Obesity Influences the Expression of Key Immunomodulators in Normal Human Breast Tissue, Basal-like Breast Cancer Patients, and Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Triple-Negative Breast Cancer Cell Lines and Culturing Procedures
2.2. Adipocyte-Conditioned Media (ACM) Collection
2.3. Adipocyte-Conditioned Media (ACM) Sample Treatments
2.4. Bioinformatics and RNA Sequencing Data (Novogene, USA) GSEA Data Analysis
3. Results
3.1. Obesity Influences the Expression of Immunomodulators in Normal Breast Tissue Samples
3.2. Obesity Influences the Expression of Immunomodulators in African-American and European-American Breast Cancer Patients
3.3. Human Visceral Adipocyte-Conditioned Media Significantly Upregulated Key Immunomodulators in MDA MB 231 and MDA MB 468 Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Voduc, K.D.; Cheang, M.C.U.; Tyldesley, S.; Gelmon, K.; Nielsen, T.O.; Kennecke, H. Breast Cancer Subtypes and the Risk of Local and Regional Relapse. J. Clin. Oncol. 2010, 28, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.K.; Edmiston, S.; et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Marra, A.; Viale, G.; Curigliano, G. Recent advances in triple negative breast cancer: The immunotherapy era. BMC Med. 2019, 17, 90. [Google Scholar] [CrossRef]
- Critchley-Thorne, R.J.; Simons, D.L.; Yan, N.; Miyahira, A.K.; Dirbas, F.M.; Johnson, D.L.; Swetter, S.M.; Carlson, R.W.; Fisher, G.A.; Koong, A.; et al. Impaired interferon signaling is a common immune defect in human cancer. Proc. Natl. Acad. Sci. 2009, 106, 9010–9015. [Google Scholar] [CrossRef] [PubMed]
- Del Cornò, M.; D’Archivio, M.; Conti, L.; Scazzocchio, B.; Varì, R.; Donninelli, G.; Varano, B.; Giammarioli, S.; De Meo, S.; Silecchia, G.; et al. Visceral fat adipocytes from obese and colorectal cancer subjects exhibit distinct secretory and ω6 polyunsaturated fatty acid profiles and deliver immunosuppressive signals to innate immunity cells. Oncotarget 2016, 7, 63093–63105. [Google Scholar] [CrossRef] [PubMed]
- Faria, S.S.; Corrêa, L.H.; Heyn, G.S.; de Sant’Ana, L.P.; Almeida, R.D.N.; Magalhães, K.G. Obesity and Breast Cancer: The Role of Crown-Like Structures in Breast Adipose Tissue in Tumor Progression, Prognosis, and Therapy. J. Breast Cancer 2020, 23, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y. M1 and M2 polarization of macrophages: A mini-review. Med Biol. Sci. Eng. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Schuchardt, M.; Prüfer, N.; Tu, Y.; Herrmann, J.; Hu, X.-P.; Chebli, S.; Dahlke, K.; Zidek, W.; van der Giet, M.; Tölle, M. Dysfunctional high-density lipoprotein activates toll-like receptors via serum amyloid A in vascular smooth muscle cells. Sci. Rep. 2019, 9, 3421. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, H.; Sun, R.; Chen, B.; Han, F.; Li, Q.; Ni, Y.; Li, X.; Liu, J.; Mou, X.; et al. Serum amyloid a induces M2b-like macrophage polarization during liver inflammation. Oncotarget 2017, 8, 109238–109246. [Google Scholar] [CrossRef] [PubMed]
- Medrek, C.; Pontén, F.; Jirström, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, B.; Li, J.; Sun, S.; Yuan, J.; Sun, S. Cancer-associated adipocytes as immunomodulators in cancer. Biomark. Res. 2021, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Blank, A.; Cui, C.; Schoenfelt, K.Q.; Zhou, G.; Xu, Y.; Khramtsova, G.; Olopade, F.; Shah, A.M.; Khan, S.A.; et al. Metabolically activated adipose tissue macrophages link obesity to triple-negative breast cancer. J. Exp. Med. 2019, 216, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- Urs, S.; Smith, C.; Campbell, B.; Saxton, A.M.; Taylor, J.; Zhang, B.; Snoddy, J.; Voy, B.J.; Moustaid-Moussa, N. Gene Expression Profiling in Human Preadipocytes and Adipocytes by Microarray Analysis. J. Nutr. 2004, 134, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Ignacio, R.M.C.; Gibbs, C.R.; Kim, S.; Lee, E.-S.; Adunyah, S.E.; Son, D.-S. Serum amyloid A predisposes inflammatory tumor microenvironment in triple negative breast cancer. Oncotarget 2019, 10, 511–526. [Google Scholar] [CrossRef]
- Cozzo, A.J.; Fuller, A.M.; Makowski, L. Contribution of Adipose Tissue to Development of Cancer. Compr. Physiol. 2017, 8, 237–282. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Németh, J.; Angel, P.; Hess, J. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 2006, 72, 1622–1631. [Google Scholar] [CrossRef] [PubMed]
- Comen, E.A.; Bowman, R.L.; Kleppe, M. Underlying Causes and Therapeutic Targeting of the Inflammatory Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
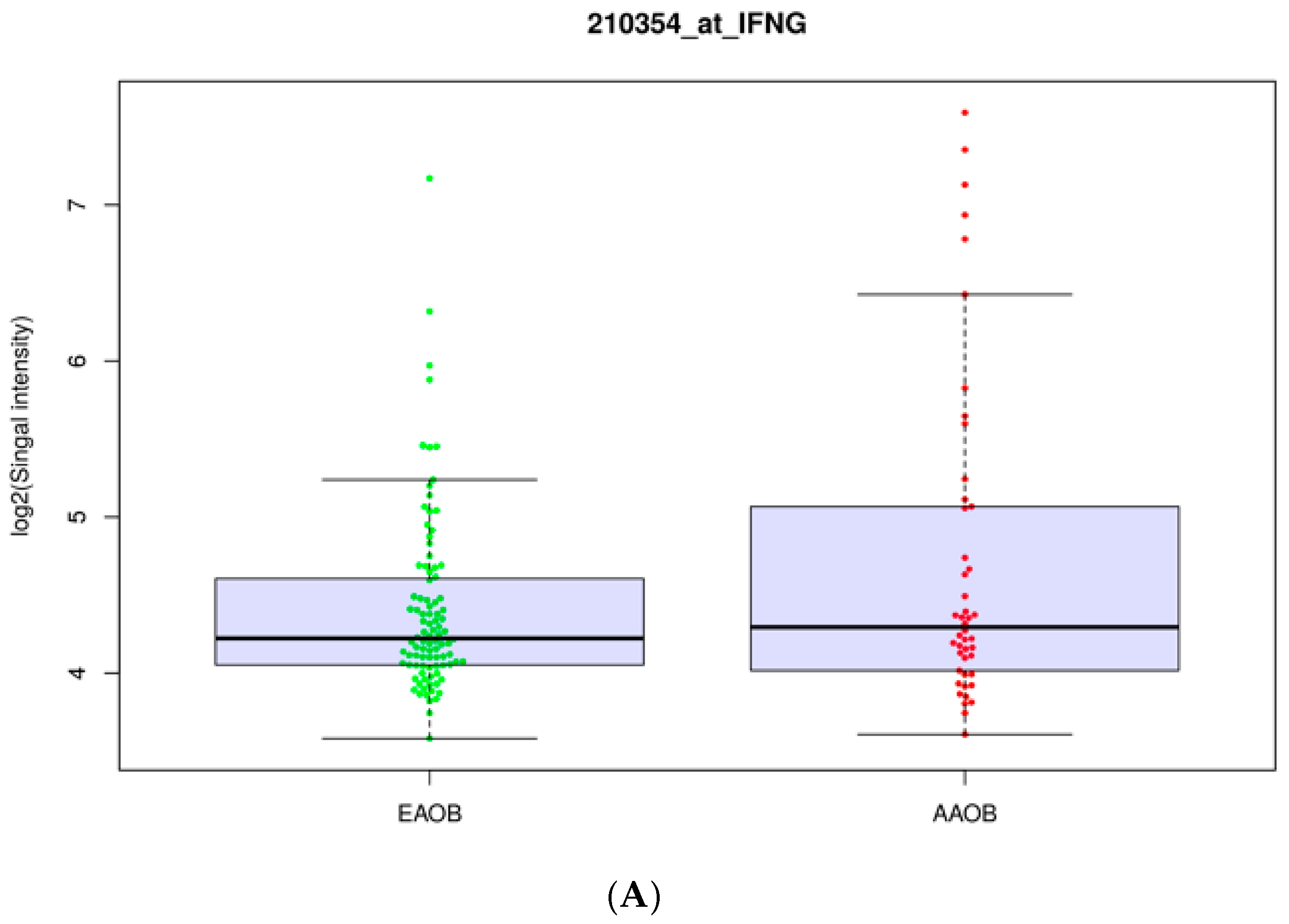
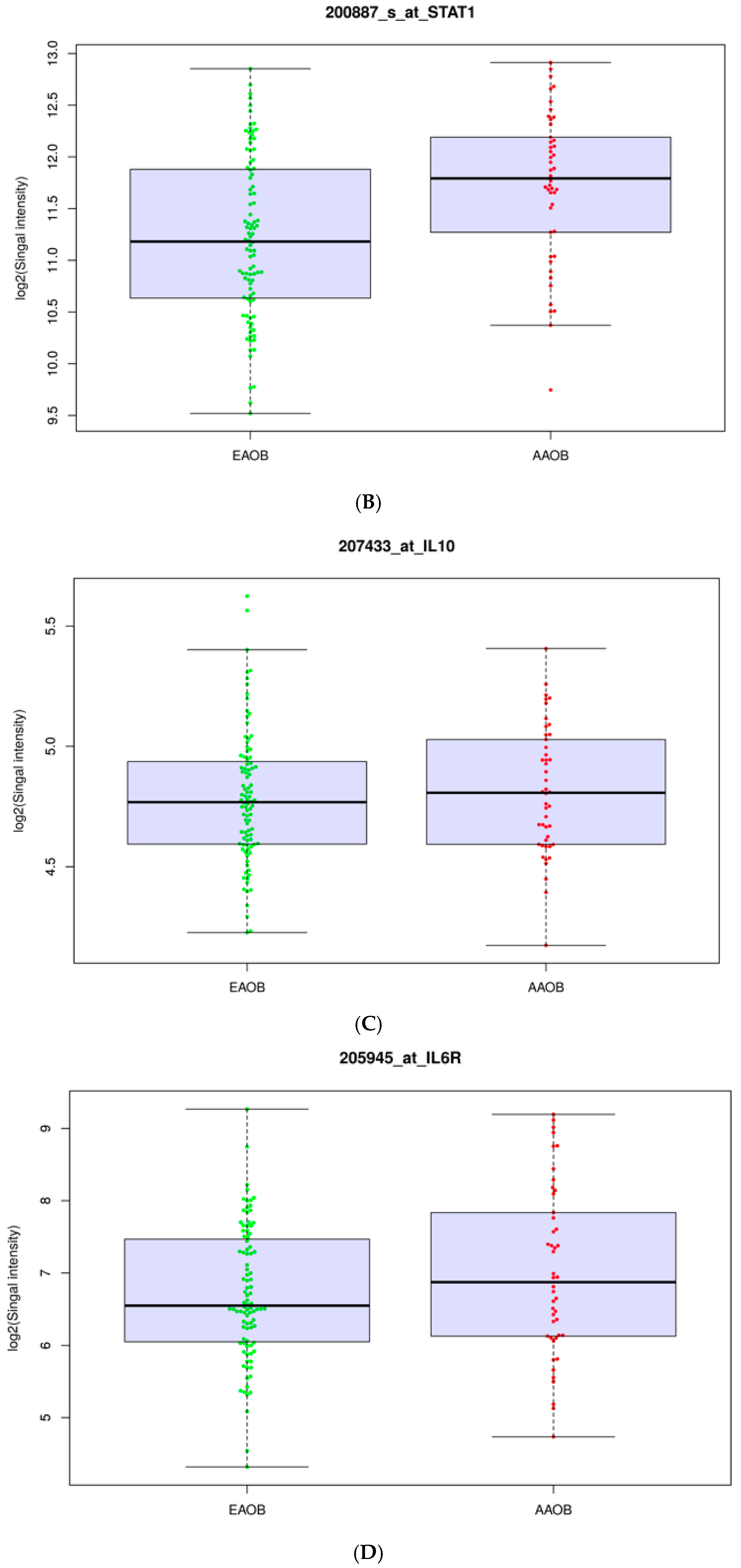
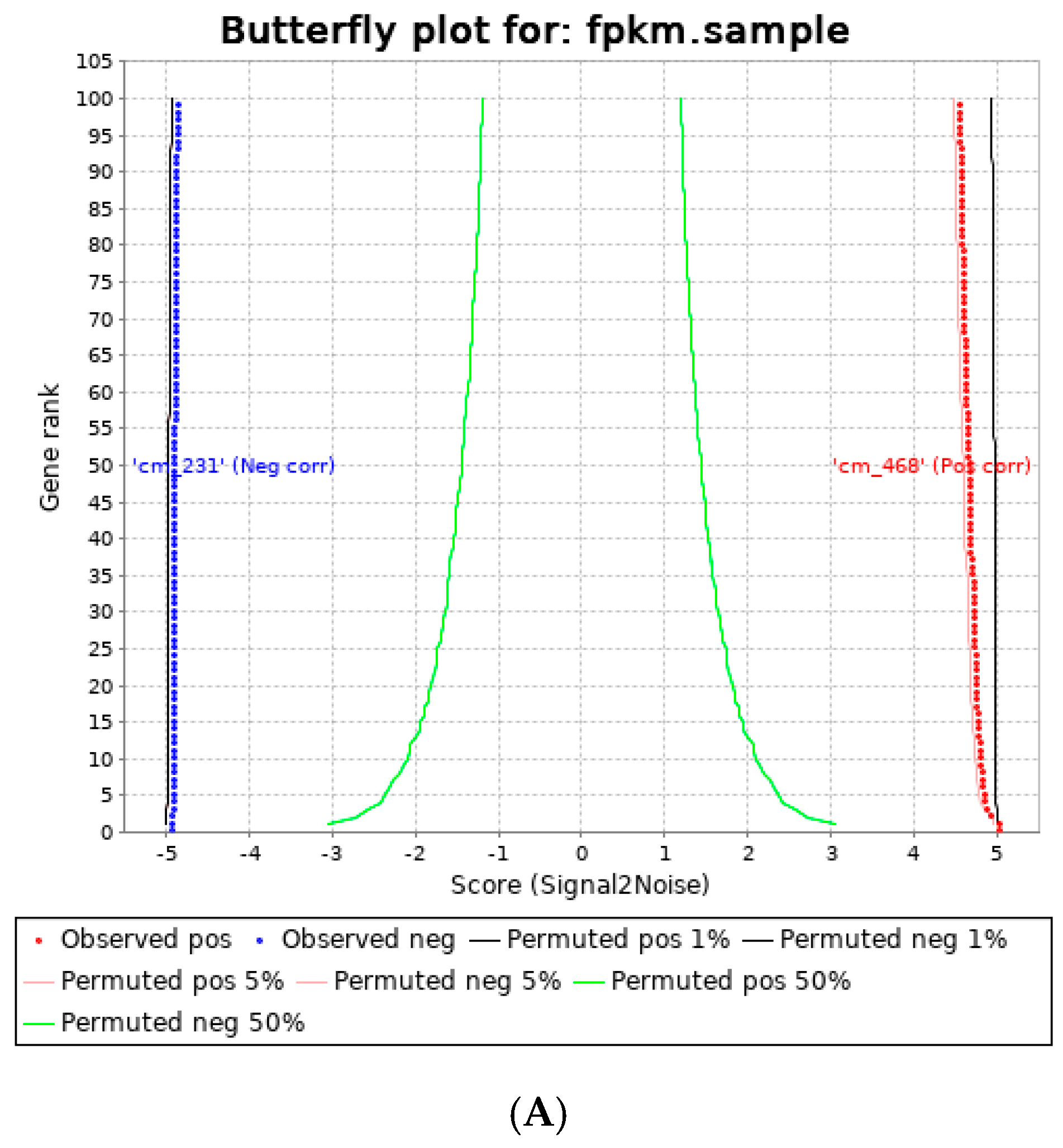
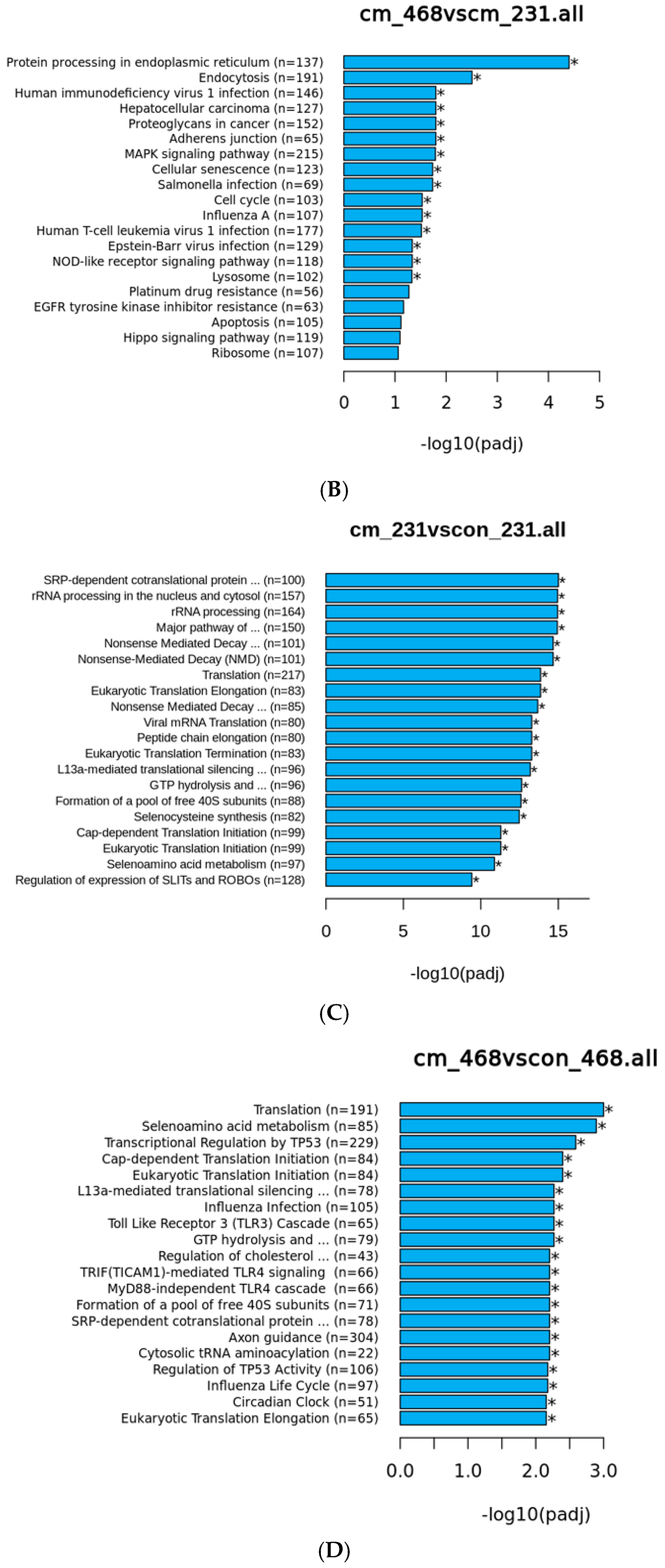
| Gene | Log2FC | p-Value |
|---|---|---|
| CCL2 | 0.37490968 | 0.0670578 |
| *** SAA1 | 0.82442043 | 0.04869515 |
| *** IL10 | 0.81295484 | 0.00218818 |
| IL6 | 0.72002903 | 0.06783646 |
| S100A8 | 0.55815376 | 0.12251586 |
| CXCL10 | 0.37490968 | 0.0670578 |
| S100A9 | 0.33124946 | 0.25984809 |
| *** IL6R | 0.22257634 | 0.04639715 |
| FPR1 | 0.32516237 | 0.25288916 |
| ARG1 | 0.19437957 | 0.26976972 |
| VTCN1 | −1.1666333 | 0.06078488 |
| a. BMI by Patient Ethnicity | |||||
| <25 | 25–29.99 | 30+ | statistic | p-value | |
| African-American | 22 (16.8%) | 26 (19.8%) | 46 (32.4%) | Fisher Exact | 0.0127 |
| Asian | 6 (4.6%) | 3 (2.3%) | 1 (0.7%) | ||
| European | 99 (75.6%) | 96 (73.3%) | 93 (65.5%) | ||
| Hispanic | 3 (2.3%) | 3 (2.3%) | 1 (0.7%) | ||
| Other | 1 (0.8%) | 3 (2.3%) | 1 (0.7%) | ||
| b. BMI by Tumor Grade | |||||
| <25 | 25–29.99 | 30+ | statistic | p-value | |
| Moderately Diff. | 4 (41.2%) | 47 (35.9%) | 45 (31.7%) | Fisher Exact | 0.0689 |
| Poorly Diff. | 47 (35.9%) | 57 (43.5%) | 70 (49.3%) | ||
| Well Diff. | 29 (22.1%) | 27 (20.6%) | 27 (19%) | ||
| c. BMI by Tumor Subtypes (Via Breastprs) | |||||
| <25 | 25–29.99 | 30+ | statistic | p-value | |
| Basal-like | 30 (22.9%) | 34 (26%) | 34 (23.9%) | Fisher Exact | 0.1785 |
| Her2-enriched | 15 (11.5%) | 21 (16%) | 12 (8.5%) | ||
| Luminal A | 68 (51.9%) | 64 (48.9%) | 77 (54.2%) | ||
| Luminal B | 17 (13%) | 10 (7.6%) | 16 (11.3%) | ||
| Normal-Like | 1 (0.8%) | 2 (1.5%) | 3 (2.1%) | ||
| Gene Set Name | Genes in Gene Set (K) | p-Value | FDR q-Value |
|---|---|---|---|
| HALLMARK_ALLOGRAFT_REJECTION | 200 | 4.78 × 10−11 | 8.79 × 10−8 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 200 | 8.77 × 10−9 | 8.07 × 10−6 |
| REACTOME_ADAPTIVE_IMMUNE_SYSTEM | 825 | 6.92 × 10−8 | 4.25 × 10−5 |
| REACTOME_INTERFERON_GAMMA_SIGNALING | 93 | 5.73 × 10−7 | 2.11 × 10−4 |
| HALLMARK_INFLAMMATORY_RESPONSE | 200 | 1.1 × 10−6 | 2.49 × 10−4 |
| REACTOME_CYTOKINE_SIGNALING_IN_IMMUNE_SYSTEM | 719 | 1.22 × 10−6 | 2.49 × 10−4 |
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | 108 | 1.58 × 10−6 | 2.9 × 10−4 |
| Gene | Log2FC | p-Value |
|---|---|---|
| CXCL8 | 1.2785982 | 5.12 × 10−6 |
| *** VTCN1 | 1.07774616 | 1.15 × 10−2 |
| *** CXADR | 0.94494262 | 6.27 × 10−4 |
| *** SAA2/SAA1 | 0.8243711 | 2.14 × 10−3 |
| *** STAT1 | 0.79720255 | 6.02 × 10−5 |
| *** IFNG | 0.66885529 | 5.29 × 10−6 |
| IL6R | 0.56846815 | 3.03 × 10−3 |
| CXCR4 | 0.29696438 | 9.08 × 10−3 |
| IL1B | 0.28383292 | 0.178386 |
| Gene | Log2FC | p-Value |
|---|---|---|
| *** SAA1 | 5.65381571 | N/A |
| *** CCL3 | 4.18500109 | 0.0072492 |
| *** IL1A | 3.42284939 | 3.04 × 10−13 |
| *** CXCL8 | 3.37438802 | 1.29 × 10−240 |
| *** VSIR | 3.15589864 | 5.00 × 10−285 |
| *** CCL5 | 2.71757585 | 1.93 × 10−24 |
| *** IL6 | 2.68749081 | 2.71 × 10−173 |
| VTCN1 | 2.629806 | 0.413524 |
| CXCL2 | 2.53842935 | 1.05 × 10−164 |
| *** TLR2 | 1.94916144 | 3.89 × 10−150 |
| *** IL6R | 1.41025426 | 1.02 × 10−23 |
| BCL3 | 1.03829151 | 3.12 × 10−25 |
| NFKB2 | 0.6914094 | 1.62 × 10−77 |
| *** TLR4 | 0.29417262 | 0.01207222 |
| Gene | Log2FC | p-Value |
|---|---|---|
| *** SAA1 | 6.8688814 | 1.84 × 10−34 |
| *** VSIR | 4.28271469 | 6.55 × 10−56 |
| *** SAA2 | 2.87594067 | 0.01984081 |
| *** BCL3 | 1.18211525 | 2.44 × 10−42 |
| *** JAK3 | 1.04059295 | 9.89 × 10−14 |
| *** STAT3 | 0.88316438 | 1.29 × 10−108 |
| IL6R | 0.75315446 | 1.19 × 10−13 |
| *** STAT1 | 0.73475986 | 9.26 × 10−76 |
| *** HIF1A | 0.3168464 | 5.28 × 10−11 |
| JAK1 | 0.22501884 | 2.52 × 10−9 |
| STAT2 | 0.16981301 | 0.01922399 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
King, D.; Khodary, M.G.; Samuel, T.; Yates, C.; Bedi, D. Obesity Influences the Expression of Key Immunomodulators in Normal Human Breast Tissue, Basal-like Breast Cancer Patients, and Cell Lines. Cancers 2022, 14, 5599. https://doi.org/10.3390/cancers14225599
King D, Khodary MG, Samuel T, Yates C, Bedi D. Obesity Influences the Expression of Key Immunomodulators in Normal Human Breast Tissue, Basal-like Breast Cancer Patients, and Cell Lines. Cancers. 2022; 14(22):5599. https://doi.org/10.3390/cancers14225599
Chicago/Turabian StyleKing, DeQuarius, Muhammad G. Khodary, Temesgen Samuel, Clayton Yates, and Deepa Bedi. 2022. "Obesity Influences the Expression of Key Immunomodulators in Normal Human Breast Tissue, Basal-like Breast Cancer Patients, and Cell Lines" Cancers 14, no. 22: 5599. https://doi.org/10.3390/cancers14225599
APA StyleKing, D., Khodary, M. G., Samuel, T., Yates, C., & Bedi, D. (2022). Obesity Influences the Expression of Key Immunomodulators in Normal Human Breast Tissue, Basal-like Breast Cancer Patients, and Cell Lines. Cancers, 14(22), 5599. https://doi.org/10.3390/cancers14225599







