CD155 Cooperates with PD-1/PD-L1 to Promote Proliferation of Esophageal Squamous Cancer Cells via PI3K/Akt and MAPK Signaling Pathways
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of Gene Expression and Immune Infiltration
2.2. Analysis of Gene and Protein Interaction
2.3. ESCA Sample Collection
2.4. Immunohistochemistry
2.5. Immunofluorescence Analysis
2.6. Cell Culture
2.7. CD155 and Nectin3 Knockdown
2.8. Xenograft Tumor Experiment
2.9. Flow Cytometry
2.10. Western Blotting
2.11. Cell Proliferation Analysis
2.12. mRNA Sequencing and Analysis
2.13. Statistical Analysis
3. Results
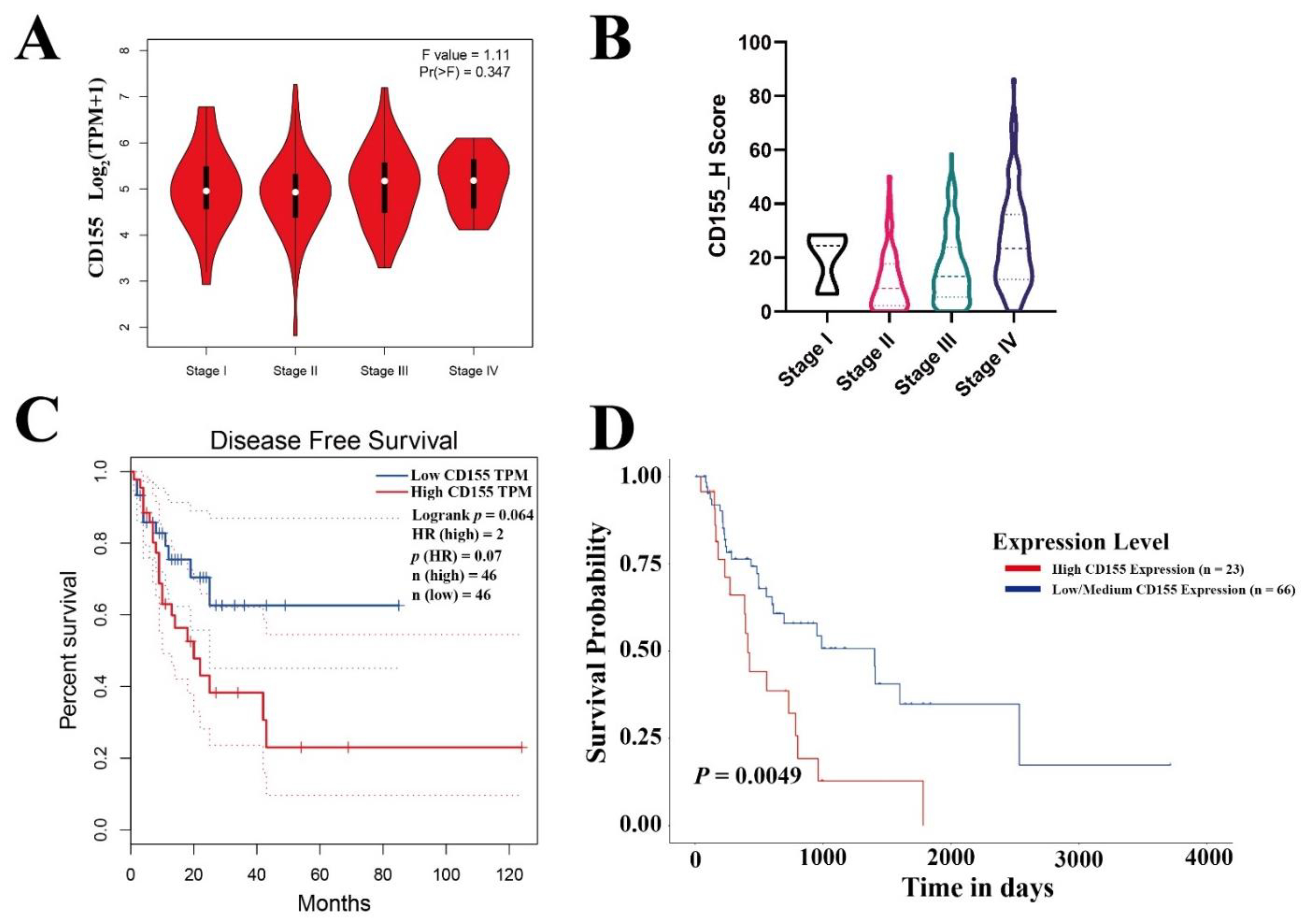
3.1. CD155 Is Highly Expressed in ESCA and Is Associated with Poor Patient Prognosis
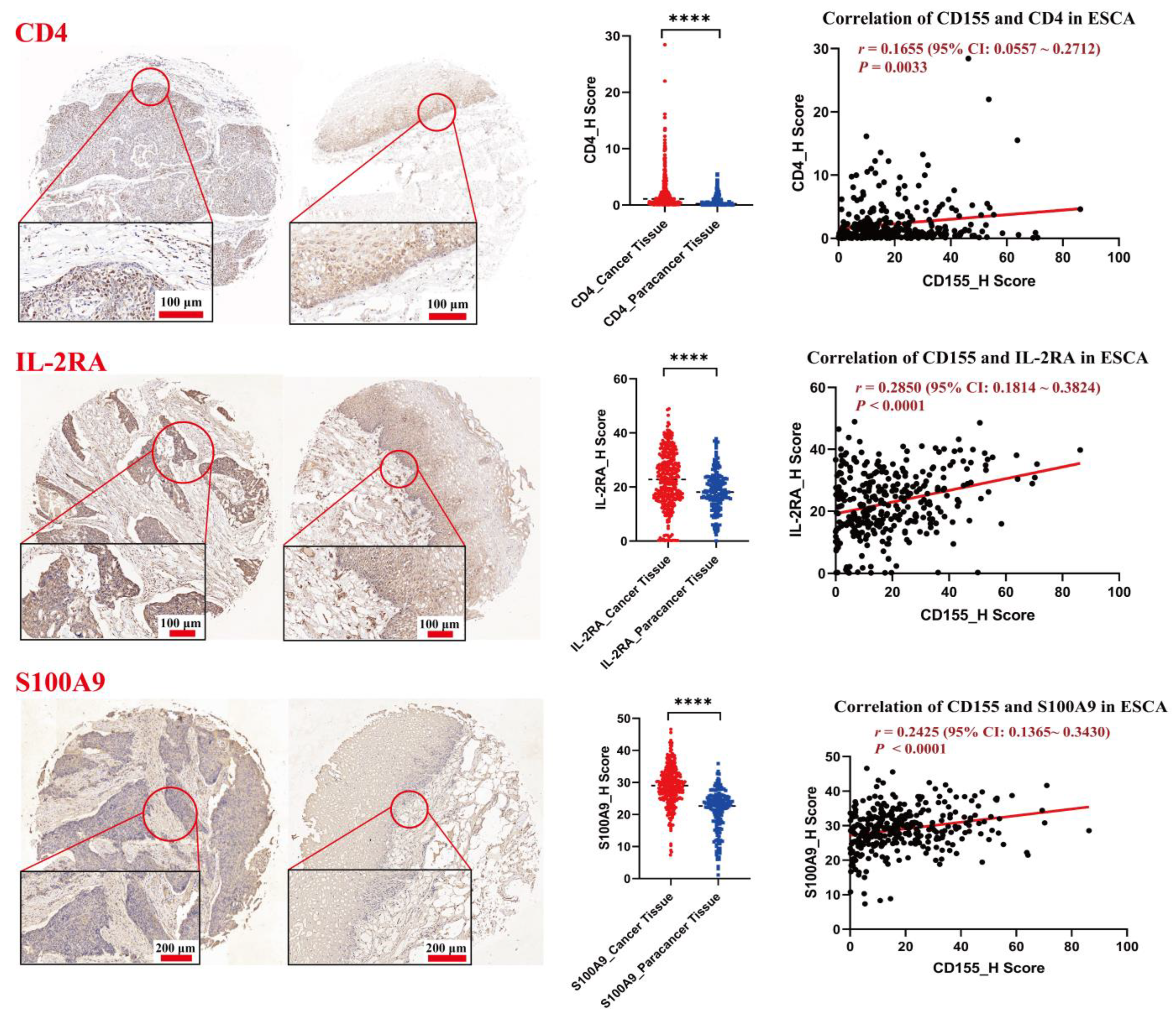
3.2. CD155 Is Positively Associated with the Expression of CD4, IL-2Rα and S100A9 in ESCA
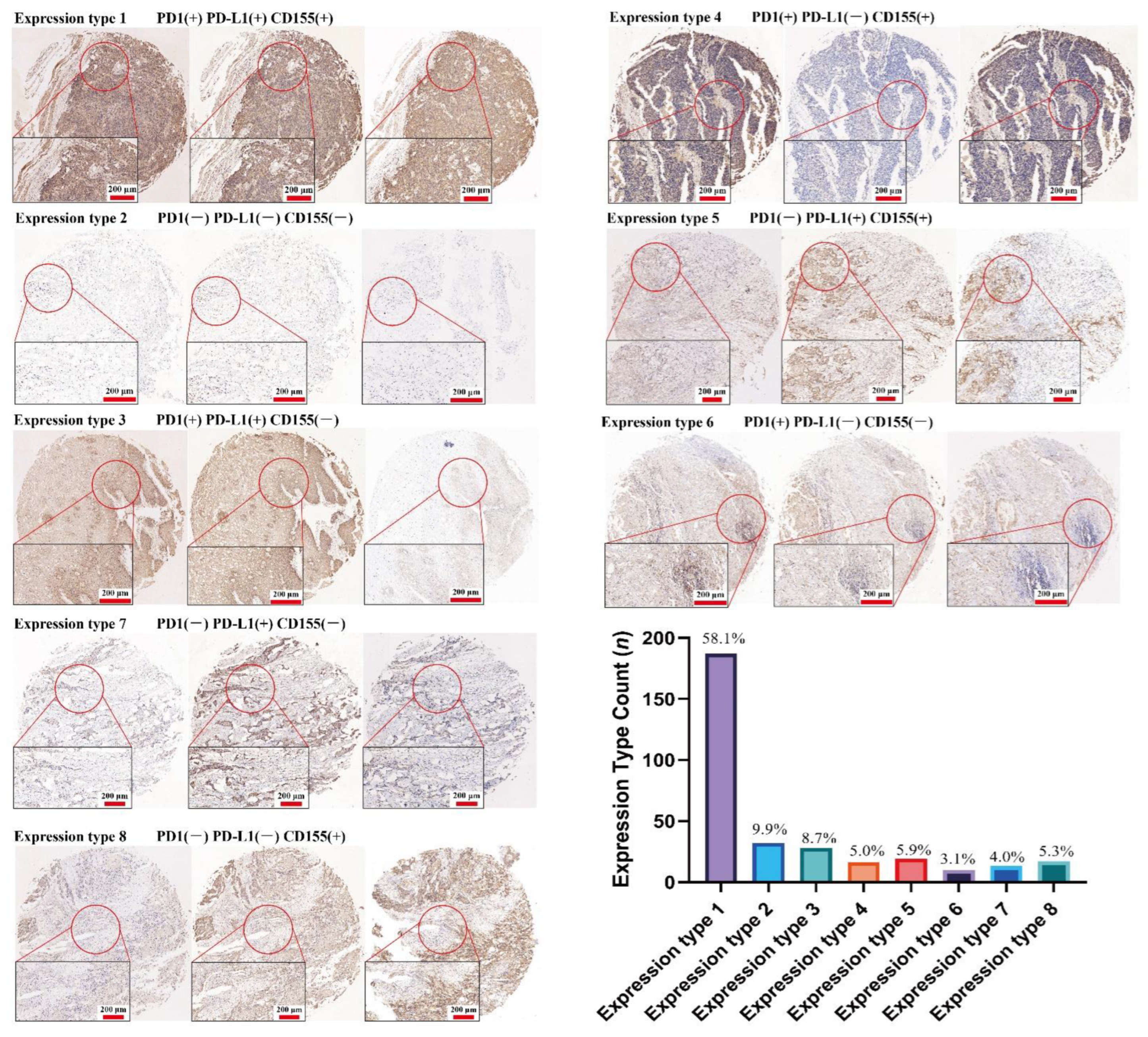
3.3. CD155 Can Cooperate with PD-1/PD-L1
3.4. CD155 Can Regulate the PI3K/Akt and MAPK Pathways in ESCA
3.5. CD155 Downregulation Inhibits ESCA Cell Proliferation by Impairing Cell Cycle and Inducing Cell Apoptosis
3.6. CD155 May Interact with Nectin3 and Regulate ESCA Proliferation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ESCA | Esophageal squamous cell cancer |
| IL | Interleukin |
| K520 | KYSE-520 |
| KD | Knockdown |
| NC | Non-targeting control |
| PD-1 | Programmed death protein 1 |
| PD-L1 | Programmed death-ligand 1 |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Fan, J.H.; Qiao, Y.L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol. Med. 2017, 14, 33–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forde, P.M.; Chaft, J.E.; Smith, K.N.; Anagnostou, V.; Cottrell, T.R.; Hellmann, M.D.; Zahurak, M.; Yang, S.C.; Jones, D.R.; Broderick, S.; et al. Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N. Engl. J. Med. 2018, 378, 1976–1986. [Google Scholar] [CrossRef]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.Y.; Choo, S.P.; Trojan, J.; Welling, T.H.R.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet (Lond. Engl.) 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Schadendorf, D.; Lipson, E.J.; Ascierto, P.A.; Matamala, L.; Castillo Gutiérrez, E.; Rutkowski, P.; Gogas, H.J.; Lao, C.D.; De Menezes, J.J.; et al. Relatlimab Nivolumab Versus Nivolumab Untreated Advanced. Melanoma. N. Engl. J. Med. 2022, 386, 24–34. [Google Scholar] [CrossRef]
- de Klerk, L.K.; Patel, A.K.; Derks, S.; Pectasides, E.; Augustin, J.; Uduman, M.; Raman, N.; Akarca, F.G.; McCleary, N.J.; Cleary, J.M.; et al. Phase II study of pembrolizumab in refractory esophageal cancer with correlates of response and survival. J. Immunother. Cancer 2021, 9, e002472. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, C.; Gao, X.; Li, X.; Cao, F.; Zhao, G.; Zhao, J.; Er, P.; Zhang, T.; Chen, X.; et al. Safety and Feasibility of Radiotherapy Plus Camrelizumab for Locally Advanced Esophageal Squamous Cell Carcinoma. Oncologist 2021, 26, e1110–e1124. [Google Scholar] [CrossRef]
- Agata, Y.; Kawasaki, A.; Nishimura, H.; Ishida, Y.; Tsubata, T.; Yagita, H.; Honjo, T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 1996, 8, 765–772. [Google Scholar] [CrossRef] [Green Version]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Yi, M.; Jiao, D.; Xu, H.; Liu, Q.; Zhao, W.; Han, X.; Wu, K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer 2018, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yan, Y.; Dong, J.; Duan, L. PD-1 expression on uveal melanoma induces tumor proliferation and predicts poor patient survival. Int. J. Biol. Markers 2020, 35, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Pawelczyk, K.; Piotrowska, A.; Ciesielska, U.; Jablonska, K.; Gletzel-Plucinska, N.; Grzegrzolka, J.; Podhorska-Okolow, M.; Dziegiel, P.; Nowinska, K. Role of PD-L1 Expression in Non-Small Cell Lung Cancer and Their Prognostic Significance according to Clinicopathological Factors and Diagnostic Markers. Int. J. Mol. Sci. 2019, 20, 824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Koo, B.S.; Kim, J.M.; Huang, S.; Rho, Y.S.; Bae, W.J.; Kang, H.J.; Kim, Y.S.; Moon, J.H.; Lim, Y.C. Wnt/β-catenin signalling maintains self-renewal and tumourigenicity of head and neck squamous cell carcinoma stem-like cells by activating Oct4. J. Pathol. 2014, 234, 99–107. [Google Scholar] [CrossRef]
- Tufano, M.; D’Arrigo, P.; D’Agostino, M.; Giordano, C.; Marrone, L.; Cesaro, E.; Romano, M.F.; Romano, S. PD-L1 Expression Fluctuates Concurrently with Cyclin D in Glioblastoma Cells. Cells 2021, 10, 2366. [Google Scholar] [CrossRef]
- Liu, M.Y.; Klement, J.D.; Langan, C.J.; van Riggelen, J.; Liu, K. Expression regulation and function of PD-1 and PD-L1 in T lymphoma cells. Cell. Immunol. 2021, 366, 104397. [Google Scholar] [CrossRef]
- Davern, M.; RM, O.B.; McGrath, J.; Donlon, N.E.; Melo, A.M.; Buckley, C.E.; Sheppard, A.D.; Reynolds, J.V.; Lynam-Lennon, N.; Maher, S.G.; et al. PD-1 blockade enhances chemotherapy toxicity in oesophageal adenocarcinoma. Sci. Rep. 2022, 12, 3259. [Google Scholar] [CrossRef]
- Bronte, V. The expanding constellation of immune checkpoints: A DNAMic control by CD155. J. Clin. Investig. 2018, 128, 2199–2201. [Google Scholar] [CrossRef]
- Freed-Pastor, W.A.; Lambert, L.J.; Ely, Z.A.; Pattada, N.B.; Bhutkar, A.; Eng, G.; Mercer, K.L.; Garcia, A.P.; Lin, L.; Rideout, W.M., 3rd; et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell 2021, 39, 1342–1360. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Li, M.; Hu, D.; Li, C.; Ge, B.; Jin, B.; Fan, Z. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. 2013, 20, 456–464. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xia, P.; Du, Y.; Liu, S.; Huang, G.; Chen, J.; Zhang, H.; Hou, N.; Cheng, X.; Zhou, L.; et al. T-cell immunoglobulin and ITIM domain (TIGIT) receptor/poliovirus receptor (PVR) ligand engagement suppresses interferon-γ production of natural killer cells via β-arrestin 2-mediated negative signaling. J. Biol. Chem. 2014, 289, 17647–17657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, W.; Zhang, H.; Han, F.; Chen, X.; Lin, R.; Wang, W.; Qiu, H.; Zhuang, Z.; Liao, Q.; Zhang, W.; et al. CD155T/TIGIT Signaling Regulates CD8(+) T-cell Metabolism and Promotes Tumor Progression in Human Gastric Cancer. Cancer Res. 2017, 77, 6375–6388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabre, S.; Reymond, N.; Cocchi, F.; Menotti, L.; Dubreuil, P.; Campadelli-Fiume, G.; Lopez, M. Prominent role of the Ig-like V domain in trans-interactions of nectins. Nectin3 and nectin 4 bind to the predicted C-C′-C′′-D beta-strands of the nectin1 V domain. J. Biol. Chem. 2002, 277, 27006–27013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujito, T.; Ikeda, W.; Kakunaga, S.; Minami, Y.; Kajita, M.; Sakamoto, Y.; Monden, M.; Takai, Y. Inhibition of cell movement and proliferation by cell-cell contact-induced interaction of Necl-5 with nectin-3. J. Cell Biol. 2005, 171, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.S.; Gao, Y.; Liu, C.; Chen, X.Q.; Zhou, L.M.; Yang, J.W.; Kui, X.Y.; Pei, Z.J. Comprehensive Analysis of Prognostic and Immune Infiltrates for E2F Transcription Factors in Human Pancreatic Adenocarcinoma. Front. Oncol. 2020, 10, 606735. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Tang, X.Y.; Xiong, Y.L.; Shi, A.P.; Sun, Y.; Han, Q.; Lv, Y.; Shi, X.G.; Frattini, M.; Malhotra, J.; Zheng, K.F.; et al. The downregulation of fibrinogen-like protein 1 inhibits the proliferation of lung adenocarcinoma via regulating MYC-target genes. Transl. Lung Cancer Res. 2022, 11, 404–419. [Google Scholar] [CrossRef]
- Maclean, A.; Bunni, E.; Makrydima, S.; Withington, A.; Kamal, A.M.; Valentijn, A.J.; Hapangama, D.K. Fallopian tube epithelial cells express androgen receptor and have a distinct hormonal responsiveness when compared with endometrial epithelium. Hum. Reprod. (Oxf. Engl.) 2020, 35, 2097–2106. [Google Scholar] [CrossRef]
- Dogan, S.; Vasudevaraja, V.; Xu, B.; Serrano, J.; Ptashkin, R.N.; Jung, H.J.; Chiang, S.; Jungbluth, A.A.; Cohen, M.A.; Ganly, I.; et al. DNA methylation-based classification of sinonasal undifferentiated carcinoma. Mod. Pathol. Off. J. U.S. Can. Acad. Pathol. Inc. 2019, 32, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Paschalis, A.; Sheehan, B.; Riisnaes, R.; Rodrigues, D.N.; Gurel, B.; Bertan, C.; Ferreira, A.; Lambros, M.B.K.; Seed, G.; Yuan, W.; et al. Prostate-specific Membrane Antigen Heterogeneity and DNA Repair Defects in Prostate Cancer. Eur. Urol. 2019, 76, 469–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, R.; Wang, B.; Cai, X.R.; Chen, Z.S.; Du, Q.; Zhou, L.Y.; Ye, J.M.; Chen, Y.L. CD276 Promotes Vasculogenic Mimicry Formation in Hepatocellular Carcinoma via the PI3K/AKT/MMPs Pathway. OncoTargets Ther. 2020, 13, 11485–11498. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, G.; Song, J.; Cai, Z.; Yang, J.; Chen, Z.; Wang, Y.; Huang, Y.; Gao, Q. B7-H3 Promotes the Migration and Invasion of Human Bladder Cancer Cells via the PI3K/Akt/STAT3 Signaling Pathway. J. Cancer 2017, 8, 816–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, C.; Fan, J.; Zhu, Q.; Feng, Y.; Pan, J.; Peng, J.; Shi, J.; Qi, S.; Liu, Y. CD47 promotes the proliferation and migration of adamantinomatous craniopharyngioma cells by activating the MAPK/ERK pathway, and CD47 blockade facilitates microglia-mediated phagocytosis. Neuropathol. Appl. Neurobiol. 2022, 48, e12795. [Google Scholar] [CrossRef] [PubMed]
- Molfetta, R.; Zitti, B.; Lecce, M.; Milito, N.D.; Stabile, H.; Fionda, C.; Cippitelli, M.; Gismondi, A.; Santoni, A.; Paolini, R. CD155: A Multi-Functional Molecule in Tumor Progression. Int. J. Mol. Sci. 2020, 21, 922. [Google Scholar] [CrossRef] [Green Version]
- Bryceson, Y.T.; March, M.E.; Ljunggren, H.G.; Long, E.O. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood 2006, 107, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Lozano, E.; Dominguez-Villar, M.; Kuchroo, V.; Hafler, D.A. The TIGIT/CD226 axis regulates human T cell function. J. Immunol. 2012, 188, 3869–3875. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Yang, J.; Shi, A.; Xiong, Y.; Wen, M.; Luo, Z.; Tian, H.; Zheng, K.; Liu, Y.; Shu, C.; et al. CD155 Cooperates with PD-1/PD-L1 to Promote Proliferation of Esophageal Squamous Cancer Cells via PI3K/Akt and MAPK Signaling Pathways. Cancers 2022, 14, 5610. https://doi.org/10.3390/cancers14225610
Tang X, Yang J, Shi A, Xiong Y, Wen M, Luo Z, Tian H, Zheng K, Liu Y, Shu C, et al. CD155 Cooperates with PD-1/PD-L1 to Promote Proliferation of Esophageal Squamous Cancer Cells via PI3K/Akt and MAPK Signaling Pathways. Cancers. 2022; 14(22):5610. https://doi.org/10.3390/cancers14225610
Chicago/Turabian StyleTang, Xiyang, Jie Yang, Anping Shi, Yanlu Xiong, Miaomiao Wen, Zhonglin Luo, Huanhuan Tian, Kaifu Zheng, Yujian Liu, Chen Shu, and et al. 2022. "CD155 Cooperates with PD-1/PD-L1 to Promote Proliferation of Esophageal Squamous Cancer Cells via PI3K/Akt and MAPK Signaling Pathways" Cancers 14, no. 22: 5610. https://doi.org/10.3390/cancers14225610
APA StyleTang, X., Yang, J., Shi, A., Xiong, Y., Wen, M., Luo, Z., Tian, H., Zheng, K., Liu, Y., Shu, C., Ma, N., Wang, R., & Zhao, J. (2022). CD155 Cooperates with PD-1/PD-L1 to Promote Proliferation of Esophageal Squamous Cancer Cells via PI3K/Akt and MAPK Signaling Pathways. Cancers, 14(22), 5610. https://doi.org/10.3390/cancers14225610






