostate biopsy of 109 patients | Random forest Neural network Ctree Support vector machine | 0.83 0.74 0.74 0.72 | [104] |
| 3. | Dataset of 551 patient including age, BMI, hypertension, diabetes, total PSA (tPSA), free PSA (fPSA), the ratio of serum fPSA to tPSA (f/tPSA), prostate volume (PV), PSA density (PSAD), neutrophil-to-lymphocyte ratio (NLR), and pathology reports of prostate biopsy | Tpsa logisticregression Multivariate logistic regression Decision tree Random forest Support vector machine | 0.84 0.91 0.92 1.00 0.88 | [105] |
| 4. | dataset of 315 patients available with preoperative T2WI, DWI, ADCMR images. Also, Trus-guided 12-needle puncture was performed within 3 months after MRI and provided P504S and P63 status | Random forest Gradient boosting Decision tree Logistic regression AdaBoost K-nearest neighbours. | 0.92 0.91 0.89 0.89 0.89 | [106] |
| 5. | 356 patients undergoing transrectal ultrasound-guided prostate biopsy | Logistic regression Decision tree classifier Dense neural network | 0.80 0.78 0.94 | [107] |
| 6. | 103 patients with mpMRI scan, PI-RADS V2 score was 4/5 and Prostatic biopsy results confirmed prostatic hyperplasia or PC | R-logistic R-SVM R-AdaBoost | 0.93 0.84 0.73 | [108] |
| 7. | 438 men with metastatic prostate cancer | Gradient boosting machine Model1 Model2 Model3 Model4 Model5 Model6 | 0.76 0.73 0.86 0.82 0.79 0.79 | [109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabaan, A.A.; Bakhrebah, M.A.; AlSaihati, H.; Alhumaid, S.; Alsubki, R.A.; Turkistani, S.A.; Al-Abdulhadi, S.; Aldawood, Y.; Alsaleh, A.A.; Alhashem, Y.N.; et al. Artificial Intelligence for Clinical Diagnosis and Treatment of Prostate Cancer. Cancers 2022, 14, 5595. https://doi.org/10.3390/cancers14225595
Rabaan AA, Bakhrebah MA, AlSaihati H, Alhumaid S, Alsubki RA, Turkistani SA, Al-Abdulhadi S, Aldawood Y, Alsaleh AA, Alhashem YN, et al. Artificial Intelligence for Clinical Diagnosis and Treatment of Prostate Cancer. Cancers. 2022; 14(22):5595. https://doi.org/10.3390/cancers14225595
Chicago/Turabian StyleRabaan, Ali A., Muhammed A. Bakhrebah, Hajir AlSaihati, Saad Alhumaid, Roua A. Alsubki, Safaa A. Turkistani, Saleh Al-Abdulhadi, Yahya Aldawood, Abdulmonem A. Alsaleh, Yousef N. Alhashem, and et al. 2022. "Artificial Intelligence for Clinical Diagnosis and Treatment of Prostate Cancer" Cancers 14, no. 22: 5595. https://doi.org/10.3390/cancers14225595
APA StyleRabaan, A. A., Bakhrebah, M. A., AlSaihati, H., Alhumaid, S., Alsubki, R. A., Turkistani, S. A., Al-Abdulhadi, S., Aldawood, Y., Alsaleh, A. A., Alhashem, Y. N., Almatouq, J. A., Alqatari, A. A., Alahmed, H. E., Sharbini, D. A., Alahmadi, A. F., Alsalman, F., Alsayyah, A., & Mutair, A. A. (2022). Artificial Intelligence for Clinical Diagnosis and Treatment of Prostate Cancer. Cancers, 14(22), 5595. https://doi.org/10.3390/cancers14225595