Implementing the Point Spread Function Deconvolution for Better Molecular Characterization of Newly Diagnosed Gliomas: A Dynamic 18F-FDOPA PET Radiomics Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. PET Acquisition and Image Reconstruction
2.3. Segmentation and Image Pre-Processing
2.4. Feature Extraction
2.4.1. Voxel-Based
2.4.2. Region-Based
2.5. Model Training and Validation
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. PSFd Impact on Radiomics Features
3.3. IDH Mutation Prediction
3.4. 1p/19q Codeletion Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teo, B.-K.; Seo, Y.; Bacharach, S.L.; Carrasquillo, J.A.; Libutti, S.K.; Shukla, H.; Hasegawa, B.H.; Hawkins, R.A.; Franc, B.L. Partial-Volume Correction in PET: Validation of an Iterative Postreconstruction Method with Phantom and Patient Data. J. Nucl. Med. 2007, 48, 802–810. [Google Scholar] [CrossRef]
- Tohka, J.; Reilhac, A. Deconvolution-Based Partial Volume Correction in Raclopride-PET and Monte Carlo Comparison to MR-Based Method. Neuroimage 2008, 39, 1570–1584. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Alessio, A.M.; Kinahan, P.E. Noise and Signal Properties in PSF-Based Fully 3D PET Image Reconstruction: An Experimental Evaluation. Phys. Med. Biol. 2010, 55, 1453–1473. [Google Scholar] [CrossRef] [PubMed]
- Prieto, E.; Martí-Climent, J.M.; Morán, V.; Sancho, L.; Barbés, B.; Arbizu, J.; Richter, J.A. Brain PET Imaging Optimization with Time of Flight and Point Spread Function Modelling. Phys. Med. 2015, 31, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Golla, S.S.V.; Lubberink, M.; van Berckel, B.N.M.; Lammertsma, A.A.; Boellaard, R. Partial Volume Correction of Brain PET Studies Using Iterative Deconvolution in Combination with HYPR Denoising. EJNMMI Res. 2017, 7, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verger, A.; Kas, A.; Darcourt, J.; Guedj, E. PET Imaging in Neuro-Oncology: An Update and Overview of a Rapidly Growing Area. Cancers 2022, 14, 1103. [Google Scholar] [CrossRef]
- Verger, A.; Imbert, L.; Zaragori, T. Dynamic Amino-Acid PET in Neuro-Oncology: A Prognostic Tool Becomes Essential. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4129–4132. [Google Scholar] [CrossRef]
- Lohmann, P.; Galldiks, N.; Kocher, M.; Heinzel, A.; Filss, C.P.; Stegmayr, C.; Mottaghy, F.M.; Fink, G.R.; Jon Shah, N.; Langen, K.-J. Radiomics in Neuro-Oncology: Basics, Workflow, and Applications. Methods 2021, 188, 112–121. [Google Scholar] [CrossRef]
- Zaragori, T.; Oster, J.; Roch, V.; Hossu, G.; Chawki, M.B.; Grignon, R.; Pouget, C.; Gauchotte, G.; Rech, F.; Blonski, M.; et al. 18F-FDOPA PET for the Non-Invasive Prediction of Glioma Molecular Parameters: A Radiomics Study. J. Nucl. Med. 2021, 63, 147–157. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahrari, S.; Zaragori, T.; Rozenblum, L.; Oster, J.; Imbert, L.; Kas, A.; Verger, A. Relevance of Dynamic 18F-DOPA PET Radiomics for Differentiation of High-Grade Glioma Progression from Treatment-Related Changes. Biomedicines 2021, 9, 1924. [Google Scholar] [CrossRef] [PubMed]
- Rogasch, J.M.M.; Albers, J.; Steinkrüger, F.L.; Lukas, M.; Wedel, F.; Amthauer, H.; Furth, C. Point Spread Function Reconstruction for Integrated 18F-FET PET/MRI in Patients with Glioma: Does It Affect SUVs and Respective Tumor-to-Background Ratios? Clin. Nucl. Med. 2019, 44, e280. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; François, M.; Chaboub, N.; Le Reste, P.-J.; Devillers, A.; Saint-Jalmes, H.; Le Jeune, F.; Palard-Novello, X. Impact of Point-Spread Function Reconstruction on Dynamic and Static 18F-DOPA PET/CT Quantitative Parameters in Glioma. Quant. Imaging Med. Surg. 2022, 12, 1397–1404. [Google Scholar] [CrossRef]
- Law, I.; Albert, N.L.; Arbizu, J.; Boellaard, R.; Drzezga, A.; Galldiks, N.; la Fougère, C.; Langen, K.-J.; Lopci, E.; Lowe, V.; et al. Joint EANM/EANO/RANO Practice Guidelines/SNMMI Procedure Standards for Imaging of Gliomas Using PET with Radiolabelled Amino Acids and [18F]FDG: Version 1.0. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 540–557. [Google Scholar] [CrossRef] [Green Version]
- Perizzolo, M.; Winkfein, B.; Hui, S.; Krulicki, W.; Chan, J.A.; Demetrick, D.J. IDH Mutation Detection in Formalin-Fixed Paraffin-Embedded Gliomas Using Multiplex PCR and Single-Base Extension: IDH1/2 Mutation Detection by SNaPshot®. Brain Pathol. 2012, 22, 619–624. [Google Scholar] [CrossRef]
- Ginet, M.; Zaragori, T.; Marie, P.-Y.; Roch, V.; Gauchotte, G.; Rech, F.; Blonski, M.; Lamiral, Z.; Taillandier, L.; Imbert, L.; et al. Integration of Dynamic Parameters in the Analysis of 18F-FDopa PET Imaging Improves the Prediction of Molecular Features of Gliomas. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1381–1390. [Google Scholar] [CrossRef]
- Schiepers, C.; Chen, W.; Cloughesy, T.; Dahlbom, M.; Huang, S.-C. 18F-FDOPA Kinetics in Brain Tumors. J. Nucl. Med. 2007, 48, 1651–1661. [Google Scholar] [CrossRef] [Green Version]
- Salvadori, J.; Imbert, L.; Perrin, M.; Karcher, G.; Lamiral, Z.; Marie, P.-Y.; Verger, A. Head-to-Head Comparison of Image Quality between Brain 18F-FDG Images Recorded with a Fully Digital versus a Last-Generation Analog PET Camera. EJNMMI Res. 2019, 9, 61. [Google Scholar] [CrossRef]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterization of Tumor Heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floberg, J.M.; Mistretta, C.A.; Weichert, J.P.; Hall, L.T.; Holden, J.E.; Christian, B.T. Improved Kinetic Analysis of Dynamic PET Data with Optimized HYPR-LR. Med. Phys. 2012, 39, 3319–3331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bros, M.; Zaragori, T.; Rech, F.; Blonski, M.; Hossu, G.; Taillandier, L.; Marie, P.-Y.; Verger, A. Effects of Carbidopa Premedication on 18F-FDOPA PET Imaging of Glioma: A Multiparametric Analysis. Cancers 2021, 13, 5340. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-Based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Beare, R.; Lowekamp, B.; Yaniv, Z. Image Segmentation, Registration and Characterization in R with SimpleITK. J. Stat. Softw. 2018, 86, 8. [Google Scholar] [CrossRef] [Green Version]
- Leger, S.; Zwanenburg, A.; Pilz, K.; Lohaus, F.; Linge, A.; Zöphel, K.; Kotzerke, J.; Schreiber, A.; Tinhofer, I.; Budach, V.; et al. A Comparative Study of Machine Learning Methods for Time-to-Event Survival Data for Radiomics Risk Modelling. Sci. Rep. 2017, 7, 13206. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ojala, M.; Garriga, G.C. Permutation Tests for Studying Classifier Performance. J. Mach. Learn. Res. 2010, 11, 1833–1863. [Google Scholar]
- Escobar, T.; Vauclin, S.; Orlhac, F.; Nioche, C.; Pineau, P.; Champion, L.; Brisse, H.; Buvat, I. Voxel-Wise Supervised Analysis of Tumors with Multimodal Engineered Features to Highlight Interpretable Biological Patterns. Med. Phys. 2022, 49, 3816–3829. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Unterrainer, M.; Ruf, V.; von Rohr, K.; Suchorska, B.; Mittlmeier, L.M.; Beyer, L.; Brendel, M.; Wenter, V.; Kunz, W.G.; Bartenstein, P.; et al. TERT-Promoter Mutational Status in Glioblastoma—Is There an Association with Amino Acid Uptake on Dynamic 18F-FET PET? Front. Oncol. 2021, 11, 645316. [Google Scholar] [CrossRef]
- Li, Z.; Kaiser, L.; Holzgreve, A.; Ruf, V.C.; Suchorska, B.; Wenter, V.; Quach, S.; Herms, J.; Bartenstein, P.; Tonn, J.-C.; et al. Prediction of TERTp-Mutation Status in IDH-Wildtype High-Grade Gliomas Using Pre-Treatment Dynamic [18F]FET PET Radiomics. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4415–4425. [Google Scholar] [CrossRef] [PubMed]
- Zaragori, T.; Ginet, M.; Marie, P.-Y.; Roch, V.; Grignon, R.; Gauchotte, G.; Rech, F.; Blonski, M.; Lamiral, Z.; Taillandier, L.; et al. Use of Static and Dynamic [18F]-F-DOPA PET Parameters for Detecting Patients with Glioma Recurrence or Progression. EJNMMI Res. 2020, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Munk, O.L.; Tolbod, L.P.; Hansen, S.B.; Bogsrud, T.V. Point-Spread Function Reconstructed PET Images of Sub-Centimeter Lesions Are Not Quantitative. EJNMMI Phys. 2017, 4, 5. [Google Scholar] [CrossRef] [PubMed]
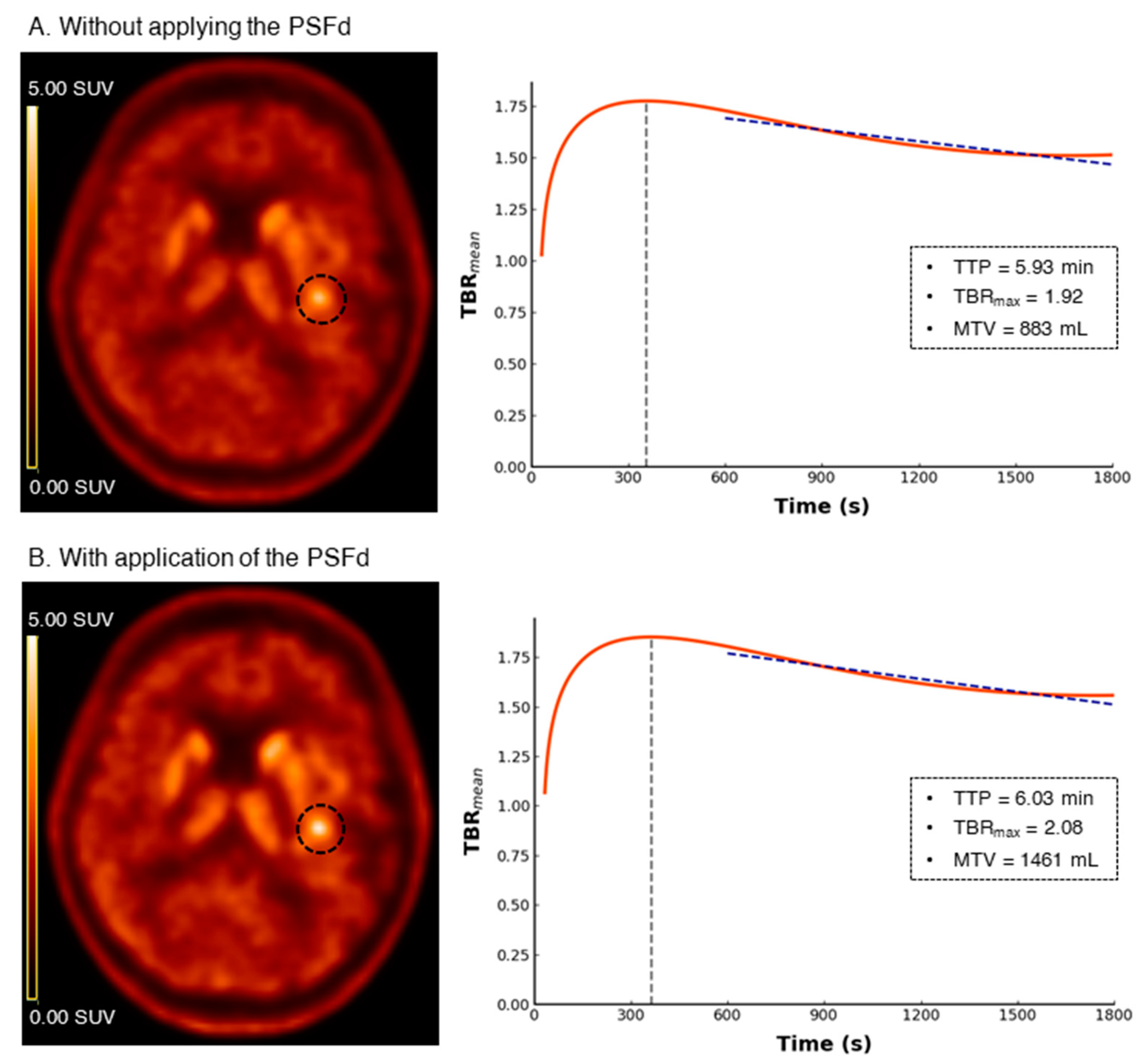



| Astrocytoma IDH-Mutant and 1p/19q Non-Codeleted | Oligodendroglioma IDH-Mutant and 1p/19q Codeleted | Glioblastoma IDH-Wildtype | p | |
|---|---|---|---|---|
| N = 12 | N = 12 | N = 33 | ||
| Age, median (IQR) | 41 (27–57) | 48 (41–62) | 62 (54–71) | 0.003 # |
| Sex, n (%) | ||||
| Female | 7 (58) | 6 (50) | 15 (45) | 0.745 |
| Male | 5 (42) | 6 (50) | 18 (55) | |
| Tumor resection, n (%) | ||||
| Surgery | 7 (58) | 6 (50) | 5 (15) | 0.007 # |
| Biopsy | 5 (42) | 6 (50) | 28 (85) | |
| Histopathological WHO grade, n (%) | ||||
| Grade II | 8 (66) | 12 (100) | - | <0.001 # |
| Grade III | 2 (17) | - | - | |
| Grade IV | 2 (17) | - | 33 (100) | |
| Carbidopa premedication, n (%) | 10 (83) | 11 (92) | 25 (76) | 0.473 |
| TBRmean *, median (IQR) | 1.92 (1.8–2.0) | 1.95 (1.8–2.1) | 2.12 (1.9–2.3) | 0.023 # |
| TBRmax *, median (IQR) | 2.84 (2.5–4.0) | 2.83 (2.6–3.6) | 3.58 (2.9–4.4) | 0.068 |
| Without PSFd | With PSFd | |||||||
|---|---|---|---|---|---|---|---|---|
| Features/Metrics | AUC | Sensitivity | Specificity | B_ACC | AUC | Sensitivity | Specificity | B_ACC |
| Voxel-based analysis | ||||||||
| Static | 0.686 ξ (0.656, 0.715) | 0.797 (0.760, 0.831) | 0.496 (0.458, 0.532) | 0.686 (0.656, 0.715) | 0.785 *,ξ (0.756, 0.815) | 0.869 (0.838, 0.897) | 0.542 (0.509, 0.572) | 0.706 (0.684, 0.728) |
| Dynamic | 0.759 ¥,ξ (0.730, 0.787) | 0.733 (0.697, 0.771) | 0.640 (0.605, 0.675) | 0.686 (0.661, 0.710) | 0.764 ξ (0.737, 0.791) | 0.743 (0.709, 0.777) | 0.653 (0.622, 0.686) | 0.698 (0.676, 0.719) |
| Static/Dynamic | 0.791 ¥,§,ξ (0.765, 0.813) | 0.755 (0.720, 0.791) | 0.630 (0.593, 0.666) | 0.693 (0.666, 0.718) | 0.831 *,¥,§,ξ (0.804, 0.854) | 0.810 (0.777, 0.843) | 0.672 (0.636, 0.707) | 0.741 (0.719, 0.763) |
| Region-based analysis | ||||||||
| Static/Dynamic | 0.827 ‡,ξ (0.806, 0.848) | 0.667 (0.620, 0.718) | 0.760 (0.727, 0.792) | 0.714 (0.689, 0.739) | 0.883 *,‡,ξ (0.863, 0.903) | 0.666 (0.626, 0.709) | 0.858 (0.828, 0.887) | 0.762 (0.740, 0.785) |
| Without PSFd | With PSFd | |||||||
|---|---|---|---|---|---|---|---|---|
| Features/Metrics | AUC | Sensitivity | Specificity | B_ACC | AUC | Sensitivity | Specificity | B_ACC |
| Voxel-based analysis | ||||||||
| Static | 0.664 ξ (0.633, 0.693) | 0.564 (0.505, 0.627) | 0.604 (0.569, 0.636) | 0.584 (0.551, 0.617) | 0.681 ξ (0.652, 0.710) | 0.552 (0.492, 0.607) | 0.623 (0.594, 0.656) | 0.588 (0.556, 0.618) |
| Dynamic | 0.688 ξ (0.647, 0.727) | 0.617 (0.555, 0.678) | 0.695 (0.663, 0.728) | 0.656 (0.624, 0.688) | 0.721 *,¥,ξ (0.686, 0.756) | 0.628 (0.567, 0.692) | 0.686 (0.656, 0.716) | 0.657 (0.624, 0.690) |
| Static/Dynamic | 0.683 ξ (0.648, 0.716) | 0.650 (0.595, 0.708) | 0.688 (0.653, 0.718) | 0.669 (0.636, 0.702) | 0.755 *,¥,ξ (0.725, 0.786) | 0.590 (0.532, 0.653) | 0.728 (0.697, 0.759) | 0.659 (0.624, 0.691) |
| Region-based analysis | ||||||||
| Static/Dynamic | 0.787 ‡,ξ (0.753, 0.815) | 0.679 (0.615, 0.733) | 0.726 (0.698, 0.754) | 0.703 (0.672, 0.729) | 0.828 *,‡,ξ (0.791, 0.860) | 0.750 (0.692, 0.802) | 0.790 (0.760, 0.820) | 0.770 (0.741, 0.797) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahrari, S.; Zaragori, T.; Bros, M.; Oster, J.; Imbert, L.; Verger, A. Implementing the Point Spread Function Deconvolution for Better Molecular Characterization of Newly Diagnosed Gliomas: A Dynamic 18F-FDOPA PET Radiomics Study. Cancers 2022, 14, 5765. https://doi.org/10.3390/cancers14235765
Ahrari S, Zaragori T, Bros M, Oster J, Imbert L, Verger A. Implementing the Point Spread Function Deconvolution for Better Molecular Characterization of Newly Diagnosed Gliomas: A Dynamic 18F-FDOPA PET Radiomics Study. Cancers. 2022; 14(23):5765. https://doi.org/10.3390/cancers14235765
Chicago/Turabian StyleAhrari, Shamimeh, Timothée Zaragori, Marie Bros, Julien Oster, Laetitia Imbert, and Antoine Verger. 2022. "Implementing the Point Spread Function Deconvolution for Better Molecular Characterization of Newly Diagnosed Gliomas: A Dynamic 18F-FDOPA PET Radiomics Study" Cancers 14, no. 23: 5765. https://doi.org/10.3390/cancers14235765
APA StyleAhrari, S., Zaragori, T., Bros, M., Oster, J., Imbert, L., & Verger, A. (2022). Implementing the Point Spread Function Deconvolution for Better Molecular Characterization of Newly Diagnosed Gliomas: A Dynamic 18F-FDOPA PET Radiomics Study. Cancers, 14(23), 5765. https://doi.org/10.3390/cancers14235765







