STIL Promotes Tumorigenesis of Bladder Cancer by Activating PI3K/AKT/mTOR Signaling Pathway and Targeting C-Myc
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Microarray and Immunohistochemistry
2.2. Cell Culture
2.3. STIL Knockout in UMUC3 and T24 Cells
2.4. CCK-8 Assay
2.5. Cell Colony Formation
2.6. Transwell Migration Assays
2.7. Soft Agar Assay
2.8. Cell Cycle Assay
2.9. The EDU (5-Ethynyl-2-deoxyuridine) Assay
2.10. Immunofluorescence Assay
2.11. RT-qPCR
2.12. Western Blotting
2.13. Tumor Xenografts
2.14. Statistical Analysis
2.15. RNA-Seq Analysis of N-butyl-N-(4-hydroxybutyl) Nitrosamine (BBN)-Treated Bladder Cancer Mouse Models
3. Results
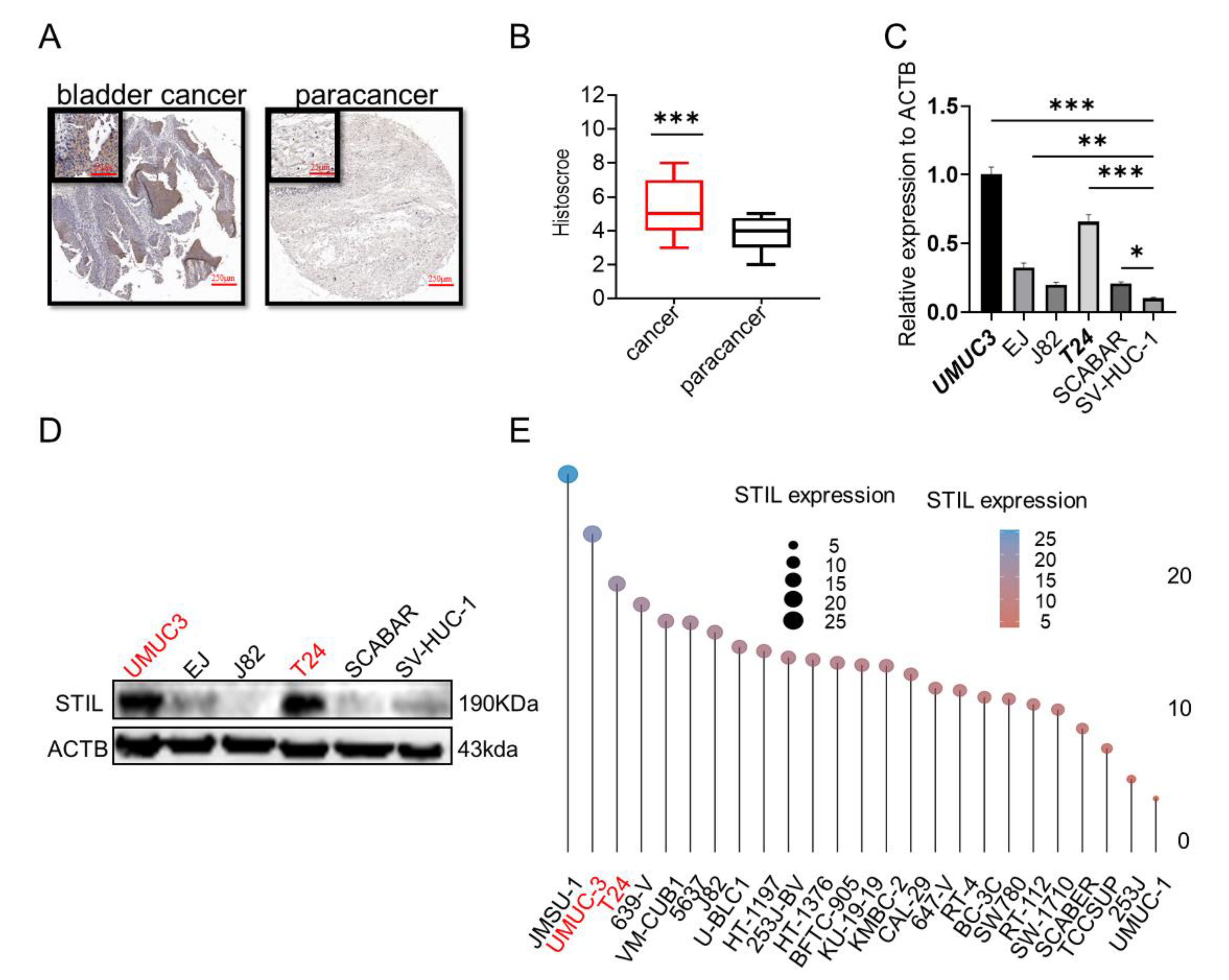
3.1. STIL Was Highly Expressed in BC and Was Inextricably Intertwined with the Cell Cycle
3.2. Expression of STIL Was Significantly Up-Regulated in BC
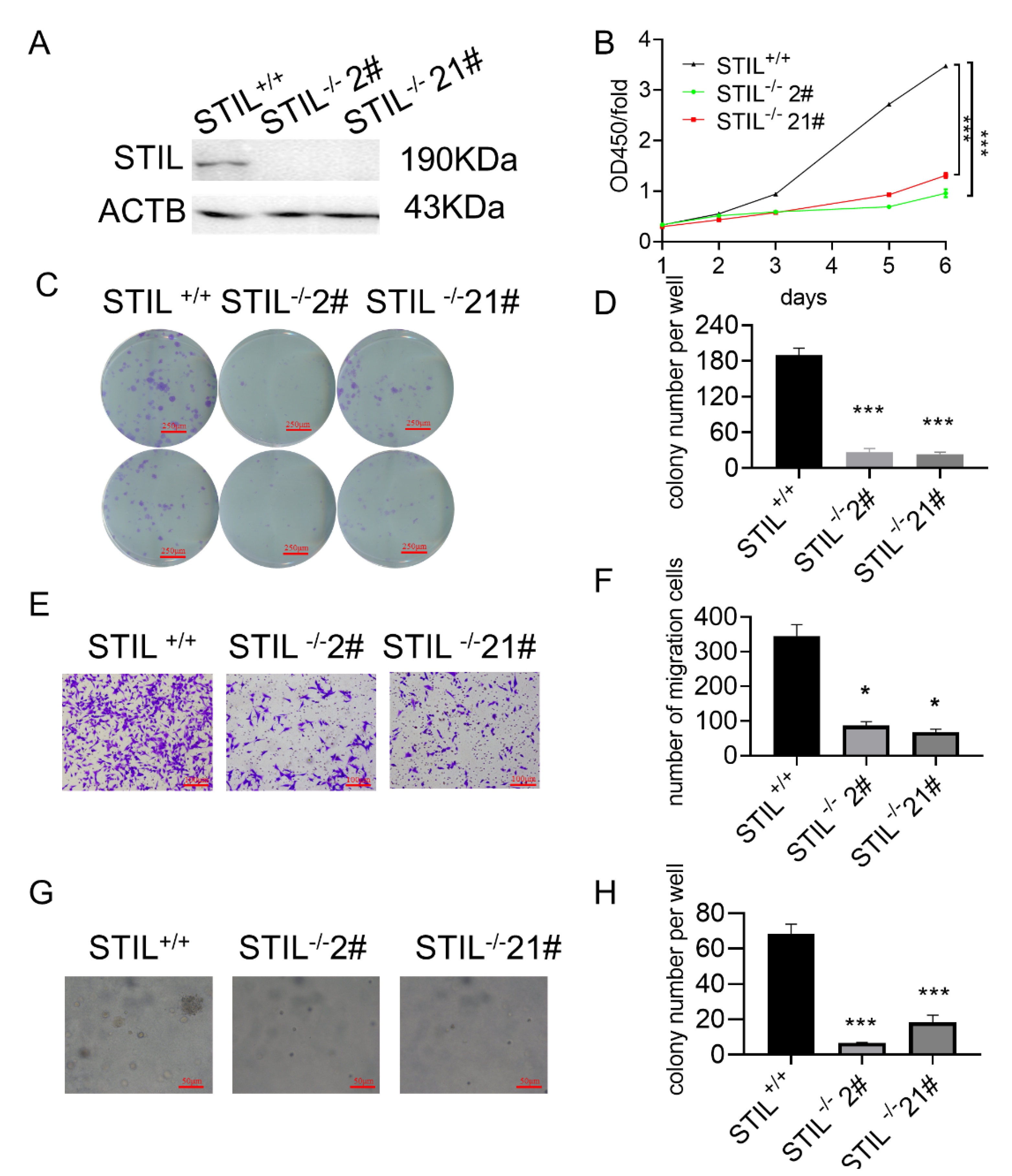
3.3. Knockout of STIL Inhibited BC Tumorigenesis In Vitro
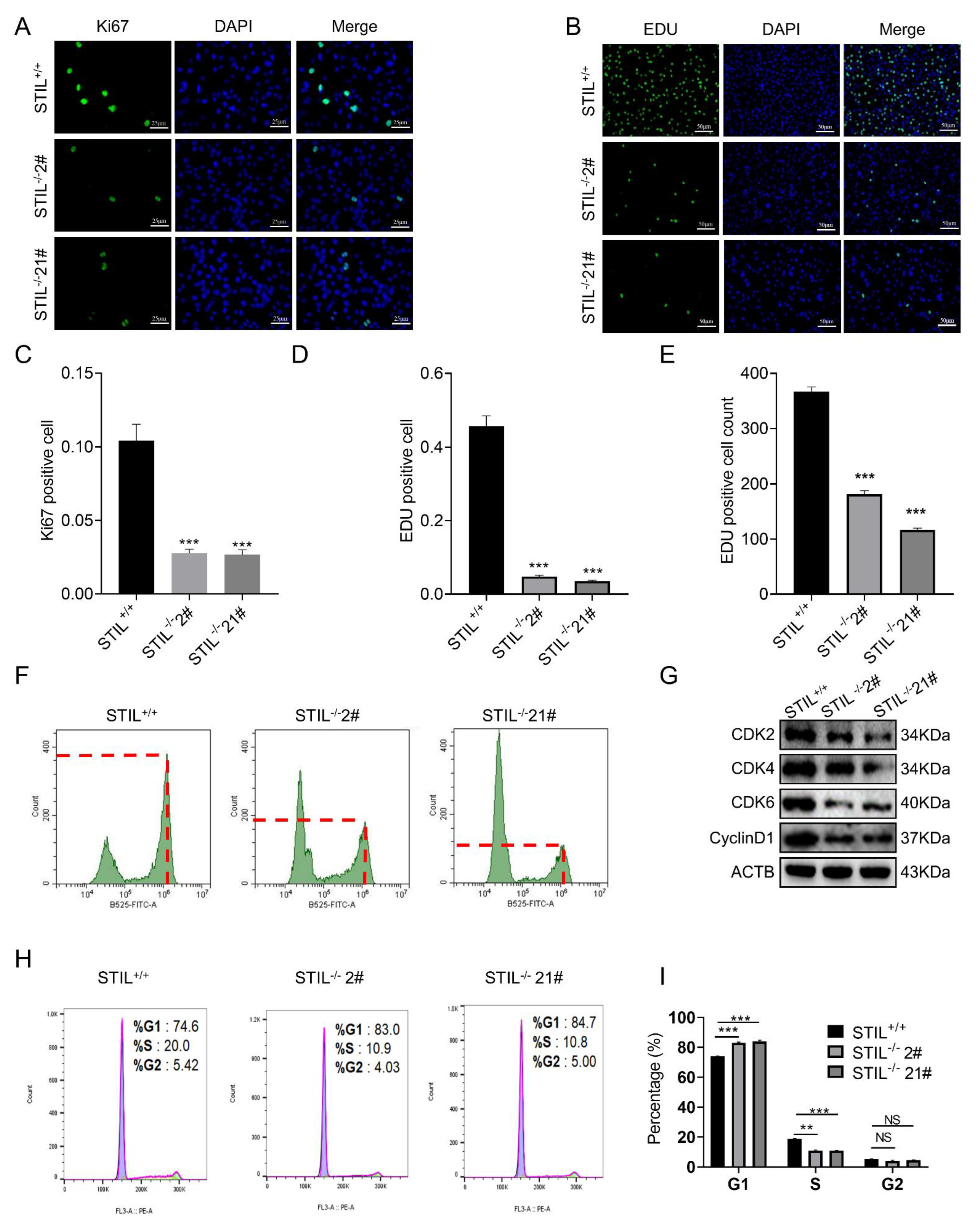
3.4. Cell Cycle of STIL Knockout Cells Was Arrested in the G0/G1 Phase
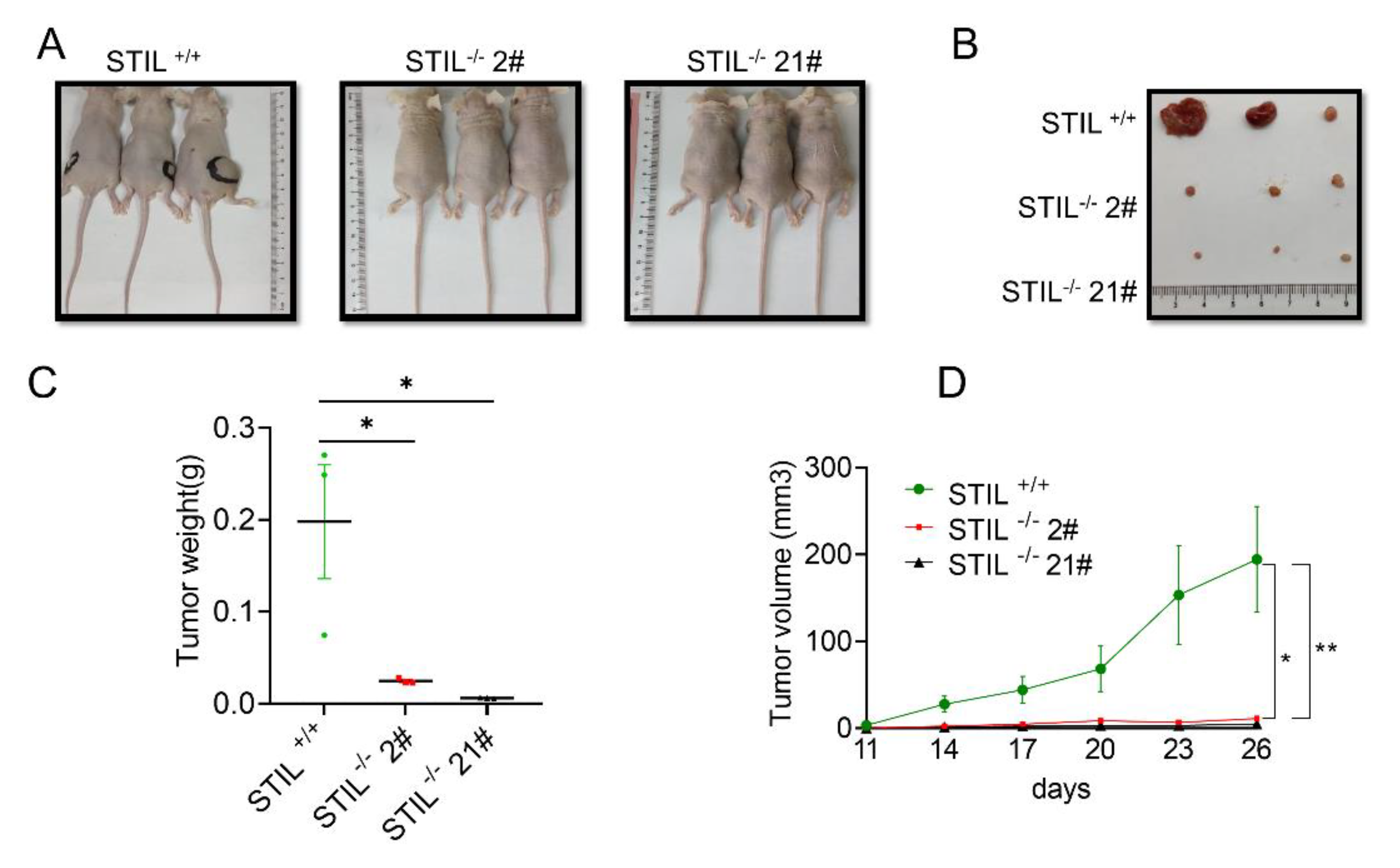
3.5. The Proliferative Function of STIL Was Confirmed by the Xenotransplantation Model
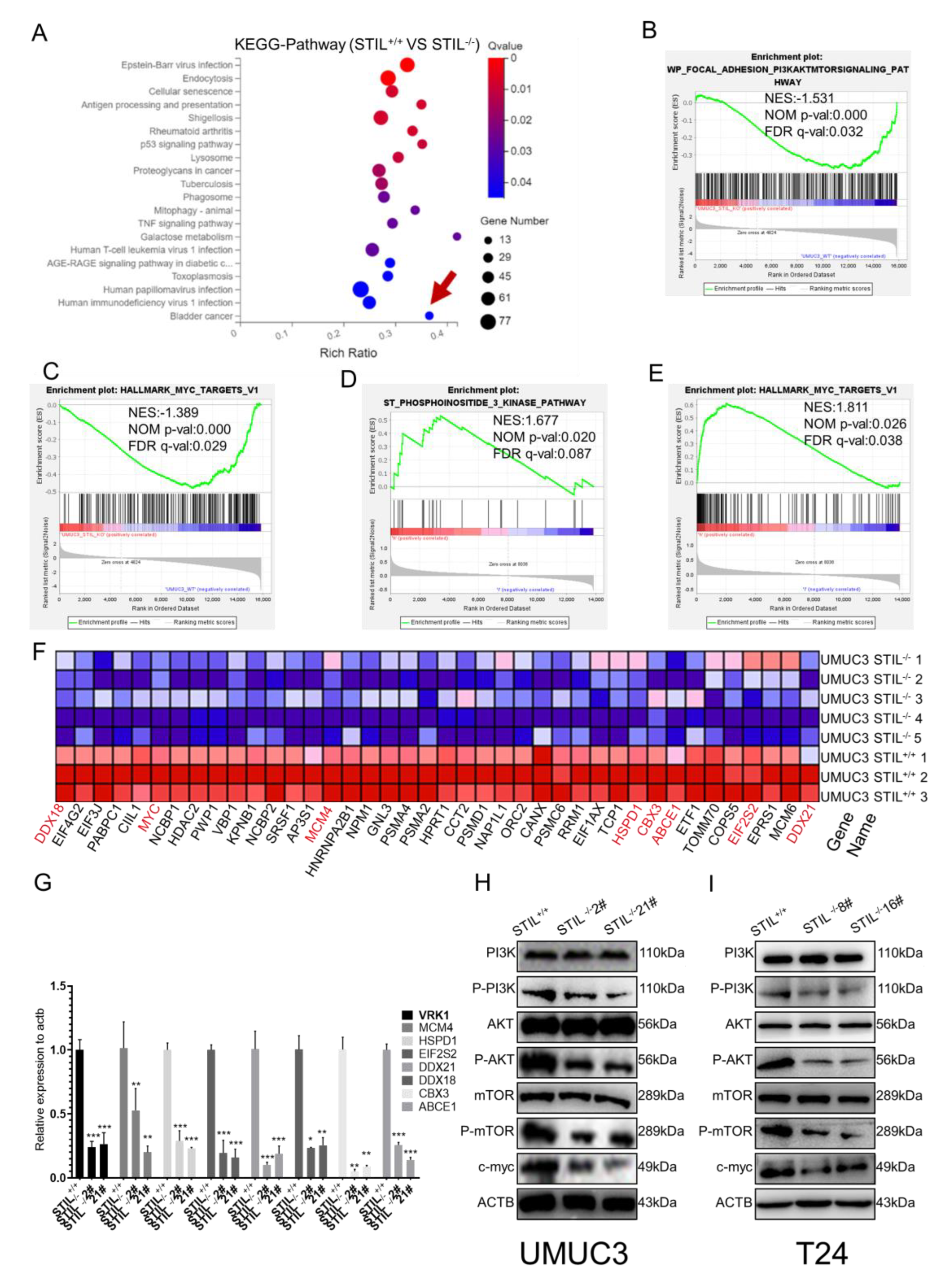
3.6. STIL Knockout Down-Regulated the PI3K/AKT/mTOR/c-myc Signaling Pathway
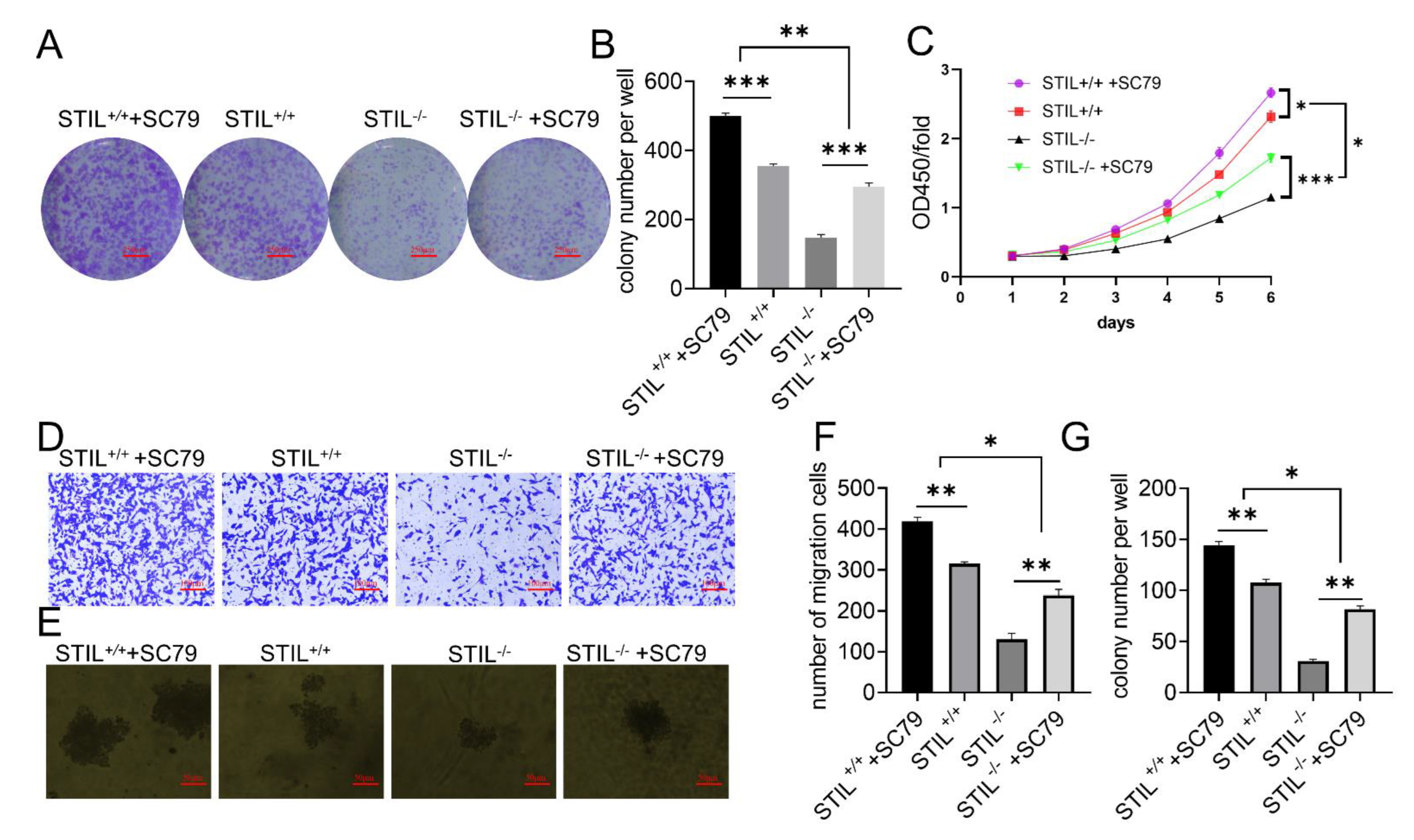
3.7. SC79 Treatment Partially Reversed the Effects of STIL Knockout on Cell Tumorigenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- China National Highways. Chinese guidelines for diagnosis and treatment of urothelial carcinoma of bladder 2018 (English version). Chin. J. Cancer Res. 2019, 31, 49–66. [Google Scholar] [CrossRef]
- Colombel, M.; Soloway, M.; Akaza, H.; Böhle, A.; Palou, J.; Buckley, R.; Lamm, D.; Brausi, M.; Witjes, J.A.; Persad, R. Epidemiology, Staging, Grading, and Risk Stratification of Bladder Cancer. Eur. Urol. Suppl. 2008, 7, 618–626. [Google Scholar] [CrossRef]
- Prout, G.R., Jr.; Barton, B.A.; Griffin, P.P.; Friedell, G.H. Treated history of noninvasive grade 1 transitional cell carcinoma. The National Bladder Cancer Group. J. Urol. 1992, 148, 1413–1419. [Google Scholar] [CrossRef]
- Messing, E.M.; Tangen, C.M.; Lerner, S.P.; Sahasrabudhe, D.M.; Koppie, T.M.; Wood, D.P., Jr.; Mack, P.C.; Svatek, R.S.; Evans, C.P.; Hafez, K.S.; et al. Effect of Intravesical Instillation of Gemcitabine vs Saline Immediately Following Resection of Suspected Low-Grade Non-Muscle-Invasive Bladder Cancer on Tumor Recurrence: SWOG S0337 Randomized Clinical Trial. JAMA 2018, 319, 1880–1888. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef]
- Soloway, M.S. Bladder cancer: Lack of progress in bladder cancer--what are the obstacles? Nat. Rev. Urol. 2013, 10, 5–6. [Google Scholar] [CrossRef]
- Arquint, C.; Nigg, E.A. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans. 2016, 44, 1253–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arquint, C.; Sonnen, K.F.; Stierhof, Y.D.; Nigg, E.A. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 2012, 125, 1342–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradhan, T.; Kumar, V.; Surya, H.E.; Krishna, R.; John, S.; Jissa, V.T.; Anjana, S.; Chandramohan, K.; Nair, S.A. STIL Endows Oncogenic and Stem-Like Attributes to Colorectal Cancer Plausibly by Shh and Wnt Signaling. Front. Oncol. 2021, 11, 581671. [Google Scholar] [CrossRef]
- Kasai, K.; Inaguma, S.; Yoneyama, A.; Yoshikawa, K.; Ikeda, H. SCL/TAL1 interrupting locus derepresses GLI1 from the negative control of suppressor-of-fused in pancreatic cancer cell. Cancer Res. 2008, 68, 7723–7729. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Dou, Z.; Jiang, H.; Wang, Y.; Gao, X.; Xin, X. Knockdown of STIL suppresses the progression of gastric cancer by down-regulating the IGF-1/PI3K/AKT pathway. J. Cell. Mol. Med. 2019, 23, 5566–5575. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, Y.; Yan, W.; Ji, Z.; Zheng, G. The human oncogene SCL/TAL1 interrupting locus (STIL) promotes tumor growth through MAPK/ERK, PI3K/Akt and AMPK pathways in prostate cancer. Gene 2019, 686, 220–227. [Google Scholar] [CrossRef]
- Erez, A.; Perelman, M.; Hewitt, S.M.; Cojacaru, G.; Goldberg, I.; Shahar, I.; Yaron, P.; Muler, I.; Campaner, S.; Amariglio, N.; et al. Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene 2004, 23, 5371–5377. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, Y.; Jin, Y.B.; Chen, X.P.; Zhang, G.Y.; Mao, S.L.; Ling, F.; Luo, W. STIL is upregulated in nasopharyngeal carcinoma tissues and promotes nasopharyngeal carcinoma proliferation, migration and invasion. Neoplasma 2020, 67, 37–45. [Google Scholar] [CrossRef]
- Degoricija, M.; Korac-Prlic, J.; Vilovic, K.; Ivanisevic, T.; Haupt, B.; Palada, V.; Petkovic, M.; Karaman, I.; Terzic, J. The dynamics of the inflammatory response during BBN-induced bladder carcinogenesis in mice. J. Transl. Med. 2019, 17, 394. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Tsunoda, T.; Riku, M.; Inaguma, S.; Inoko, A.; Murakami, H.; Ikeda, H.; Matsuda, M.; Kasai, K. Indispensable role of STIL in the regulation of cancer cell motility through the lamellipodial accumulation of ARHGEF7-PAK1 complex. Oncogene 2020, 39, 1931–1943. [Google Scholar] [CrossRef] [PubMed]
- Izraeli, S.; Colaizzo-Anas, T.; Bertness, V.L.; Mani, K.; Aplan, P.D.; Kirsch, I.R. Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 1997, 8, 1171–1179. [Google Scholar] [PubMed]
- Toyoshima, H.; Hunter, T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 1994, 78, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Hume, S.; Dianov, G.L.; Ramadan, K. A unified model for the G1/S cell cycle transition. Nucleic Acids Res. 2020, 48, 12483–12501. [Google Scholar] [CrossRef]
- Youn, M.J.; Kim, J.K.; Park, S.Y.; Kim, Y.; Kim, S.J.; Lee, J.S.; Chai, K.Y.; Kim, H.J.; Cui, M.X.; So, H.S.; et al. Chaga mushroom (Inonotus obliquus) induces G0/G1 arrest and apoptosis in human hepatoma HepG2 cells. World J. Gastroenterol. 2008, 14, 511–517. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Zhou, H.; Jia, G.; Liu, J.; Han, B.; Cheng, Z.; Jiang, H.; Pan, S.; Sun, B. Pristimerin causes G1 arrest, induces apoptosis, and enhances the chemosensitivity to gemcitabine in pancreatic cancer cells. PLoS ONE 2012, 7, e43826. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yu, Y.; Zhao, W.; You, S.; Feng, M.; Xie, C.; Chi, X.; Zhang, Y.; Wang, X. As a downstream target of the AKT pathway, NPTX1 inhibits proliferation and promotes apoptosis in hepatocellular carcinoma. Biosci. Rep. 2019, 39, BSR20181662. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Tsuchiya, Y.; Ohta, M.; Shiratsuchi, G.; Kitagawa, D. NEK7 is required for G1 progression and procentriole formation. Mol. Biol. Cell. 2017, 28, 2123–2134. [Google Scholar] [CrossRef] [PubMed]
- Erez, A.; Castiel, A.; Trakhtenbrot, L.; Perelman, M.; Rosenthal, E.; Goldstein, I.; Stettner, N.; Harmelin, A.; Eldar-Finkelman, H.; Campaner, S.; et al. The SIL gene is essential for mitotic entry and survival of cancer cells. Cancer Res. 2007, 67, 4022–4027. [Google Scholar] [CrossRef] [Green Version]
- Campaner, S.; Kaldis, P.; Izraeli, S.; Kirsch, I.R. Sil phosphorylation in a Pin1 binding domain affects the duration of the spindle checkpoint. Mol. Cell. Biol. 2005, 25, 6660–6672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Cheng, X.; Yan, J.; Jiang, S. CTHRC1 facilitates bladder cancer cell proliferation and invasion through regulating the PI3K/Akt signaling pathway. Arch. Med. Sci. 2022, 18, 183–194. [Google Scholar] [CrossRef]
- Yu, X.; Li, S.; Pang, M.; Du, Y.; Xu, T.; Bai, T.; Yang, K.; Hu, J.; Zhu, S.; Wang, L.; et al. TSPAN7 Exerts Anti-Tumor Effects in Bladder Cancer Through the PTEN/PI3K/AKT Pathway. Front. Oncol. 2020, 10, 613869. [Google Scholar] [CrossRef]
- Liu, H.; Liu, N.; Zhao, Y.; Zhu, X.; Wang, C.; Liu, Q.; Gao, C.; Zhao, X.; Li, J. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging (Albany NY) 2019, 11, 9643–9660. [Google Scholar] [CrossRef]
- Borzi, C.; Calzolari, L.; Ferretti, A.M.; Caleca, L.; Pastorino, U.; Sozzi, G.; Fortunato, O. c-Myc shuttled by tumour-derived extracellular vesicles promotes lung bronchial cell proliferation through miR-19b and miR-92a. Cell Death Dis. 2019, 10, 759. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Su, L.; Cai, M.; Yao, B.; Xiao, S.; He, Q.; Xu, L.; Yang, L.; Zhao, C.; Wan, T.; et al. Downregulation of CPA4 inhibits non small-cell lung cancer growth by suppressing the AKT/c-MYC pathway. Mol. Carcinog. 2019, 58, 2026–2039. [Google Scholar] [CrossRef] [Green Version]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [Green Version]
- Bautista, S.J.; Boras, I.; Vissa, A.; Mecica, N.; Yip, C.M.; Kim, P.K.; Antonescu, C.N. mTOR complex 1 controls the nuclear localization and function of glycogen synthase kinase 3beta. J. Biol. Chem. 2018, 293, 14723–14739. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Dong, X.; Lu, H.; Tong, F.; Chen, L.; Zhang, R.; Dong, J.; Hu, Y.; Wu, G.; Dong, X. LPCAT1 promotes brain metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC pathway. J. Exp. Clin. Cancer Res. 2019, 38, 95. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Russell, A.J.; Liu, B.; George, J.; Liu, P.Y.; Liu, T.; DeFazio, A.; Bowtell, D.D.; Oberthuer, A.; London, W.B.; et al. A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers. Cancer Res. 2017, 77, 971–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.Y.; Varthi, M.; Sykes, S.M.; Phillips, C.; Warzecha, C.; Zhu, W.; Wyce, A.; Thorne, A.W.; Berger, S.L.; McMahon, S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 2008, 29, 102–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penn, L.J.; Laufer, E.M.; Land, H. C-MYC: Evidence for multiple regulatory functions. Semin. Cancer Biol. 1990, 1, 69–80. [Google Scholar]
- Facchini, L.M.; Penn, L.Z. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998, 12, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Buchholz, M.A.; Chrest, F.J.; Nordin, A.A. Up-regulation of c-myc induces the gene expression of the murine homologues of p34cdc2 and cyclin-dependent kinase-2 in T lymphocytes. J. Immunol. 1994, 152, 4328–4335. [Google Scholar] [PubMed]
- Haas, K.; Staller, P.; Geisen, C.; Bartek, J.; Eilers, M.; Moroy, T. Mutual requirement of CDK4 and Myc in malignant transformation: Evidence for cyclin D1/CDK4 and p16INK4A as upstream regulators of Myc. Oncogene 1997, 15, 179–192. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Chen, L.; Wang, X.; Tang, F.; Wan, Z.; Wang, H.; Fu, Q.; Chen, Z.; Shi, J.; Hu, X.; et al. STIL Promotes Tumorigenesis of Bladder Cancer by Activating PI3K/AKT/mTOR Signaling Pathway and Targeting C-Myc. Cancers 2022, 14, 5777. https://doi.org/10.3390/cancers14235777
Yu H, Chen L, Wang X, Tang F, Wan Z, Wang H, Fu Q, Chen Z, Shi J, Hu X, et al. STIL Promotes Tumorigenesis of Bladder Cancer by Activating PI3K/AKT/mTOR Signaling Pathway and Targeting C-Myc. Cancers. 2022; 14(23):5777. https://doi.org/10.3390/cancers14235777
Chicago/Turabian StyleYu, Hua, Liang Chen, Xia Wang, Feng Tang, Ziyu Wan, Hao Wang, Qiqi Fu, Zhizhuang Chen, Jiageng Shi, Xuan Hu, and et al. 2022. "STIL Promotes Tumorigenesis of Bladder Cancer by Activating PI3K/AKT/mTOR Signaling Pathway and Targeting C-Myc" Cancers 14, no. 23: 5777. https://doi.org/10.3390/cancers14235777
APA StyleYu, H., Chen, L., Wang, X., Tang, F., Wan, Z., Wang, H., Fu, Q., Chen, Z., Shi, J., Hu, X., Zuhaer, Y., Aersi, M., Liu, T., Tao, H., & Peng, J. (2022). STIL Promotes Tumorigenesis of Bladder Cancer by Activating PI3K/AKT/mTOR Signaling Pathway and Targeting C-Myc. Cancers, 14(23), 5777. https://doi.org/10.3390/cancers14235777





