Epigenetic Regulation in Chromium-, Nickel- and Cadmium-Induced Carcinogenesis
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Epigenetic Mechanisms
1.1.1. DNA Methylation and Demethylation
1.1.2. Histone Modification
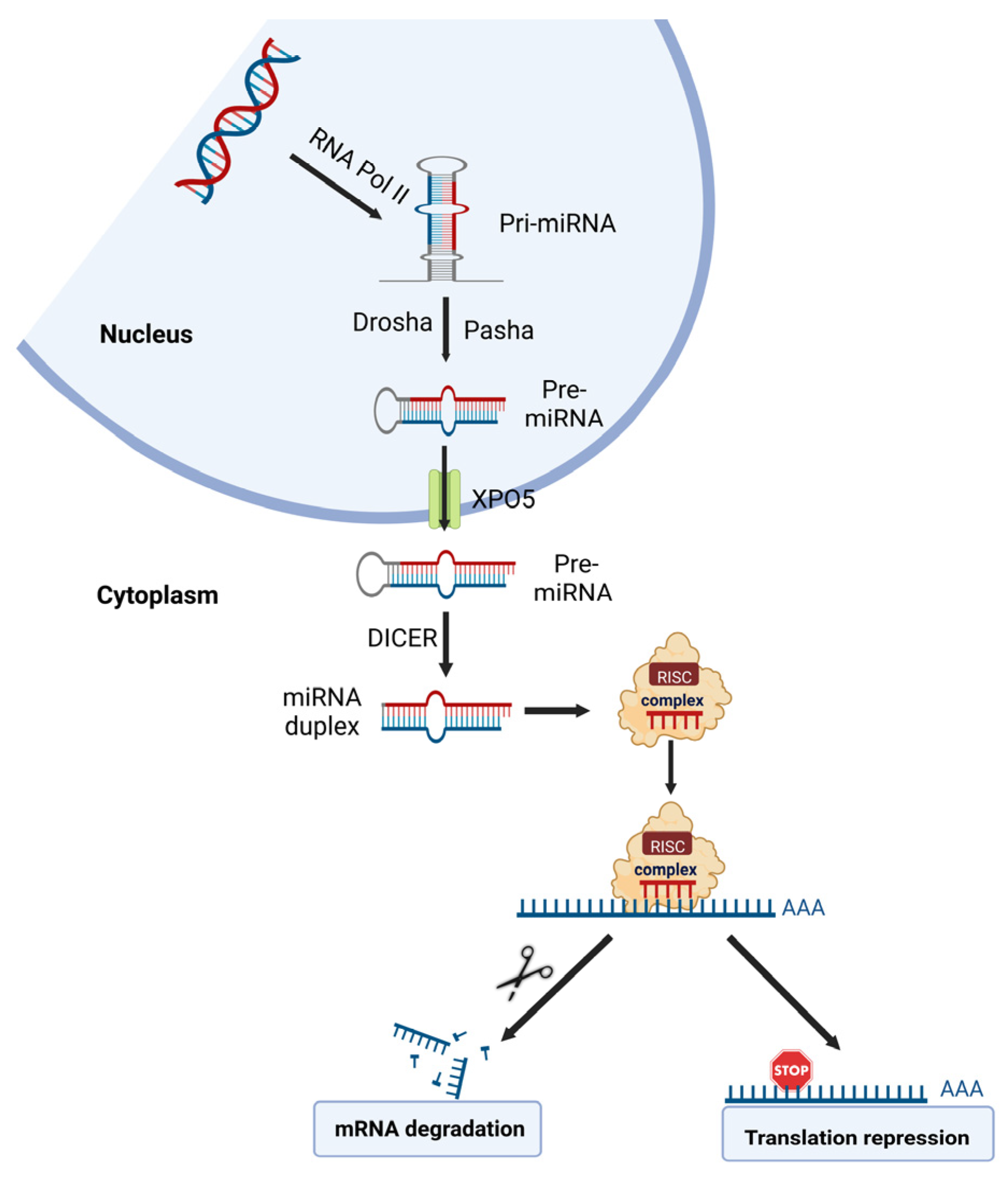
1.1.3. Non-Coding RNA
miRNA
lncRNA
2. Chromium (Cr)
2.1. DNA Methylation
2.2. Histone Modification
2.2.1. Histone Methylation
2.2.2. Histone Acetylation
2.3. Non-Coding RNA
2.3.1. miRNA
2.3.2. lncRNA
3. Nickel (Ni)
3.1. Ni and DNA Methylation
3.2. Histone Modification
3.2.1. Histone Methylation
3.2.2. Histone Acetylation
3.3. Non-Coding RNA
3.3.1. miRNA
3.3.2. LncRNA
4. Cadmium (Cd)
4.1. DNA Methylation
4.2. Histone Modification
4.3. Non-Coding RNAs
4.3.1. miRNAs
4.3.2. LncRNA
4.3.3. CircRNAs
4.3.4. RNA N6-Methyladenosine (m6A) Modification
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paithankar, J.G.; Saini, S.; Dwivedi, S.; Sharma, A.; Chowdhuri, D.K. Heavy metal associated health hazards: An interplay of oxidative stress and signal transduction. Chemosphere 2021, 262, 128350. [Google Scholar] [CrossRef] [PubMed]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renu, K.; Chakraborty, R.; Myakala, H.; Koti, R.; Famurewa, A.C.; Madhyastha, H.; Vellingiri, B.; George, A.; Valsala Gopalakrishnan, A. Molecular mechanism of heavy metals (Lead, Chromium, Arsenic, Mercury, Nickel and Cadmium)-induced hepatotoxicity—A review. Chemosphere 2021, 271, 129735. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Re-Evaluation of Some Organic Chemicals, Hydrazine and Hydrogen Peroxide; International Agency for Research on Cancer: Lyou, France, 1999. [Google Scholar]
- Islam, R.; Zhao, L.; Wang, Y.; Lu-Yao, G.; Liu, L.-Z. Epigenetic Dysregulations in Arsenic-Induced Carcinogenesis. Cancers 2022, 14, 4502. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, Y.J.; Seo, Y.R. An Overview of Carcinogenic Heavy Metal: Molecular Toxicity Mechanism and Prevention. J. Cancer Prev. 2015, 20, 232–240. [Google Scholar] [CrossRef]
- Martinez-Zamudio, R.; Ha, H.C. Environmental epigenetics in metal exposure. Epigenetics 2011, 6, 820–827. [Google Scholar] [CrossRef] [Green Version]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Della Chiara, G.; Gervasoni, F.; Fakiola, M.; Godano, C.; D’Oria, C.; Azzolin, L.; Bonnal, R.J.P.; Moreni, G.; Drufuca, L.; Rossetti, G.; et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nat. Commun. 2021, 12, 2340. [Google Scholar] [CrossRef]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef] [Green Version]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, N.; Takenaga, K. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin. Exp. Metastasis 1998, 16, 471–479. [Google Scholar] [CrossRef]
- Long, C.; Yin, B.; Lu, Q.; Zhou, X.; Hu, J.; Yang, Y.; Yu, F.; Yuan, Y. Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Investig. 2007, 25, 685–690. [Google Scholar] [CrossRef]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [Green Version]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Sasidharan Nair, V.; El Salhat, H.; Taha, R.Z.; John, A.; Ali, B.R.; Elkord, E. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin. Epigenetics 2018, 10, 78. [Google Scholar] [CrossRef]
- Cao, M.; Wang, Y.; Xiao, Y.; Zheng, D.; Zhi, C.; Xia, X.; Yuan, X. Activation of the clock gene TIMELESS by H3k27 acetylation promotes colorectal cancer tumorigenesis by binding to Myosin-9. J. Exp. Clin. Cancer Res. 2021, 40, 162. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasetti, M.; Gaetani, S.; Monaco, F.; Neuzil, J.; Santarelli, L. Epigenetic Regulation of miRNA Expression in Malignant Mesothelioma: miRNAs as Biomarkers of Early Diagnosis and Therapy. Front. Oncol. 2019, 9, 1293. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Li, J.; Wang, H.Q.; Li, X.; Wen, B.; Wang, Y.J. MiR-141-3p promotes prostate cancer cell proliferation through inhibiting kruppel-like factor-9 expression. Biochem. Biophys. Res. Commun. 2017, 482, 1381–1386. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, L.Z.; Qian, X.; Chen, Q.; Jiang, Y.; Li, D.; Lai, L.; Jiang, B.H. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012, 40, 761–774. [Google Scholar] [CrossRef] [Green Version]
- Xia, C.; Yang, Y.; Kong, F.; Kong, Q.; Shan, C. MiR-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie 2018, 147, 98–104. [Google Scholar] [CrossRef]
- Gao, Y.; Fan, X.; Li, W.; Ping, W.; Deng, Y.; Fu, X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem. Biophys. Res. Commun. 2014, 446, 179–186. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Guzel, E.; Okyay, T.M.; Yalcinkaya, B.; Karacaoglu, S.; Gocmen, M.; Akcakuyu, M.H. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North. Clin. Istanb. 2020, 7, 81–86. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiang, C.; Wang, Y.; Duan, Y.; Liu, C.; Jin, Y.; Zhang, Y. lncRNA LINC00152 knockdown had effects to suppress biological activity of lung cancer via EGFR/PI3K/AKT pathway. Biomed. Pharmacother. 2017, 94, 644–651. [Google Scholar] [CrossRef]
- Ma, C.; Shi, X.; Zhu, Q.; Li, Q.; Liu, Y.; Yao, Y.; Song, Y. The growth arrest-specific transcript 5 (GAS5): A pivotal tumor suppressor long noncoding RNA in human cancers. Tumor Biol. 2016, 37, 1437–1444. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Murphy, A.; Sun, H.; Costa, M. Molecular and epigenetic mechanisms of Cr(VI)-induced carcinogenesis. Toxicol. Appl. Pharmacol. 2019, 377, 114636. [Google Scholar] [CrossRef]
- den Braver-Sewradj, S.P.; van Benthem, J.; Staal, Y.C.M.; Ezendam, J.; Piersma, A.H.; Hessel, E.V.S. Occupational exposure to hexavalent chromium. Part II. Hazard assessment of carcinogenic effects. Regul. Toxicol. Pharmacol. 2021, 126, 105045. [Google Scholar] [CrossRef]
- Yang, J.; Black, J. Competitive binding of chromium, cobalt and nickel to serum proteins. Biomaterials 1994, 15, 262–268. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on Dietary Reference Values for chromium. EFSA J. 2014, 12, 3845. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Hatakeyama, E.; Hosomi, R.; Kanda, S.; Nishiyama, T.; Fukunaga, K. Tissue accumulation and urinary excretion of chromium in rats fed diets containing graded levels of chromium chloride or chromium picolinate. J. Toxicol. Sci. 2010, 35, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Kiilunen, M.; Kivistö, H.; Ala-Laurila, P.; Tossavainen, A.; Aitio, A. Exceptional pharmacokinetics of trivalent chromium during occupational exposure to chromium lignosulfonate dust. Scand. J. Work Environ. Health 1983, 9, 265–271. [Google Scholar] [CrossRef] [Green Version]
- Gargas, M.L.; Norton, R.L.; Harris, M.A.; Paustenbach, D.J.; Finley, B.L. Urinary Excretion of Chromium Following Ingestion of Chromite-Ore Processing Residues in Humans: Implications for Biomonitoring. Risk Anal. 1994, 14, 1019–1024. [Google Scholar] [CrossRef]
- Bradberry, S.M.; Vale, A. Chromium intoxication: Features and management. J. Toxicol. Clin. Toxicol. 2001, 39, 233–235. [Google Scholar]
- Stoss, F.; Blackburn, K.; Harris, B.; Neal, M. Health Assessment Document for Chronium; US Environmental Protection Agency, Office of Research and Development: Research Triangle Park, NC, USA, 1983; p. 328.
- Gibb, H.J.; Lees, P.S.; Pinsky, P.F.; Rooney, B.C. Lung cancer among workers in chromium chemical production. Am. J. Ind. Med. 2000, 38, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Gibb, H.J.; Lees, P.S.; Wang, J.; Grace O’Leary, K. Extended followup of a cohort of chromium production workers. Am. J. Ind. Med. 2015, 58, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, J.; Humphries, B.; Kondo, K.; Jiang, Y.; Shi, X.; Yang, C. Upregulation of histone-lysine methyltransferases plays a causal role in hexavalent chromium-induced cancer stem cell-like property and cell transformation. Toxicol. Appl. Pharmacol. 2018, 342, 22–30. [Google Scholar] [CrossRef] [PubMed]
- DesMarais, T.L.; Costa, M. Mechanisms of Chromium-Induced Toxicity. Curr. Opin. Toxicol. 2019, 14, 1–7. [Google Scholar] [CrossRef]
- Guo, X.; Feng, L.; Lemos, B.; Lou, J. DNA methylation modifications induced by hexavalent chromium. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2019, 37, 133–145. [Google Scholar] [CrossRef]
- Feng, L.; Guo, X.; Li, T.; Yao, C.; Xia, H.; Jiang, Z.; Jia, J.; Fang, Y.; Shi, L.; Lu, C.A.; et al. Novel DNA methylation biomarkers for hexavalent chromium exposure: An epigenome-wide analysis. Epigenomics 2020, 12, 221–233. [Google Scholar] [CrossRef]
- Hu, G.; Li, P.; Li, Y.; Wang, T.; Gao, X.; Zhang, W.; Jia, G. Methylation levels of P16 and TP53 that are involved in DNA strand breakage of 16HBE cells treated by hexavalent chromium. Toxicol. Lett. 2016, 249, 15–21. [Google Scholar] [CrossRef]
- Kondo, K.; Takahashi, Y.; Hirose, Y.; Nagao, T.; Tsuyuguchi, M.; Hashimoto, M.; Ochiai, A.; Monden, Y.; Tangoku, A. The reduced expression and aberrant methylation of p16(INK4a) in chromate workers with lung cancer. Lung Cancer 2006, 53, 295–302. [Google Scholar] [CrossRef]
- Hu, G.; Li, P.; Cui, X.; Li, Y.; Zhang, J.; Zhai, X.; Yu, S.; Tang, S.; Zhao, Z.; Wang, J.; et al. Cr(VI)-induced methylation and down-regulation of DNA repair genes and its association with markers of genetic damage in workers and 16HBE cells. Environ. Pollut. 2018, 238, 833–843. [Google Scholar] [CrossRef]
- Hirose, T.; Kondo, K.; Takahashi, Y.; Ishikura, H.; Fujino, H.; Tsuyuguchi, M.; Hashimoto, M.; Yokose, T.; Mukai, K.; Kodama, T.; et al. Frequent microsatellite instability in lung cancer from chromate-exposed workers. Mol. Carcinog. 2002, 33, 172–180. [Google Scholar] [CrossRef]
- Takahashi, Y.; Kondo, K.; Hirose, T.; Nakagawa, H.; Tsuyuguchi, M.; Hashimoto, M.; Sano, T.; Ochiai, A.; Monden, Y. Microsatellite instability and protein expression of the DNA mismatch repair gene, hMLH1, of lung cancer in chromate-exposed workers. Mol. Carcinog. 2005, 42, 150–158. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Tsuboi, M.; Kondo, K.; Soejima, S.; Kajiura, K.; Kawakita, N.; Toba, H.; Kawakami, Y.; Yoshida, M.; Takizawa, H.; Tangoku, A. Chromate exposure induces DNA hypermethylation of the mismatch repair gene MLH1 in lung cancer. Mol. Carcinog. 2020, 59, 24–31. [Google Scholar] [CrossRef]
- Ali, A.H.; Kondo, K.; Namura, T.; Senba, Y.; Takizawa, H.; Nakagawa, Y.; Toba, H.; Kenzaki, K.; Sakiyama, S.; Tangoku, A. Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol. Carcinog. 2011, 50, 89–99. [Google Scholar] [CrossRef]
- Sun, H.; Zhou, X.; Chen, H.; Li, Q.; Costa, M. Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium. Toxicol. Appl. Pharmacol. 2009, 237, 258–266. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Li, Q.; Arita, A.; Sun, H.; Costa, M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol. Appl. Pharmacol. 2009, 236, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Zhang, X.; Murphy, A.J.; Costa, M.; Zhao, X.; Sun, H. Downregulation of hedgehog-interacting protein (HHIP) contributes to hexavalent chromium-induced malignant transformation of human bronchial epithelial cells. Carcinogenesis 2021, 42, 136–147. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, H.; Sun, M.; Deng, X.; Wu, X.; Ma, Y.; Li, M.; Shuoa, S.M.; You, Q.; Miao, L. CXCL5/CXCR2 axis in tumor microenvironment as potential diagnostic biomarker and therapeutic target. Cancer Commun. 2020, 40, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; He, J.; Wang, L.; Zhao, L.; Wang, Y.; Wu, G.; Liu, W.; Shu, Y.; Gong, W.; Ma, X.-L.; et al. Epigenetic alterations of CXCL5 in Cr(VI)-induced carcinogenesis. Sci. Total Environ. 2022, 838, 155713. [Google Scholar] [CrossRef]
- Chen, D.; Kluz, T.; Fang, L.; Zhang, X.; Sun, H.; Jin, C.; Costa, M. Hexavalent Chromium (Cr(VI)) Down-Regulates Acetylation of Histone H4 at Lysine 16 through Induction of Stressor Protein Nupr1. PLoS ONE 2016, 11, e0157317. [Google Scholar] [CrossRef] [PubMed]
- Morales, V.; Straub, T.; Neumann, M.F.; Mengus, G.; Akhtar, A.; Becker, P.B. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 2004, 23, 2258–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Wang, W.; Hu, J.; Feng, K.; Pan, Y.; Zhang, L.; Feng, Y. Lentivirus-mediated RNAi knockdown of NUPR1 inhibits human nonsmall cell lung cancer growth in vitro and in vivo. Anat. Rec. 2012, 295, 2114–2121. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Long, J.; Chen, X.; Tan, M.D. NUPR1 promotes the proliferation and migration of breast cancer cells by activating TFE3 transcription to induce autophagy. Exp. Cell Res. 2022, 418, 113234. [Google Scholar] [CrossRef] [PubMed]
- Santofimia-Castaño, P.; Xia, Y.; Peng, L.; Velázquez-Campoy, A.; Abián, O.; Lan, W.; Lomberk, G.; Urrutia, R.; Rizzuti, B.; Soubeyran, P.; et al. Targeting the Stress-Induced Protein NUPR1 to Treat Pancreatic Adenocarcinoma. Cells 2019, 8, 1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, D.M.; Thomas, S.D.; Islam, A.; Muench, D.; Sedoris, K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012, 18, 5546–5553. [Google Scholar] [CrossRef] [Green Version]
- Clementino, M.; Xie, J.; Yang, P.; Li, Y.; Lin, H.P.; Fenske, W.K.; Tao, H.; Kondo, K.; Yang, C.; Wang, Z. A Positive Feedback Loop Between c-Myc Upregulation, Glycolytic Shift, and Histone Acetylation Enhances Cancer Stem Cell-like Property and Tumorigenicity of Cr(VI)-transformed Cells. Toxicol. Sci. 2020, 177, 71–83. [Google Scholar] [CrossRef]
- Chen, Z.; Zhong, J.; Ren, X.; Liu, W.; Wu, D.; Chen, C.; Huang, H.; Huang, X.; Liu, Y.; Liu, J. Involvement of a novel regulatory cascade consisting of SET-H3K18ac/H3K27ac-53BP1 in Cr(VI)-induced malignant transformation of 16HBE cells. Toxicol. Lett. 2021, 339, 70–77. [Google Scholar] [CrossRef]
- Ren, X.; Xia, B.; Chen, Z.; Chen, X.; Wu, D.; Lu, W.; Luo, N.; Zhou, L.; Liu, W.; Yang, X.; et al. Short-term and long-term exposure to hexavalent chromium alters 53BP1 via H3K18ac and H3K27ac. Chemosphere 2019, 229, 284–294. [Google Scholar] [CrossRef]
- Ward, I.M.; Minn, K.; Van Deursen, J.; Chen, J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 2003, 23, 2556–2563. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Li, X.; Zhao, Y.; Yang, Q.; Kong, B. 53BP1 suppresses tumor growth and promotes susceptibility to apoptosis of ovarian cancer cells through modulation of the Akt pathway. Oncol. Rep. 2012, 27, 1251–1257. [Google Scholar]
- Speer, R.M.; Meaza, I.; Toyoda, J.H.; Lu, Y.; Xu, Q.; Walter, R.B.; Kong, M.; Lu, H.; Kouokam, J.C.; Wise, J.P., Sr. Particulate hexavalent chromium alters microRNAs in human lung cells that target key carcinogenic pathways. Toxicol. Appl. Pharmacol. 2022, 438, 115890. [Google Scholar] [CrossRef]
- Wang, L.; Bayanbold, K.; Zhao, L.; Wang, Y.; Adamcakova-Dodd, A.; Thorne, P.S.; Yang, H.; Jiang, B.H.; Liu, L.Z. Redox sensitive miR-27a/b/Nrf2 signaling in Cr(VI)-induced carcinogenesis. Sci. Total Environ. 2022, 809, 151118. [Google Scholar] [CrossRef]
- Kumar, H.; Kumar, R.M.; Bhattacharjee, D.; Somanna, P.; Jain, V. Role of Nrf2 Signaling Cascade in Breast Cancer: Strategies and Treatment. Front. Pharmacol. 2022, 13, 720076. [Google Scholar] [CrossRef]
- Pratheeshkumar, P.; Son, Y.O.; Divya, S.P.; Turcios, L.; Roy, R.V.; Hitron, J.A.; Wang, L.; Kim, D.; Dai, J.; Asha, P.; et al. Hexavalent chromium induces malignant transformation of human lung bronchial epithelial cells via ROS-dependent activation of miR-21-PDCD4 signaling. Oncotarget 2016, 7, 51193–51210. [Google Scholar] [CrossRef] [Green Version]
- Pratheeshkumar, P.; Son, Y.O.; Divya, S.P.; Wang, L.; Turcios, L.; Roy, R.V.; Hitron, J.A.; Kim, D.; Dai, J.; Asha, P.; et al. Quercetin inhibits Cr(VI)-induced malignant cell transformation by targeting miR-21-PDCD4 signaling pathway. Oncotarget 2017, 8, 52118–52131. [Google Scholar] [CrossRef] [Green Version]
- Sato, M.; Larsen, J.E.; Lee, W.; Sun, H.; Shames, D.S.; Dalvi, M.P.; Ramirez, R.D.; Tang, H.; DiMaio, J.M.; Gao, B.; et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol. Cancer Res. 2013, 11, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Raghu, H.; Gondi, C.S.; Dinh, D.H.; Gujrati, M.; Rao, J.S. Specific knockdown of uPA/uPAR attenuates invasion in glioblastoma cells and xenografts by inhibition of cleavage and trafficking of Notch -1 receptor. Mol. Cancer 2011, 10, 130. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xiao, Y.; Ma, Y.; Liang, N.; Liang, Y.; Lu, C.; Xiao, F. ROS-mediated miR-21-5p regulates the proliferation and apoptosis of Cr(VI)-exposed L02 hepatocytes via targeting PDCD4. Ecotoxicol. Environ. Saf. 2020, 191, 110160. [Google Scholar] [CrossRef]
- Yan, X.; Chen, X.; Liang, H.; Deng, T.; Chen, W.; Zhang, S.; Liu, M.; Gao, X.; Liu, Y.; Zhao, C.; et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer 2014, 13, 220. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Qian, X.; Carpenter, R.; Xu, Q.; Wang, L.; Qi, Y.; Wang, Z.X.; Liu, L.Z.; Jiang, B.H. Repression of miR-143 mediates Cr (VI)-induced tumor angiogenesis via IGF-IR/IRS1/ERK/IL-8 pathway. Toxicol. Sci. 2013, 134, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Qiu, J.G.; He, J.; Liu, W.J.; Ge, X.; Zhou, F.M.; Huang, Y.X.; Jiang, B.H.; Liu, L.Z. Suppression of miR-143 contributes to overexpression of IL-6, HIF-1α and NF-κB p65 in Cr(VI)-induced human exposure and tumor growth. Toxicol. Appl. Pharmacol. 2019, 378, 114603. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Yu, S.; Zhang, J.; Wang, T.; Jia, G. miR-3940-5p associated with genetic damage in workers exposed to hexavalent chromium. Toxicol. Lett. 2014, 229, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, G.; Li, P.; Tang, S.; Zhang, J.; Jia, G. miR-3940-5p enhances homologous recombination after DSB in Cr(VI) exposed 16HBE cell. Toxicology 2016, 344–346, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Feng, L.; Tong, Y.; Jia, J.; Li, T.; Wang, J.; Jiang, Z.; Yu, M.; Xia, H.; Jin, Q.; et al. Genome wide profiling of miRNAs relevant to the DNA damage response induced by hexavalent chromium exposure (DDR-related miRNAs in response to Cr (VI) exposure). Environ. Int. 2021, 157, 106782. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, H.-P.; Li, Y.; Tao, H.; Yang, P.; Xie, J.; Maddy, D.; Kondo, K.; Yang, C. Chronic Hexavalent Chromium Exposure Induces Cancer Stem Cell-Like Property and Tumorigenesis by Increasing c-Myc Expression. Toxicol. Sci. Off. J. Soc. Toxicol. 2019, 172, 252–264. [Google Scholar] [CrossRef]
- Hu, G.; Feng, H.; Long, C.; Zhou, D.; Li, P.; Gao, X.; Chen, Z.; Wang, T.; Jia, G. LncRNA expression profiling and its relationship with DNA damage in Cr(VI)-treated 16HBE cells. Sci. Total Environ. 2019, 655, 622–632. [Google Scholar] [CrossRef]
- Genchi, G.; Carocci, A.; Lauria, G.; Sinicropi, M.S.; Catalano, A. Nickel: Human Health and Environmental Toxicology. Int. J. Environ. Res. Public Health 2020, 17, 679. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Trivedi, A. A review on role of nickel in the biological system. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 719–727. [Google Scholar] [CrossRef]
- Grimsrud, T.K.; Berge, S.R.; Haldorsen, T.; Andersen, A. Exposure to Different Forms of Nickel and Risk of Lung Cancer. Am. J. Epidemiol. 2002, 156, 1123–1132. [Google Scholar] [CrossRef]
- Sinicropi, M.S.; Caruso, A.; Capasso, A.; Palladino, C.; Panno, A.; Saturnino, C. Heavy metals: Toxicity and carcinogenicity. Pharmacologyonline 2010, 2, 329–333. [Google Scholar]
- Das, K.K.; Reddy, R.C.; Bagoji, I.B.; Das, S.; Bagali, S.; Mullur, L.; Khodnapur, J.P.; Biradar, M.S. Primary concept of nickel toxicity—An overview. J. Basic Clin. Physiol. Pharmacol. 2019, 30, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Silverberg, N.B.; Pelletier, J.L.; Jacob, S.E.; Schneider, L.C. Nickel Allergic Contact Dermatitis: Identification, Treatment, and Prevention. Pediatrics 2020, 145, e20200628. [Google Scholar] [CrossRef]
- Sunderman, F.W., Sr. Chelation therapy in nickel poisoning. Ann. Clin. Lab. Sci. 1981, 11, 1–8. [Google Scholar]
- Prueitt, R.L.; Li, W.; Chang, Y.C.; Boffetta, P.; Goodman, J.E. Systematic review of the potential respiratory carcinogenicity of metallic nickel in humans. Crit. Rev. Toxicol. 2020, 50, 605–639. [Google Scholar] [CrossRef]
- Lee, Y.W.; Klein, C.B.; Kargacin, B.; Salnikow, K.; Kitahara, J.; Dowjat, K.; Zhitkovich, A.; Christie, N.T.; Costa, M. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: A new model for epigenetic carcinogens. Mol. Cell. Biol. 1995, 15, 2547–2557. [Google Scholar] [CrossRef] [Green Version]
- Ellen, T.P.; Kluz, T.; Harder, M.E.; Xiong, J.; Costa, M. Heterochromatinization as a potential mechanism of nickel-induced carcinogenesis. Biochemistry 2009, 48, 4626–4632. [Google Scholar] [CrossRef] [Green Version]
- Ji, W.; Yang, L.; Yu, L.; Yuan, J.; Hu, D.; Zhang, W.; Yang, J.; Pang, Y.; Li, W.; Lu, J.; et al. Epigenetic silencing of O6-methylguanine DNA methyltransferase gene in NiS-transformed cells. Carcinogenesis 2008, 29, 1267–1275. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Jose, C.C.; Cuddapah, S. Epithelial-mesenchymal transition: Insights into nickel-induced lung diseases. Semin. Cancer Biol. 2021, 76, 99–109. [Google Scholar] [CrossRef]
- Wu, C.H.; Tang, S.C.; Wang, P.H.; Lee, H.; Ko, J.L. Nickel-induced epithelial-mesenchymal transition by reactive oxygen species generation and E-cadherin promoter hypermethylation. J. Biol. Chem. 2012, 287, 25292–25302. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, J.; Li, M.; Wu, Y.; Fan, Y.; Zhou, Y.; Tan, L.; Shao, Z.; Shi, H. Methylation of RAR-β2, RASSF1A, and CDKN2A genes induced by nickel subsulfide and nickel-carcinogenesis in rats. Biomed. Environ. Sci. 2011, 24, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, B.; Klafter, R.; Miller, M.S.; Mansur, C.; Mizesko, M.; Bai, X.; LaMontagne, K., Jr.; Arbiser, J.L. Reactive oxygen-induced carcinogenesis causes hypermethylation of p16(Ink4a) and activation of MAP kinase. Mol. Med. 2002, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasaei, H.; Gilham, E.; Pickles, J.C.; Roberts, T.P.; O’Donovan, M.; Newbold, R.F. Carcinogen-specific mutational and epigenetic alterations in INK4A, INK4B and p53 tumour-suppressor genes drive induced senescence bypass in normal diploid mammalian cells. Oncogene 2013, 32, 171–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.-C.; Yang, H.; Wang, K.-F.; Chen, T.-H.; Jiang, W.-Q.; Shi, Y.-X. ANGPTL4 overexpression inhibits tumor cell adhesion and migration and predicts favorable prognosis of triple-negative breast cancer. BMC Cancer 2020, 20, 878. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.J.; Teo, Z.; Sng, M.K.; Zhu, P.; Tan, N.S. Emerging Roles of Angiopoietin-like 4 in Human Cancer. Mol. Cancer Res. 2012, 10, 677–688. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.-T.; Hsu, W.-C.; Ou, C.-C.; Tai, H.-C.; Hsu, H.-T.; Yeh, K.-T.; Ko, J.-L. Metformin Mitigates Nickel-Elicited Angiopoietin-Like Protein 4 Expression via HIF-1α for Lung Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 619. [Google Scholar] [CrossRef] [Green Version]
- Mariani, C.J.; Vasanthakumar, A.; Madzo, J.; Yesilkanal, A.; Bhagat, T.; Yu, Y.; Bhattacharyya, S.; Wenger, R.H.; Cohn, S.L.; Nanduri, J.; et al. TET1-Mediated Hydroxymethylation Facilitates Hypoxic Gene Induction in Neuroblastoma. Cell Rep. 2014, 7, 1343–1352. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.T.; Li, C.T.; Tang, S.C.; Hsin, I.L.; Lai, Y.C.; Hsiao, Y.P.; Ko, J.L. Nickel chloride regulates ANGPTL4 via the HIF-1α-mediated TET1 expression in lung cells. Toxicol. Lett. 2021, 352, 17–25. [Google Scholar] [CrossRef]
- Jose, C.C.; Wang, Z.; Tanwar, V.S.; Zhang, X.; Zang, C.; Cuddapah, S. Nickel-induced transcriptional changes persist post exposure through epigenetic reprogramming. Epigenetics Chromatin 2019, 12, 75. [Google Scholar] [CrossRef]
- Jose, C.C.; Xu, B.; Jagannathan, L.; Trac, C.; Mallela, R.K.; Hattori, T.; Lai, D.; Koide, S.; Schones, D.E.; Cuddapah, S. Epigenetic dysregulation by nickel through repressive chromatin domain disruption. Proc. Natl. Acad. Sci. USA 2014, 111, 14631–14636. [Google Scholar] [CrossRef] [Green Version]
- Ke, Q.; Davidson, T.; Chen, H.; Kluz, T.; Costa, M. Alterations of histone modifications and transgene silencing by nickel chloride. Carcinogenesis 2006, 27, 1481–1488. [Google Scholar] [CrossRef] [Green Version]
- Tchou-Wong, K.M.; Kiok, K.; Tang, Z.; Kluz, T.; Arita, A.; Smith, P.R.; Brown, S.; Costa, M. Effects of nickel treatment on H3K4 trimethylation and gene expression. PLoS ONE 2011, 6, e17728. [Google Scholar] [CrossRef]
- Jose, C.C.; Jagannathan, L.; Tanwar, V.S.; Zhang, X.; Zang, C.; Cuddapah, S. Nickel exposure induces persistent mesenchymal phenotype in human lung epithelial cells through epigenetic activation of ZEB1. Mol. Carcinog. 2018, 57, 794–806. [Google Scholar] [CrossRef]
- Chen, H.; Kluz, T.; Zhang, R.; Costa, M. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis 2010, 31, 2136–2144. [Google Scholar] [CrossRef]
- Chen, H.; Costa, M. Iron- and 2-oxoglutarate-dependent dioxygenases: An emerging group of molecular targets for nickel toxicity and carcinogenicity. Biometals Int. J. Role Met. Ions Biol. Biochem. Med. 2009, 22, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Goda, S.; Isagawa, T.; Chikaoka, Y.; Kawamura, T.; Aburatani, H. Control of Histone H3 Lysine 9 (H3K9) Methylation State via Cooperative Two-step Demethylation by Jumonji Domain Containing 1A (JMJD1A) Homodimer. J. Biol. Chem. 2013, 288, 36948–36956. [Google Scholar] [CrossRef] [Green Version]
- Beyer, S.; Kristensen, M.M.; Jensen, K.S.; Johansen, J.V.; Staller, P. The Histone Demethylases JMJD1A and JMJD2B Are Transcriptional Targets of Hypoxia-inducible Factor HIF. J. Biol. Chem. 2008, 283, 36542–36552. [Google Scholar] [CrossRef] [Green Version]
- Jeon, H.Y.; Ryu, H.; Pornour, M.; Qi, J. Histone demethylase JMJD1A in cancer progression and therapeutic resistance. Mol. Carcinog. 2022, 61, 392–396. [Google Scholar] [CrossRef]
- Chen, H.; Giri, N.C.; Zhang, R.; Yamane, K.; Zhang, Y.; Maroney, M.; Costa, M. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J. Biol. Chem. 2010, 285, 7374–7383. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Lu, J.; Wang, Y.; Gui, Y.; Duan, X.; Cai, Z. Ascorbate antagonizes nickel ion to regulate JMJD1A expression in kidney cancer cells. Acta Biochim. Biophys. Sin. 2012, 44, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhang, Y.; Zhang, Q.; Fa, P.; Gui, Y.; Gao, G.; Cai, Z. The regulatory role of nickel on H3K27 demethylase JMJD3 in kidney cancer cells. Toxicol. Ind. Health 2016, 32, 1286–1292. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ke, Q.; Kluz, T.; Yan, Y.; Costa, M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell. Biol. 2006, 26, 3728–3737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, M.; Shen, H.; Jin, Y.; Lin, T.; Cai, Q.; Pinard, M.A.; Biswas, S.; Tran, Q.; Li, G.; Shenoy, A.K.; et al. The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition. J. Biol. Chem. 2013, 288, 27680–27691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; He, M.D.; Mao, L.; Wang, X.; Jiang, Y.L.; Li, M.; Lu, Y.H.; Yu, Z.P.; Zhou, Z. Nicotinamide N-Methyltransferase Suppression Participates in Nickel-Induced Histone H3 Lysine9 Dimethylation in BEAS-2B Cells. Cell. Physiol. Biochem. 2017, 41, 2016–2026. [Google Scholar] [CrossRef]
- Golebiowski, F.; Kasprzak, K.S. Inhibition of core histones acetylation by carcinogenic nickel(II). Mol. Cell. Biochem. 2005, 279, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Broday, L.; Peng, W.; Kuo, M.H.; Salnikow, K.; Zoroddu, M.; Costa, M. Nickel compounds are novel inhibitors of histone H4 acetylation. Cancer Res. 2000, 60, 238–241. [Google Scholar]
- Yatouji, S.; El-Khoury, V.; Trentesaux, C.; Trussardi-Regnier, A.; Benabid, R.; Bontems, F.; Dufer, J. Differential modulation of nuclear texture, histone acetylation, and MDR1 gene expression in human drug-sensitive and -resistant OV1 cell lines. Int. J. Oncol. 2007, 30, 1003–1009. [Google Scholar] [CrossRef] [Green Version]
- Zoroddu, M.A.; Schinocca, L.; Kowalik-Jankowska, T.; Kozlowski, H.; Salnikow, K.; Costa, M. Molecular mechanisms in nickel carcinogenesis: Modeling Ni(II) binding site in histone H4. Environ. Health Perspect. 2002, 110 (Suppl. 5), 719–723. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Zhang, Y.; Chen, J.; Chen, H.; Lin, C.; Wang, Q.; Ou, Y. Nickel-induced histone hypoacetylation: The role of reactive oxygen species. Toxicol. Sci. 2003, 74, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Zhang, D.; Chen, J.; Lin, C.; Liu, Q. Involvement of histone hypoacetylation in Ni2+-induced bcl-2 down-regulation and human hepatoma cell apoptosis. J. Biol. Inorg. Chem. 2004, 9, 713–723. [Google Scholar] [CrossRef]
- Zhang, Q.; Salnikow, K.; Kluz, T.; Chen, L.C.; Su, W.C.; Costa, M. Inhibition and reversal of nickel-induced transformation by the histone deacetylase inhibitor trichostatin A. Toxicol. Appl. Pharmacol. 2003, 192, 201–211. [Google Scholar] [CrossRef]
- Ke, Q.; Li, Q.; Ellen, T.P.; Sun, H.; Costa, M. Nickel compounds induce phosphorylation of histone H3 at serine 10 by activating JNK-MAPK pathway. Carcinogenesis 2008, 29, 1276–1281. [Google Scholar] [CrossRef] [Green Version]
- Komar, D.; Juszczynski, P. Rebelled epigenome: Histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin. Epigenetics 2020, 12, 147. [Google Scholar] [CrossRef]
- Qi, H.; Yang, Z.; Dai, C.; Wang, R.; Ke, X.; Zhang, S.; Xiang, X.; Chen, K.; Li, C.; Luo, J.; et al. STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis. Oncogenesis 2020, 9, 15. [Google Scholar] [CrossRef]
- Ke, Q.; Ellen, T.P.; Costa, M. Nickel compounds induce histone ubiquitination by inhibiting histone deubiquitinating enzyme activity. Toxicol. Appl. Pharmacol. 2008, 228, 190–199. [Google Scholar] [CrossRef] [Green Version]
- Karaczyn, A.A.; Golebiowski, F.; Kasprzak, K.S. Ni(II) affects ubiquitination of core histones H2B and H2A. Exp. Cell Res. 2006, 312, 3252–3259. [Google Scholar] [CrossRef]
- Karaczyn, A.A.; Bal, W.; North, S.L.; Bare, R.M.; Hoang, V.M.; Fisher, R.J.; Kasprzak, K.S. The octapeptidic end of the C-terminal tail of histone H2A is cleaved off in cells exposed to carcinogenic nickel (II). Chem. Res. Toxicol. 2003, 16, 1555–1559. [Google Scholar] [CrossRef]
- Bal, W.; Liang, R.; Lukszo, J.; Lee, S.-H.; Dizdaroglu, M.; Kasprzak, K.S. Ni (II) specifically cleaves the C-terminal tail of the major variant of histone H2A and forms an oxidative damage-mediating complex with the cleaved-off octapeptide. Chem. Res. Toxicol. 2000, 13, 616–624. [Google Scholar] [CrossRef]
- Karaczyn, A.A.; Golebiowski, F.; Kasprzak, K.S. Truncation, deamidation, and oxidation of histone H2B in cells cultured with nickel(II). Chem. Res. Toxicol. 2005, 18, 1934–1942. [Google Scholar] [CrossRef]
- Wu, C.H.; Hsiao, Y.M.; Yeh, K.T.; Tsou, T.C.; Chen, C.Y.; Wu, M.F.; Ko, J.L. Upregulation of microRNA-4417 and Its Target Genes Contribute to Nickel Chloride-promoted Lung Epithelial Cell Fibrogenesis and Tumorigenesis. Sci. Rep. 2017, 7, 15320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Chen, Q.Y.; Jordan, A.; Sun, H.; Roy, N.; Costa, M. RUNX2/miR-31/SATB2 pathway in nickel-induced BEAS-2B cell transformation. Oncol. Rep. 2021, 46, 154. [Google Scholar] [CrossRef]
- Wu, F.; Jordan, A.; Kluz, T.; Shen, S.; Sun, H.; Cartularo, L.A.; Costa, M. SATB2 expression increased anchorage-independent growth and cell migration in human bronchial epithelial cells. Toxicol. Appl. Pharmacol. 2016, 293, 30–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, Y.H.; Liou, S.H.; Wong, R.H.; Chen, C.Y.; Lee, H. Nickel may contribute to EGFR mutation and synergistically promotes tumor invasion in EGFR-mutated lung cancer via nickel-induced microRNA-21 expression. Toxicol. Lett. 2015, 237, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, Y.; Ma, L.; Huang, S.; Wang, R.; Gao, R.; Wu, Y.; Shi, H.; Zhang, J. The alteration of miR-222 and its target genes in nickel-induced tumor. Biol. Trace Elem. Res. 2013, 152, 267–274. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Wu, Y.J.; Li, M.J.; Wang, R.J.; Huang, S.Q.; Gao, R.R.; Ma, L.; Shi, H.J.; Zhang, J. Hyper-methylated miR-203 dysregulates ABL1 and contributes to the nickel-induced tumorigenesis. Toxicol. Lett. 2013, 223, 42–51. [Google Scholar] [CrossRef]
- Ji, W.; Yang, L.; Yuan, J.; Yang, L.; Zhang, M.; Qi, D.; Duan, X.; Xuan, A.; Zhang, W.; Lu, J.; et al. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis 2013, 34, 446–453. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Lu, Y.; Xu, S.; Mao, L.; Zhang, L.; Duan, W.; Liu, C.; Pi, H.; Zhang, Y.; Zhong, M.; et al. MiRNA-210 modulates a nickel-induced cellular energy metabolism shift by repressing the iron-sulfur cluster assembly proteins ISCU1/2 in Neuro-2a cells. Cell Death Dis. 2014, 5, e1090. [Google Scholar] [CrossRef] [Green Version]
- Tong, W.H.; Rouault, T.A. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006, 3, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Crooks, D.R.; Maio, N.; Lane, A.N.; Jarnik, M.; Higashi, R.M.; Haller, R.G.; Yang, Y.; Fan, T.W.; Linehan, W.M.; Rouault, T.A. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J. Biol. Chem. 2018, 293, 8297–8311. [Google Scholar] [CrossRef] [Green Version]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Li, Y.; Zhang, H.; Huang, P.; Luthra, R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 2010, 29, 4362–4368. [Google Scholar] [CrossRef] [Green Version]
- Favaro, E.; Ramachandran, A.; McCormick, R.; Gee, H.; Blancher, C.; Crosby, M.; Devlin, C.; Blick, C.; Buffa, F.; Li, J.L.; et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE 2010, 5, e10345. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Zhou, C.; Lu, Y.; Mao, L.; Xi, Y.; Mei, X.; Wang, X.; Zhang, L.; Yu, Z.; Zhou, Z. Melatonin Antagonizes Nickel-Induced Aerobic Glycolysis by Blocking ROS-Mediated HIF-1α/miR210/ISCU Axis Activation. Oxid. Med. Cell. Longev. 2020, 2020, 5406284. [Google Scholar] [CrossRef]
- Saquib, Q.; Xia, P.; Siddiqui, M.A.; Zhang, J.; Xie, Y.; Faisal, M.; Ansari, S.M.; Alwathnani, H.A.; Alatar, A.A.; Al-Khedhairy, A.A.; et al. High-throughput transcriptomics: An insight on the pathways affected in HepG2 cells exposed to nickel oxide nanoparticles. Chemosphere 2020, 244, 125488. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, C.; Wang, J.; Huang, H.; Li, J.; Xie, Q.; Liu, Y.; Zhu, J.; Li, Y.; Zhang, D.; et al. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1α translation. Oncogene 2017, 36, 3878–3889. [Google Scholar] [CrossRef] [Green Version]
- Faroon, O.; Ashizawa, A.; Wright, S.; Tucker, P.; Jenkins, K.; Ingerman, L.; Rudisill, C. Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological Profiles. In Toxicological Profile for Cadmium; Agency for Toxic Substances and Disease Registry (US): Atlanta, GA, USA, 2012. [Google Scholar]
- Satarug, S.; Moore, M.R. Adverse health effects of chronic exposure to low-level cadmium in foodstuffs and cigarette smoke. Environ. Health Perspect. 2004, 112, 1099–1103. [Google Scholar] [CrossRef] [Green Version]
- Godt, J.; Scheidig, F.; Grosse-Siestrup, C.; Esche, V.; Brandenburg, P.; Reich, A.; Groneberg, D.A. The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. 2006, 1, 22. [Google Scholar] [CrossRef] [Green Version]
- Staessen, J.A.; Roels, H.A.; Emelianov, D.; Kuznetsova, T.; Thijs, L.; Vangronsveld, J.; Fagard, R. Environmental exposure to cadmium, forearm bone density, and risk of fractures: Prospective population study. Lancet 1999, 353, 1140–1144. [Google Scholar] [CrossRef]
- Johri, N.; Jacquillet, G.; Unwin, R. Heavy metal poisoning: The effects of cadmium on the kidney. Biometals 2010, 23, 783–792. [Google Scholar] [CrossRef]
- Everett, C.J.; Frithsen, I.L. Association of urinary cadmium and myocardial infarction. Environ. Res. 2008, 106, 284–286. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.; Bannigan, J. Cadmium: Toxic effects on the reproductive system and the embryo. Reprod. Toxicol. 2008, 25, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Rafati Rahimzadeh, M.; Rafati Rahimzadeh, M.; Kazemi, S.; Moghadamnia, A.A. Cadmium toxicity and treatment: An update. Casp. J. Intern. Med. 2017, 8, 135–145. [Google Scholar] [CrossRef]
- Hartwig, A. Cadmium and cancer. Met. Ions Life Sci. 2013, 11, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Luevano, J.; Damodaran, C. A review of molecular events of cadmium-induced carcinogenesis. J. Environ. Pathol. Toxicol. Oncol. 2014, 33, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.-G.; Ahmed, K.; Zaidi, S.F.; Muhammad, J.S. Ins and outs of cadmium-induced carcinogenesis: Mechanism and prevention. Cancer Treat. Res. Commun. 2021, 27, 100372. [Google Scholar] [CrossRef]
- Wang, B.; Li, Y.; Tan, Y.; Miao, X.; Liu, X.D.; Shao, C.; Yang, X.H.; Turdi, S.; Ma, L.J.; Ren, J.; et al. Low-dose Cd induces hepatic gene hypermethylation, along with the persistent reduction of cell death and increase of cell proliferation in rats and mice. PLoS ONE 2012, 7, e33853. [Google Scholar] [CrossRef]
- Takiguchi, M.; Achanzar, W.E.; Qu, W.; Li, G.; Waalkes, M.P. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. 2003, 286, 355–365. [Google Scholar] [CrossRef]
- Qu, W.; Fuquay, R.; Sakurai, T.; Waalkes, M.P. Acquisition of apoptotic resistance in cadmium-induced malignant transformation: Specific perturbation of JNK signal transduction pathway and associated metallothionein overexpression. Mol. Carcinog. 2006, 45, 561–571. [Google Scholar] [CrossRef]
- Hirao-Suzuki, M.; Takeda, S.; Sakai, G.; Waalkes, M.P.; Sugihara, N.; Takiguchi, M. Cadmium-stimulated invasion of rat liver cells during malignant transformation: Evidence of the involvement of oxidative stress/TET1-sensitive machinery. Toxicology 2021, 447, 152631. [Google Scholar] [CrossRef]
- Suzuki, M.; Takeda, S.; Teraoka-Nishitani, N.; Yamagata, A.; Tanaka, T.; Sasaki, M.; Yasuda, N.; Oda, M.; Okano, T.; Yamahira, K.; et al. Cadmium-induced malignant transformation of rat liver cells: Potential key role and regulatory mechanism of altered apolipoprotein E expression in enhanced invasiveness. Toxicology 2017, 382, 16–23. [Google Scholar] [CrossRef]
- Hirao-Suzuki, M.; Takeda, S.; Kobayashi, T.; Kino, K.; Miyazawa, H.; Waalkes, M.P.; Takiguchi, M. Cadmium down-regulates apolipoprotein E (ApoE) expression during malignant transformation of rat liver cells: Direct evidence for DNA hypermethylation in the promoter region of ApoE. J. Toxicol. Sci. 2018, 43, 537–543. [Google Scholar] [CrossRef] [Green Version]
- Benbrahim-Tallaa, L.; Tokar, E.J.; Diwan, B.A.; Dill, A.L.; Coppin, J.F.; Waalkes, M.P. Cadmium malignantly transforms normal human breast epithelial cells into a basal-like phenotype. Environ. Health Perspect. 2009, 117, 1847–1852. [Google Scholar] [CrossRef] [Green Version]
- Tarhonska, K.; Lesicka, M.; Janasik, B.; Roszak, J.; Reszka, E.; Braun, M.; Kołacińska-Wow, A.; Jabłońska, E. Cadmium and breast cancer—Current state and research gaps in the underlying mechanisms. Toxicol. Lett. 2022, 361, 29–42. [Google Scholar] [CrossRef]
- Filippini, T.; Torres, D.; Lopes, C.; Carvalho, C.; Moreira, P.; Naska, A.; Kasdagli, M.I.; Malavolti, M.; Orsini, N.; Vinceti, M. Cadmium exposure and risk of breast cancer: A dose-response meta-analysis of cohort studies. Environ. Int. 2020, 142, 105879. [Google Scholar] [CrossRef]
- Liang, Z.Z.; Zhu, R.M.; Li, Y.L.; Jiang, H.M.; Li, R.B.; Tang, L.Y.; Wang, Q.; Ren, Z.F. Differential epigenetic and transcriptional profile in MCF-7 breast cancer cells exposed to cadmium. Chemosphere 2020, 261, 128148. [Google Scholar] [CrossRef]
- Li, L.-y.; Guan, Y.-d.; Chen, X.-s.; Yang, J.-m.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2021, 11, 629266. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Lei, Y.X.; Wang, C.X. Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium. Toxicol. Sci. 2012, 125, 412–417. [Google Scholar] [CrossRef] [Green Version]
- Cartularo, L.; Kluz, T.; Cohen, L.; Shen, S.S.; Costa, M. Molecular Mechanisms of Malignant Transformation by Low Dose Cadmium in Normal Human Bronchial Epithelial Cells. PLoS ONE 2016, 11, e0155002. [Google Scholar] [CrossRef] [Green Version]
- Benbrahim-Tallaa, L.; Waterland, R.A.; Dill, A.L.; Webber, M.M.; Waalkes, M.P. Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ. Health Perspect. 2007, 115, 1454–1459. [Google Scholar] [CrossRef] [Green Version]
- Venza, M.; Visalli, M.; Biondo, C.; Oteri, R.; Agliano, F.; Morabito, S.; Teti, D.; Venza, I. Epigenetic marks responsible for cadmium-induced melanoma cell overgrowth. Toxicol. Vitr. 2015, 29, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Chen, H.; Hu, Y.; Bai, Y.; Zhou, M.; Xu, A.; Shao, C. Combination effects of chronic cadmium exposure and gamma-irradiation on the genotoxicity and cytotoxicity of peripheral blood lymphocytes and bone marrow cells in rats. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2012, 743, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Lafuente, A.; Gonzalez-Carracedo, A.; Romero, A.; Esquifino, A. Effect of cadmium on lymphocyte subsets distribution in thymus and spleen. J. Physiol. Biochem. 2003, 59, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Ye, S.; Pan, Y.; Bao, Y.; Chen, H.; Shao, C. Long-term cadmium exposure leads to the enhancement of lymphocyte proliferation via down-regulating p16 by DNA hypermethylation. Mutat. Res. 2013, 757, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Zhang, Y.; Qi, Y.; Chen, C.; Ji, W. Global DNA hypomethylation, rather than reactive oxygen species (ROS), a potential facilitator of cadmium-stimulated K562 cell proliferation. Toxicol. Lett. 2008, 179, 43–47. [Google Scholar] [CrossRef]
- Pelch, K.E.; Tokar, E.J.; Merrick, B.A.; Waalkes, M.P. Differential DNA methylation profile of key genes in malignant prostate epithelial cells transformed by inorganic arsenic or cadmium. Toxicol. Appl. Pharmacol. 2015, 286, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Stopa, N.; Krebs, J.E.; Shechter, D. The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015, 72, 2041–2059. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef]
- Ghosh, K.; Chatterjee, B.; Behera, P.; Kanade, S.R. The carcinogen cadmium elevates CpG-demethylation and enrichment of NFYA and E2F1 in the promoter of oncogenic PRMT5 and EZH2 methyltransferases resulting in their elevated expression in vitro. Chemosphere 2020, 242, 125186. [Google Scholar] [CrossRef]
- Liang, Z.L.; Wu, D.D.; Yao, Y.; Yu, F.Y.; Yang, L.; Tan, H.W.; Hylkema, M.N.; Rots, M.G.; Xu, Y.M.; Lau, A.T.Y. Epiproteome profiling of cadmium-transformed human bronchial epithelial cells by quantitative histone post-translational modification-enzyme-linked immunosorbent assay. J. Appl. Toxicol. 2018, 38, 888–895. [Google Scholar] [CrossRef]
- Liang, Y.; Pi, H.; Liao, L.; Tan, M.; Deng, P.; Yue, Y.; Xi, Y.; Tian, L.; Xie, J.; Chen, M.; et al. Cadmium promotes breast cancer cell proliferation, migration and invasion by inhibiting ACSS2/ATG5-mediated autophagy. Environ. Pollut. 2021, 273, 116504. [Google Scholar] [CrossRef]
- Dhar, S.; Gursoy-Yuzugullu, O.; Parasuram, R.; Price, B.D. The tale of a tail: Histone H4 acetylation and the repair of DNA breaks. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160284. [Google Scholar] [CrossRef] [Green Version]
- Somji, S.; Garrett, S.H.; Toni, C.; Zhou, X.D.; Zheng, Y.; Ajjimaporn, A.; Sens, M.A.; Sens, D.A. Differences in the epigenetic regulation of MT-3 gene expression between parental and Cd+2 or As+3 transformed human urothelial cells. Cancer Cell Int. 2011, 11, 2. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Liu, Y.; Xie, C.; Tu, W.; Xia, Y.; Costa, M.; Zhou, X. Cadmium induces histone H3 lysine methylation by inhibiting histone demethylase activity. Toxicol. Sci. 2015, 145, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Chen, W.; Sun, Y.; Xia, P.; Liu, J.; Zhang, W. The role of microRNAs in regulating cadmium-induced apoptosis by targeting Bcl-2 in IEC-6 cells. Toxicol. Appl. Pharmacol. 2021, 432, 115737. [Google Scholar] [CrossRef]
- Hao, R.; Ge, J.; Song, X.; Li, F.; Sun-Waterhouse, D.; Li, D. Cadmium induces ferroptosis and apoptosis by modulating miR-34a-5p/Sirt1axis in PC12 cells. Environ. Toxicol. 2022, 37, 41–51. [Google Scholar] [CrossRef]
- Zheng, L.; Jiang, Y.L.; Fei, J.; Cao, P.; Zhang, C.; Xie, G.F.; Wang, L.X.; Cao, W.; Fu, L.; Zhao, H. Circulatory cadmium positively correlates with epithelial-mesenchymal transition in patients with chronic obstructive pulmonary disease. Ecotoxicol. Environ. Saf. 2021, 215, 112164. [Google Scholar] [CrossRef]
- Tanwar, V.S.; Zhang, X.; Jagannathan, L.; Jose, C.C.; Cuddapah, S. Cadmium exposure upregulates SNAIL through miR-30 repression in human lung epithelial cells. Toxicol. Appl. Pharmacol. 2019, 373, 1–9. [Google Scholar] [CrossRef]
- Urani, C.; Melchioretto, P.; Bruschi, M.; Fabbri, M.; Sacco, M.G.; Gribaldo, L. Impact of Cadmium on Intracellular Zinc Levels in HepG2 Cells: Quantitative Evaluations and Molecular Effects. BioMed Res. Int. 2015, 2015, 949514. [Google Scholar] [CrossRef] [Green Version]
- Mongroo, P.S.; Rustgi, A.K. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010, 10, 219–222. [Google Scholar] [CrossRef]
- Nie, D.; Fu, J.; Chen, H.; Cheng, J.; Fu, J. Roles of MicroRNA-34a in Epithelial to Mesenchymal Transition, Competing Endogenous RNA Sponging and Its Therapeutic Potential. Int. J. Mol. Sci. 2019, 20, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufhold, S.; Bonavida, B. Central role of Snail1 in the regulation of EMT and resistance in cancer: A target for therapeutic intervention. J. Exp. Clin. Cancer Res. 2014, 33, 62. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Pacheco, M.; Hidalgo-Miranda, A.; Romero-Córdoba, S.; Valverde, M.; Rojas, E. MRNA and miRNA expression patterns associated to pathways linked to metal mixture health effects. Gene 2014, 533, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Rojas, E.; Martínez-Pacheco, M.; Rodríguez-Sastre, M.A.; Valverde, M. As-Cd-Pb Mixture Induces Cellular Transformation via Post-Transcriptional Regulation of Rad51c by miR-222. Cell. Physiol. Biochem. 2019, 53, 910–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, Y.; Liu, L.Z.; Jiang, Y.; Zhu, Y.; Guo, N.L.; Barnett, J.; Rojanasakul, Y.; Agani, F.; Jiang, B.H. Cadmium increases HIF-1 and VEGF expression through ROS, ERK, and AKT signaling pathways and induces malignant transformation of human bronchial epithelial cells. Toxicol. Sci. 2012, 125, 10–19. [Google Scholar] [CrossRef]
- Liu, F.; Wang, B.; Li, L.; Dong, F.; Chen, X.; Li, Y.; Dong, X.; Wada, Y.; Kapron, C.M.; Liu, J. Low-Dose Cadmium Upregulates VEGF Expression in Lung Adenocarcinoma Cells. Int. J. Environ. Res. Public Health 2015, 12, 10508–10521. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lim, W.; Ko, Y.; Kwon, H.; Kim, S.; Kim, O.; Park, G.; Choi, H.; Kim, O. The effects of cadmium on VEGF-mediated angiogenesis in HUVECs. J. Appl. Toxicol. 2012, 32, 342–349. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L.; Wang, Y.; Dong, F.; Chen, X.; Liu, F.; Xu, D.; Yi, F.; Kapron, C.M.; Liu, J. NF-κB signaling maintains the survival of cadmium-exposed human renal glomerular endothelial cells. Int. J. Mol. Med. 2016, 38, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.Z.; Hu, X.W.; Xia, C.; He, J.; Zhou, Q.; Shi, X.; Fang, J.; Jiang, B.H. Reactive oxygen species regulate epidermal growth factor-induced vascular endothelial growth factor and hypoxia-inducible factor-1alpha expression through activation of AKT and P70S6K1 in human ovarian cancer cells. Free. Radic. Biol. Med. 2006, 41, 1521–1533. [Google Scholar] [CrossRef]
- Dong, F.; Zhou, X.; Li, C.; Yan, S.; Deng, X.; Cao, Z.; Li, L.; Tang, B.; Allen, T.D.; Liu, J. Dihydroartemisinin targets VEGFR2 via the NF-κB pathway in endothelial cells to inhibit angiogenesis. Cancer Biol. Ther. 2014, 15, 1479–1488. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Jia, J.; Wada, Y.; Kapron, C.M.; Liu, J. Dose dependent effects of cadmium on tumor angiogenesis. Oncotarget 2017, 8, 44944–44959. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.S.; Jeong, E.M.; Park, E.K.; Kim, Y.M.; Sohn, S.; Lee, S.H.; Baik, E.J.; Moon, C.H. Cadmium induces apoptotic cell death through p38 MAPK in brain microvessel endothelial cells. Eur. J. Pharmacol. 2008, 578, 11–18. [Google Scholar] [CrossRef]
- Che, L.; Wu, Z.L.; Huang, L.Y.; Wu, J.S.; Du, Z.B.; Lin, J.X.; Su, Y.H.; Chen, X.X.; Lin, Z.N.; Lin, Y.C. MicroRNA-101 inhibits cadmium-induced angiogenesis by targeting cyclooxygenase-2 in primary human umbilical vein endothelial cells. Biochem. Pharmacol. 2021, 189, 114192. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, S.; Wang, S.; Li, X.; Hou, D.; Li, H.; Wang, L.; Xu, Y.; Ma, B.; Wang, H.; et al. LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol. Environ. Saf. 2021, 220, 112376. [Google Scholar] [CrossRef]
- Zhou, M.; Li, L.; Chen, B.; Pan, S.; Tu, W.; Hou, Y.; Chen, P.; Hernández, R.R.; Zhou, X. Circ-SHPRH suppresses cadmium-induced transformation of human bronchial epithelial cells by regulating QKI expression via miR-224-5p. Ecotoxicol. Environ. Saf. 2021, 220, 112378. [Google Scholar] [CrossRef]
- Yue, Y.; Deng, P.; Xiao, H.; Tan, M.; Wang, H.; Tian, L.; Xie, J.; Chen, M.; Luo, Y.; Wang, L.; et al. N6-methyladenosine-mediated downregulation of miR-374c-5p promotes cadmium-induced cell proliferation and metastasis by targeting GRM3 in breast cancer cells. Ecotoxicol. Environ. Saf. 2022, 229, 113085. [Google Scholar] [CrossRef]
- García-Pérez, J.; Pérez-Abad, N.; Lope, V.; Castelló, A.; Pollán, M.; González-Sánchez, M.; Valencia, J.L.; López-Abente, G.; Fernández-Navarro, P. Breast and prostate cancer mortality and industrial pollution. Environ. Pollut. 2016, 214, 394–399. [Google Scholar] [CrossRef]
- Mullins, J.K.; Loeb, S. Environmental exposures and prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2012, 30, 216–219. [Google Scholar] [CrossRef]
- Kulkarni, P.; Dasgupta, P.; Bhat, N.S.; Hashimoto, Y.; Saini, S.; Shahryari, V.; Yamamura, S.; Shiina, M.; Tanaka, Y.; Dahiya, R.; et al. Role of the PI3K/Akt pathway in cadmium induced malignant transformation of normal prostate epithelial cells. Toxicol. Appl. Pharmacol. 2020, 409, 115308. [Google Scholar] [CrossRef]
- Huang, Q.; Lu, Q.; Chen, B.; Shen, H.; Liu, Q.; Zhou, Z.; Lei, Y. LncRNA-MALAT1 as a novel biomarker of cadmium toxicity regulates cell proliferation and apoptosis. Toxicol. Res. 2017, 6, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Liu, H.; Wang, C.; Lu, Q.; Huang, Q.; Zheng, C.; Lei, Y. Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci. Rep. 2015, 5, 15293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Huang, Z.; Chen, B.; Lu, Q.; Cao, L.; Chen, W. LncRNA-ENST00000446135 is a novel biomarker of cadmium toxicity in 16HBE cells, rats, and Cd-exposed workers and regulates DNA damage and repair. Toxicol. Res. 2020, 9, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Qi, Z.; Dong, Z.; Liu, W.; Xu, M.; Gao, M.; Liu, S. LncRNA MT1DP promotes cadmium-induced DNA replication stress by inhibiting chromatin recruitment of SMARCAL1. Sci. Total Environ. 2022, 807, 151078. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Rea, M.; Wang, Z.; Yang, C. Down-regulation of lncRNA MEG3 promotes chronic low dose cadmium exposure-induced cell transformation and cancer stem cell-like property. Toxicol. Appl. Pharmacol. 2021, 430, 115724. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.P.; Wang, Z.; Yang, C. LncRNA DUXAP10 Upregulation and the Hedgehog Pathway Activation Are Critically Involved in Chronic Cadmium Exposure-Induced Cancer Stem Cell-Like Property. Toxicol. Sci. 2021, 184, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-Y.; Cai, Z.-R.; Liu, J.; Wang, D.-S.; Ju, H.-Q.; Xu, R.-H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cui, H.; Huang, C.; Kong, F.; Yang, Q.; Miao, P.; Cao, Z.; Zhang, W.; Chang, D. Interactions of circRNAs with methylation: An important aspect of circRNA biogenesis and function (Review). Mol. Med. Rep. 2022, 25, 169. [Google Scholar] [CrossRef]
- Wang, S.; Liu, F.; Ma, H.; Cui, X.; Yang, S.; Qin, R. circCDYL Acts as a Tumor Suppressor in Triple Negative Breast Cancer by Sponging miR-190a-3p and Upregulating TP53INP1. Clin. Breast Cancer 2020, 20, 422–430. [Google Scholar] [CrossRef]
- Xue, M.; Hong, W.; Jiang, J.; Zhao, F.; Gao, X. Circular RNA circ-LDLRAD3 serves as an oncogene to promote non-small cell lung cancer progression by upregulating SLC1A5 through sponging miR-137. RNA Biol. 2020, 17, 1811–1822. [Google Scholar] [CrossRef]
- Shi, F.; Wei, D.; Zhu, Z.; Yan, F.; Wang, F.; Zhang, K.; Li, X.; Zheng, Y.; Yuan, J.; Lu, Z.; et al. The RNA-binding protein QKI suppresses tumorigenesis of clear cell renal cell carcinoma by regulating the expression of HIF-1α. J. Cancer 2020, 11, 1359–1370. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.J.; Kim, J.S.; Lee, S.; Lee, H.; Yoon, J.S.; Hong, J.H.; Chun, S.H.; Sun, S.; Won, H.S.; Hong, S.A.; et al. QKI, a miR-200 target gene, suppresses epithelial-to-mesenchymal transition and tumor growth. Int. J. Cancer 2019, 145, 1585–1595. [Google Scholar] [CrossRef]
- Wang, Q.; Pan, S.; Jiang, Q.; Li, L.; Tu, W.; Zhang, Q.; Zhou, X. CircSPAG16 suppresses cadmium-induced transformation of human bronchial epithelial cells by decoying PIP5K1α to inactivate Akt. Mol. Carcinog. 2021, 60, 582–594. [Google Scholar] [CrossRef]
- Pan, S.; Wang, Q.; Zhang, Q.; Zhou, M.; Li, L.; Zhou, X. A novel circular RNA, circPUS7 promotes cadmium-induced transformation of human bronchial epithelial cells by regulating Kirsten rat sarcoma viral oncogene homolog expression via sponging miR-770. Metallomics 2021, 13, mfab043. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef]
- Chen, Z.; Liang, Y.; Lu, Q.; Nazar, M.; Mao, Y.; Aboragah, A.; Yang, Z.; Loor, J.J. Cadmium promotes apoptosis and inflammation via the circ08409/miR-133a/TGFB2 axis in bovine mammary epithelial cells and mouse mammary gland. Ecotoxicol. Environ. Saf. 2021, 222, 112477. [Google Scholar] [CrossRef]
- Schaefer, M.; Kapoor, U.; Jantsch, M.F. Understanding RNA modifications: The promises and technological bottlenecks of the ‘epitranscriptome’. Open Biol. 2017, 7, 170077. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.; Shi, X.; Dai, C.; Shen, F.; Rocco, G.; Chen, J.; Huang, Z.; Chen, C.; He, C.; Huang, T.; et al. RNA m6A Modification in Cancers: Molecular Mechanisms and Potential Clinical Applications. Innovation 2020, 1, 100066. [Google Scholar] [CrossRef]
- Li, L.; Zhou, M.; Chen, B.; Wang, Q.; Pan, S.; Hou, Y.; Xia, J.; Zhou, X. ALKBH5 promotes cadmium-induced transformation of human bronchial epithelial cells by regulating PTEN expression in an m6A-dependent manner. Ecotoxicol. Environ. Saf. 2021, 224, 112686. [Google Scholar] [CrossRef]
- Wu, B.; Jiang, X.; Huang, Y.; Ying, X.; Zhang, H.; Liu, B.; Li, Z.; Qi, D.; Ji, W.; Cai, X. Integrated analysis of mRNA-m6A-protein profiles reveals novel insights into the mechanisms for cadmium-induced urothelial transformation. Biomarkers 2021, 26, 499–507. [Google Scholar] [CrossRef]
- Chen, A.; Wang, L.; Li, B.Y.; Sherman, J.; Ryu, J.E.; Hamamura, K.; Liu, Y.; Nakshatri, H.; Yokota, H. Reduction in Migratory Phenotype in a Metastasized Breast Cancer Cell Line via Downregulation of S100A4 and GRM3. Sci. Rep. 2017, 7, 3459. [Google Scholar] [CrossRef] [PubMed]


| Cells or Tissues | Compound | Route | Exposure Time | Change (s) | Role | Mechanism |
|---|---|---|---|---|---|---|
| 16HBE cells [50] | Dichromate (Cr2O72−) | Culture medium | 24 h | Hypermethylation at CpG1, CpG31, and CpG32 of p16INK4a promoter | Pro-oncogenic | Hypermethylation of the promoter caused lowered p16INK4a expression and accumulation of DNA damage. |
| Human lung cancer [51] | Chromate | Inhalation | >15 y | |||
| 16HBE/human lung cancer [52] | Potassium dichromate (K2Cr2O7) | Culture medium/ inhalation | 24 h | Hypermethylation of MGMT, HOGG1, RAD51, XRCC1, and ERCC3 | Pro-oncogenic | Decreased the expression of these DNA repair genes, leading to suppressed DNA repair system and resultant accumulation of genetic damages. |
| Human lung cancer [54,56] | Chromate | Inhalation | N/A | Hypermethylation of MLH1 promoter | Pro-oncogenic | Inhibit the expression of MLH1, a DNA mismatch repair gene, causing increased microsatellite instability of the genome. |
| Human lung cancer [57] | Chromate | Inhalation | N/A | Global DNA hypermethylation Hypermethylation of p16 and APC promoters | Pro-oncogenic | Globally increased genomic methylation; increased promoter methylation caused decreased expression of p16 and APC, two tumor suppressors, contributing to Cr(VI) carcinogenesis. |
| Cells or Tissues | Compound | Route | Exposure Time | Modification (s) | Role | Mechanism |
|---|---|---|---|---|---|---|
| BEAS-2B/16HBE cells [46] | K2Cr2O7 | Culture medium | 20 w/40 w | Increased global H3K9me2 and H3K27me3 | Pro-oncogenic | Cr(VI) led to up-regulated global H3K9me2 and H3K27me3 by increasing the levels of HMTs. The aberrant histone methylation contributed to Cr(VI)-induced genotoxicity and carcinogenesis. |
| Human lung cancer [57] | Chromate | Inhalation | NA | Globally increased H3K9me2/3 and H3K4me2/3, but decreased H3K27me3 and H3R2me2 Increased H3K9me2 in MLH1 promoter | Pro-oncogenic | Cr(VI) induced globally increased H3K9me2/3 and H3K4me2/3, but decreased H3K27me3 and H3R2me2. Cr(VI) also caused increased H3K9me2 in MLH1 promoter, likely via up-regulating G9a and SUV39H1, causing repressed expression of MLH1, a tumor suppressor. |
| A549 [58] | K2CrO4 | Culture medium | 1 h–24 h | |||
| A549 [59] | K2CrO4 | Culture medium | 24 h | |||
| BEAS-2B [60] | K2CrO4 | Culture medium | 6 months | Reduced H3K9ac and H3K4me3, but enriched H3K27me3 in the promoter of HHIP | Pro-oncogenic | Cr(VI) caused reduced levels of H3K9ac and H3K4me3 but enriched H3K27me3 in the promoter of HHIP, leading to decreased expression of HHIP. Decreased HHIP activated the hedgehog signaling, which contributed to Cr(VI) carcinogenesis. |
| BEAS-2B [62] | K2Cr2O7 | Culture medium | 6 months | Enhanced histone H3 acetylation in the promoter of CXCL5 | Pro-oncogenic | Cr(VI) activated the c-Myc/p300 complex, which was specifically bound to the CXCL5 promoter and enhanced H3 acetylation and eventually promoted CXCL5 transcription. Up-regulated CXCL5 contributed to Cr(VI) carcinogenesis. |
| BEAS-2B [63] | K2CrO4 | Culture medium | 24 h/1–2 w | Increased H3K9ac and H3K14ac of NUPR1 promoter | Pro-oncogenic | Cr(VI) caused increased H3K9ac and H3K14ac of NUPR1 promoter and elevated NUPR1 expression, which in turn promoted Cr(VI)-induced transformation of BEAS-2B cells. |
| BEAS-2B cells [69] | K2Cr2O7 | Culture medium | 20 w | Increased H3K9ac and H3K27ac at the promoter of ACLY | Pro-oncogenic | Cr(VI) led to a glycolytic shift, which led to increased H3K9ac, H3K27ac, and H2B and H4 acetylation. The up-regulated H3ac at the ACLY promoter promoted ACLY transcription; up-regulated ACLY in turn increased the glycolytic shift and histone acetylation, forming a positive feedback loop. The glycolytic shift played a critical role in maintaining the malignant phenotypes of Cr(VI)-transformed cells. |
| 16HBE [70,71] | K2CrO4 | Culture medium | 15 w | Global and 53BP1 promoter-specific decreases in H3K18ac and H3K27ac | Pro-oncogenic | Cr(VI) caused increased SET, resulting in global and 53BP1 promoter-specific reduction in H3K18ac and H3K27ac. The promoter histone hypoacetylation resulted in decreased expression of 53BP1, which contributed to Cr(VI) carcinogenesis by causing DNA damage accumulation and inhibition of apoptosis. |
| Cells or Tissues | Compound | Route | Exposure Time | miRNA | Alteration | Role | Target (s) | Mechanism |
|---|---|---|---|---|---|---|---|---|
| BEAS-2B/BALB/cJ mice [75] | Na2Cr2O7/ zinc chromate | Culture medium/intranasal instillation | 6 months /12 w | miR-27a miR-27b | Down | Tumor suppressor | NRF2 | MiR-27a/b were down-regulated in response to ROS production upon Cr(VI) exposure; down-regulated miR27a/b promoted Cr(VI)-induced tumorigenesis and angiogenesis by up-regulating NRF2. |
| BEAS-2B [77,78] | K2Cr2O7 | Culture medium | 24 h/ 6 months | miR-21 | Up | Oncogenic | PDCD4 | Cr(VI) promoted miR-21 transcription by activating STAT3. Up-regulated miR-21 directly targeted PDCD4, a tumor suppressor. Inhibition of PDCD4 contributed to Cr(VI)-induced cancer by suppressing the expression of E-cadherin, c-Myc, and uPAR. |
| L02 hepatocytes [81] | NA | Culture medium | 24 h | miR-21 | Down | Pro-apoptotic | PDCD4 | Cr(VI) caused a decrease in miR-21 and a subsequent increase in PDCD4, contributing to Cr(VI)-induced apoptosis and decreased proliferation of liver cells. |
| BEAS-2B [83,84] | Na2Cr2O7 | Culture medium | 6 months | miR-143 | Down | Tumor suppressor | IGF-IR IRS1 | Cr(VI) lowered miR-143 expression; decreased miR-143 promoted Cr(VI)-induced tumor growth and angiogenesis by activating IGF-IR/IRS1/ERK, mTOR/p70S6K1, HIF-1α/VEGF, and NF-κB p65 pathways. |
| Human lung cancer [85] | Na2Cr2O7 | Inhalation | 3.0–10.0 years | miR-3940-5p | Down | Oncogenic | NA | Cr(VI) led to decreased miR-3940-5p; the lowered miR-3940-5p then enhanced the HR of DSBs to mitigate the accumulation of DNA damage, playing a protective role in Cr(VI)-caused cell transformation. |
| 16HBE [86] | Na2CrO4 | Culture medium | 24 h | |||||
| BEAS-2B [88] | K2Cr2O7 | Culture medium | 20 w | miR-494 | Down | Tumor suppressor | c-Myc | Cr(VI) caused down-regulation of miR-494, which in turn led to increased c-Myc, contributing to Cr(VI)-induced transformation and acquisition of CSC-like properties in BEAS-2B cells. |
| Cells or Tissues | Compound | Route | Exposure Time | Modification (s) | Role | Mechanism |
|---|---|---|---|---|---|---|
| 16HBE cells [100] | Crystalline NiS | Culture medium | 24 h | Hypermethylation of MGMT promoter | Pro-oncogenic | MGMT was down-regulated by DNA hypermethylation, reduced histone H4-ac and H3K9ac, and up-regulated H3K9me2. Lowered expression of MGMT contributed to Ni-induced malignant transformation. |
| BEAS-2B cells [102] | NiCl2 | Culture medium | 72 h/ 6–9 d | Hypermethylation of E-cadherin promoter | Pro-oncogenic | Ni inhibited E-cadherin expression by induction of ROS-dependent promoter hypermethylation, resulting in the acquisition of EMT, which contributed to Ni carcinogenesis. |
| Wistar rats [103] | Ni3S2 | Intra-muscular injection | 32 w | Hypermethylation of RAR-β2, RASSF1A, and CDKN2A | Unknown | Ni induced hypermethylation of the 5’ region of RAR-β2, RASSF1A, and CDKN2A, resulting in decreased mRNA expressions of these genes. |
| C57BL6 mice [104] | Ni3S2 | Intra-muscular injection | 8 months | Hypermethylation of p16Ink4a | Pro-oncogenic | Ni induced hypermethylation and down-regulated expression of p16Ink4a. The p16Ink4a silence together with activation of the MAPK signaling may contribute to Ni carcinogenesis. |
| Dermal fibroblasts of Syrian hamster [105] | NiCl2 | Culture medium | N/A | Hypermethylation of p16Ink4a | Pro-oncogenic | Promoter hypermethylation-induced silencing of p16Ink4a contributed to Ni-induced immortalization of primary dermal fibroblast SHD cells. |
| Cells or Tissues | Compound | Route | Exposure Time | Modification (s) | Role | Mechanism |
|---|---|---|---|---|---|---|
| BEAS-2B cells [111] | NiCl2 | Culture medium | 6 w | Global increases in H3K4me3 and H3K27me3 | Unknown | Ni caused genome-wide increases in H3K4me3 and H3K27me3, but the role of these changes was unknown. |
| BEAS-2B [112] | NiCl2 | Culture medium | 72 h | Global H3K9me2 spreading | Unknown | Ni caused disrupted H3K9me2 (a repressive mark) domains and subsequent spreading of H3K9me2 into active chromatin regions, causing global gene silencing. |
| A549 cells [59] | K2CrO4 | Culture medium | 24 h | Global increase in H3K9me2 and H3K4me3 Increased H3K4me3 of CA9 and NDRG1 | Unknown | Ni led to increased global levels of H3K9me2 and H3K4me3. Increased H3K4me3 was also discovered in the promoter and coding regions of CA9 and NDRG1, which were up-regulated in Ni-exposed A549 cells. |
| A549 [113] | NiCl2 | |||||
| A549 [114] | NiCl2 | |||||
| BEAS-2B [115] | NiCl2 | Culture medium | 72 h/6 w | Decreased H3K27me3 at the ZEB1 promoter | Pro-oncogenic | Ni led to decreased H3K27me3 (a repressive mark) level at the ZEB1 promoter; loss of H3K27me3 likely caused increased ZEB1 expression, finally resulting in EMT in Ni-exposed cells. |
| BEAS-2B [116] | NiCl2 | Culture medium | 24 h/8 w | Increased H3K9me2 at the SPRY2 promoter | Pro-oncogenic | Ni caused increased H3K9me2 at the SPRY2 promoter by inhibiting JMJD1A, resulting in repressed SPRY2 expression. SPRY2 is a negative regulator of ERK signaling; repression of SPRY2 thus activated the ERK signaling to promote Ni carcinogenesis. |
| HEK293T/786-0 cells [123] | NiCl2 | Culture medium | 24 h | Reduced H3K27me3 level | Unknown | Ni caused a reduced level of H3K27me3, possibly caused by up-regulated JMJD3, an H3K27me3 demethylase, but the function of decreased H3K27me3 was not studied. |
| HEK293/A549 cells [125] | NiCl2 | Culture medium | 48 h | Decreased H3K4me2 of E-cadherin promoter | Pro-oncogenic | Ni caused recruitment of LSD1 complex to E-cadherin promoter to catalyze the reduction in H3K4me2 modification of E-cadherin promoter, resulting in down-regulated E-cadherin level and induction of EMT. |
| 16HBE [100] | Crystalline NiS | Culture medium | 24 h | Reduced H4ac and H3K9ac and up-regulated H3K9me2 of MGMT | Pro-oncogenic | MGMT was down-regulated by DNA hypermethylation, reduced histone H4ac, and H3K9ac and up-regulated H3K9me2. Lowered expression of MGMT led to impaired DNA repairing machinery. |
| Hep3B cells [132] | NiCl2 | Culture medium | 2–24 h | Global histone hypoacetylation Reduced H4ac in Bcl-2 promoter | Tumor suppressive | Ni induced histone hypoacetylation by inhibiting the overall HAT activity. Ni also led to reduced H4 acetylation in the Bcl-2 promoter, resulting in Bcl-2 down-regulation, which contributed to Ni-caused cell growth inhibition and apoptosis. |
| Hep3B [133] | 12–48 h | |||||
| A549 [135] | NiCl2 | Culture medium | 24–48 h | Increased H3S10 phosphorylation | Unknown | Ni induce phosphorylation of H3S10 by activating the JNK/SAPK signaling pathway, but the role of H3S10 phosphorylation was not studied. |
| Cells or Tissues | Compound | Route | Exposure Time | miRNA | Alteration | Role | Target (s) | Mechanism |
|---|---|---|---|---|---|---|---|---|
| BEAS-2B A549 cells [143] | NiCl2 | Culture medium | 15 min–72 h | miR-4417 | Up | Oncogenic | TAB2 | Activation of miR-4417/TAB2 was involved in Ni-induced EMT and carcinogenesis. |
| BEAS-2B [144] | NiSO4 | Culture medium | 4 w | miR-31 | Down | Tumor suppressor | SATB2 | RUNX2 transcriptionally inhibited miR-31, leading to up-regulation of SATB2 to contribute to Ni-induced BEAS-2B cell transformation. |
| H1355/ H23 cells [146] | NiCl2 | Culture medium | 24 h | miR-21 | Up | Oncogenic | SPRY2 RECK | Activation of the EGFR/NF-κB signaling pathway upon Ni exposure induced miR-21, resulting in down-regulation of SPRY 2 and RECK to promote the invasiveness of lung cancer cells. |
| Wistar rats [147] | Ni3S2 | Intra-muscular injection | 32 w | miR-222 | Up | Oncogenic | CDKN1B CDKN1C | In muscle tumors and lung cancer, miR-222 mediated the down-regulation of CDKN1B and CDKN1C, which contributed to Ni-induced tumorigenesis. |
| 16HBE [148] | Crystalline Ni3S2 | Culture medium | 13 rounds of chronic exposure | miR-203 | Down | Tumor suppressor | ABL1 | Down-regulation of miR-203 resulted in the up-regulation of ABL1, contributing to Ni-induced cancer. |
| 16HBE [149] | Crystalline NiS | Culture medium | NA | miR-152 | Down | Tumor suppressor | DNMT1 | Inhibition of miR-152 resulted in increased expression of DNMT1, which promotes Ni-induced cell growth and malignant transformation. |
| Neuro-2a cells [150] | NiCl2 | Culture medium | 2–8 h | miR-210 | Up | Oncogenic | ISCU1/2 | Up-regulated HIF-1α promoted the transcription of miR-210; the elevated miR-210 modulates the energy metabolism shift to aerobic glycolysis through targeting ISCU1/2. |
| BEAS-2B [156] | NiCl2 | Culture medium | 24 h |
| Cells or Tissues | Compound | Route | Exposure Time | Modification (s) | Role | Mechanism |
|---|---|---|---|---|---|---|
| Wistar rats C57BL/6 mice [170] | CdCl2 | i.p. injection | 4 w | Caspase-8 promoter hypermethylation | Tumor suppressive | Cd led to reduced caspase-8 due to promoter hypermethylation, leading to decreased hepatic apoptosis and increased preneoplastic lesions. |
| TRL 1215 cell [171] | CdCl2 | Culture medium | 24 h/ 1–10 w | Global DNA and ApoE promoter hypermethylation | Pro-oncogenic | Cd caused global DNA hypermethylation and hypermethylation of ApoE promoter, leading to lowered ApoE expression. Lowered TET1 mediated ApoE promoter hypermethylation. |
| TRL 1215 [173,174] | CdCl2 | Culture medium | 10 w | |||
| MCF-10A cell [176] | CdCl2 | Culture medium | 40 w | Global DNA hypomethylation | Unknown | Cd induced global DNA hypomethylation and c-Myc and K-Ras overexpression, but neither their correlation nor the role of global DNA hypomethylation in Cd-induced breast cancer was studied. |
| 16HBE [181] | CdCl2 | Culture medium | Chronic exposure for 35 passages | Global and hMSH2, ERCC1, XRCC1, and hOGG1 promoter hypermethylation | Pro-oncogenic | Cd induced global DNA hypermethylation and hypermethylated promoters of DNA repair genes (hMSH2, ERCC1, XRCC1, and hOGG1), which caused reduced expression of these genes and thereby accumulation of DNA damage. |
| RWPE-1 cell [183] | CdCl2 | Culture medium | 10 w | Global and RASSF1A and p16 promoter hypermethylation | Pro-oncogenic | Cd induced global DNA hypermethylation and hypermethylation of the promoter of RASSF1A and p16, which led to markedly reduced expression of these two tumor suppressors. |
| Multiple melanoma cell [184] | CdCl2 | Culture medium | 48–72 h | p16INK4A and caspase-8 hypermethylation | Pro-oncogenic | Cd induced silencing of p16INK4A and caspase-8, attributed to promoter hypermethylation caused by increased activity of DNMTs. |
| HMy2.CIR lymphoblast cell [187] | CdCl2 | Culture medium | 48 h/ 3 months | p16 promoter hypermethylation | Pro-oncogenic | Cd caused the down-regulation of p16 via hypermethylation of the CpG island in its promoter. |
| RWPE-1 cell [189] | CdCl2 | Culture medium | 8 w | Hypomethylation of HYAL1 and S100P; hypermethylation of NTM and NES | Pro-oncogenic | Expression of HYAL1 and S100P was up-regulated likely due to promoter hypomethylation; expression of NTM and NES was decreased likely due to promoter hypermethylation. Aberrant expression of these genes may contribute to Cd carcinogenesis. |
| HepG2 MCF7 cells [192] | CdCl2 | Culture medium | 24–48 h | Global DNA hypomethylation Hypomethylation of PRMT5 and EZH2 promoters | Pro-oncogenic | Cd induced global DNA hypomethylation and hypomethylation of PRMT5 and EZH2 promoters to induce their transcription. Up-regulated PRMT5 and EZH2 in turn led to the increased global level of H4R3me2 and H3K27me3 (both repressive histone marks), which potentially silenced tumor suppressors through remodeling the chromatin. |
| Cells or Tissues | Compound | Route | Exposure Time | Modification (s) | Role | Mechanism |
|---|---|---|---|---|---|---|
| BEAS-2B cell [193] | CdCl2 | Culture medium | 20 passages | Global alterations of H3K4me2 H3K36me3 H3K9acS10ph H4K5ac H4K8ac H4K12ac | Pro- oncogenic | Cd-transformed cells showed markedly decreased H3K4me2 and H3K36me3 and up-regulated H3K9acS10ph, H4K5ac, H4K8ac, and H4K12ac histone modifications. Inhibition of histone acetyltransferase activity led to suppressed cancer phenotypes of transformed cells, suggesting that histone hyperacetylation is involved in Cd carcinogenesis. |
| MCF-7 T47-D cells [194] | CdCl2 | Culture medium | 24–72 h | Decreased H3K27ac in ATG5 promoter | Pro- oncogenic | Cd caused decreased ACSS2 expression, resulting in lowered ATG5 expression by reducing the level of H3K27ac in the ATG5 promoter. The resultant inhibition of ATG5-dependent autophagic flux contributed to Cd carcinogenesis. |
| UROtsa cell [196] | CdCl2 | Culture medium | N/A | Increased H4-ac and methylation of H3K4, H3K9, and H3K27 | Pro- oncogenic | Cd or As caused increased H4 acetylation and H3K4 methylation (transcriptional activation marks) and increased H3K9 and H3K27 methylation (transcriptional repression marks) in the promoter of MT-3. This “bivalent” histone modifications status of MT-3 facilitated MTF-1 binding to MT-3 promoter to activate its transcription. Up-regulated MT-3 in turn contributed to Cd carcinogenesis. |
| BEAS-2B [197] | CdCl2 | Culture medium | 6–48 h/20 w | Increased global H3K4me3 and H3K9me2 | Unknown | Cd exposure led to increased global H3K4me3 and H3K9me2 at 24 h and 4 w by inhibiting the activities of H3K4 and H3K9 demethylases, respectively. However, global H3K4me3 and H3K9me2 had no significant change from 8 to 20 weeks, suggesting that increased global H3K4me3 and H3K9me2 are involved in early events of Cd carcinogenesis. However, the exact role of this change was not studied. |
| Cells or Tissues | Compound | Route | Exposure Time | miRNA | Alteration | Role | Target (s) | Mechanism |
|---|---|---|---|---|---|---|---|---|
| IEC-6 cell [198] | CdCl2 | Culture medium | 6–24 h | miR-124-3p miR-370-3p | Up | Tumor suppressor | Bcl-2 | Cd induced miR-124-3p and miR-370-3p, which promoted Cd-induced apoptosis by targeting Bcl-2. |
| PC12 cell [199] | CdCl2 | Culture medium | 24 h | miR-34a-5p | Up | Tumor suppressor | Sirt1 | Cd-induced miR-34a-5p expression and up-regulated miR-34a-5p contributed to Cd-induced apoptosis and ferroptosis by targeting Sirt1. |
| BEAS-2B BEP2D cells [201] | CdCl2 | Culture medium | 1–24 h/ 72 h | miR-30e | Down | Tumor suppressor | SNAIL1 | Down-regulated miR-30e led to increased SNAIL1 expression, which contributed to Cd-induced cancer through the induction of EMT. |
| HepG2 [202] | CdCl2 | Culture medium | 24 h | miR-34a miR-200a | Down | Tumor suppressor | SNAIL1 | miR-34a and miR-200a were down-regulated, causing increased SNAIL1, which contributed to Cd-induced cancer by inducing EMT. |
| Balb/c3T3 cell [207] | Mixture of NaAsO2, CdCl2, and Pb(C2H3O2)2 | Culture medium | 16 d | miR-222 | Up | Oncogenic | Rad51c | Elevated miR-222 down-regulated Rad51c expression and impaired DNA homologous recombination during the initiation stage of cell transformation. |
| HUVEC cell [216] | CdCl2 | Culture medium | 12–36 h | miR-101 | Down | Anti-angiogenic | COX-2 | Suppressed miR-101 induced COX2 expression and ER stress; up-regulated COX2 and ER stress led to increased VEGF protein levels, resulting in abnormal angiogenesis. |
| PC3 DU145 cells [217] | CdCl2 | Culture medium | 20 w | miR-128-3p | Down | Tumor suppressor | SLC7A11 | Cd induced LncRNA OIP5-AS1, which served as an endogenous sponge of miR-128-3p to up-regulate the expression of SLC7A11, thereby inhibiting ferroptosis. |
| BEAS-2B [218] | CdCl2 | Culture medium | 12–20 w | miR-224-5p | Up | Oncogenic | QKI | Circular RNA SHPRH was down-regulated, leading to up-regulated miR-224-5p and resultant down-regulation of QKI to promote proliferation, EMT, migration and invasion, and anchorage-independent growth of cells. |
| T-47D [219] | CdCl2 | Culture medium | 72 h | miR-374c-5p | Down | Tumor suppressor | GRM3 | Cd exposure caused reduced m6A modification of pri-miRNA-374c, resulting in miR-374c-5p down-regulation. Suppression of miR-374c-5p in turn promotes proliferation, migration, and invasion of cells by activating GRM3. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Islam, R.; Wang, Y.; Zhang, X.; Liu, L.-Z. Epigenetic Regulation in Chromium-, Nickel- and Cadmium-Induced Carcinogenesis. Cancers 2022, 14, 5768. https://doi.org/10.3390/cancers14235768
Zhao L, Islam R, Wang Y, Zhang X, Liu L-Z. Epigenetic Regulation in Chromium-, Nickel- and Cadmium-Induced Carcinogenesis. Cancers. 2022; 14(23):5768. https://doi.org/10.3390/cancers14235768
Chicago/Turabian StyleZhao, Lei, Ranakul Islam, Yifang Wang, Xiujuan Zhang, and Ling-Zhi Liu. 2022. "Epigenetic Regulation in Chromium-, Nickel- and Cadmium-Induced Carcinogenesis" Cancers 14, no. 23: 5768. https://doi.org/10.3390/cancers14235768






