A Critical Role of the IL-22–IL-22 Binding Protein Axis in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Samples and Studies
2.2. Animals
2.3. Chemical Induction of Liver Carcinogenesis
2.4. Choline-Deficient High-Fat Diet
2.5. Magnetic Resonance Imaging (MRI)
2.6. Measurement of Alanine Transaminase (ALT)
2.7. Leucocyte Isolation from the Liver
2.8. Fluorescent-Activated Cell Sorting (FACS)
2.9. Isolation of Primary Hepatocytes
2.10. Murine RNA Extraction
2.11. Human RNA Extraction
2.12. cDNA Synthesis and qPCR
2.13. RNA-seq
2.14. Statistical Analysis
3. Results
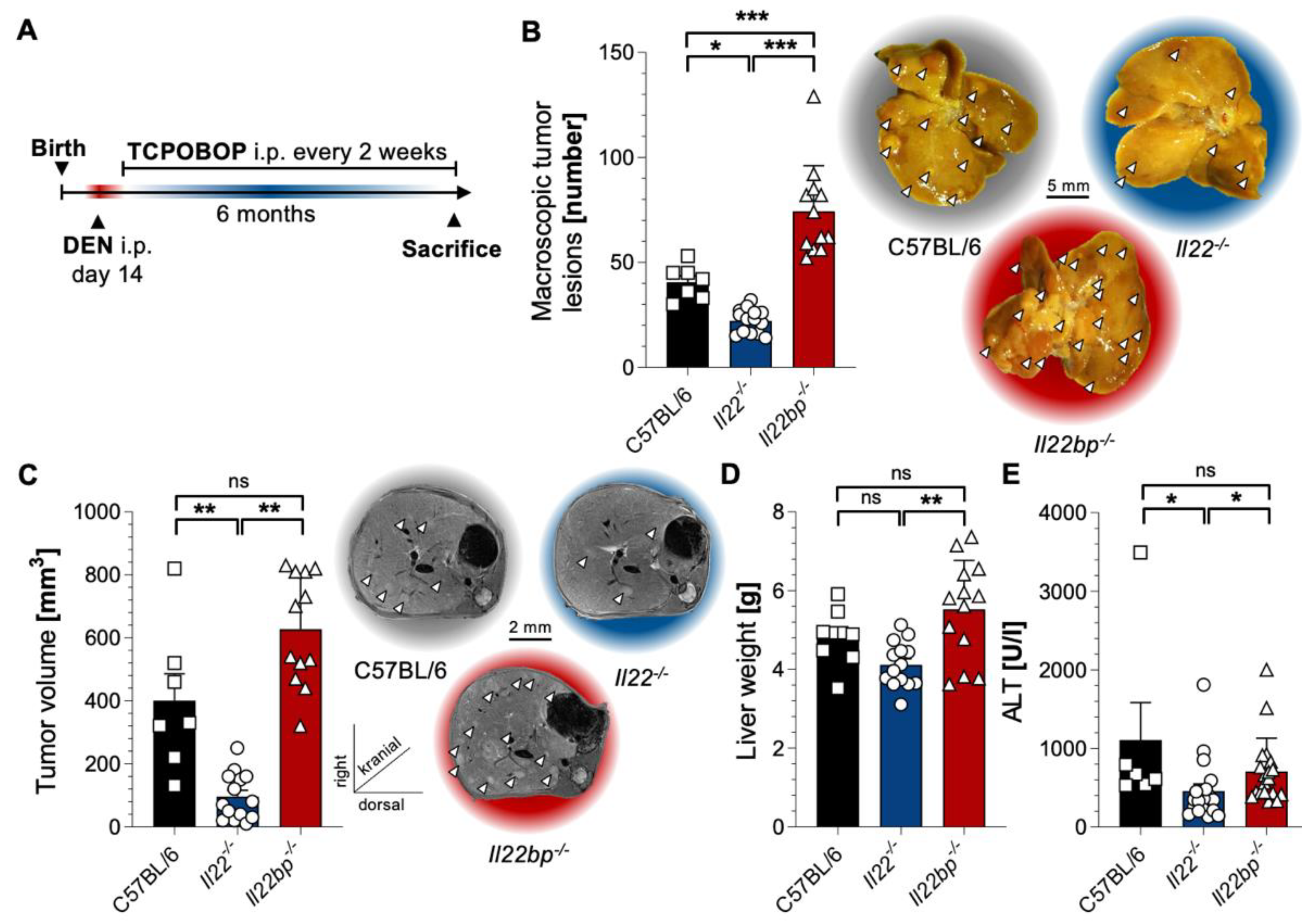
3.1. A Critical Role of the IL-22–IL-22BP Axis in HCC Mouse Models
3.2. CD4+ T Cells and Neutrophils Show a High Expression of IL-22 and IL-22BP in HCC, Respectively
3.3. IL-22 Signaling in Hepatocytes Promotes HCC Development
3.4. Deficiency of IL-22 Signaling in Hepatocytes Leads to Downregulation of STEAP4 in HCC
3.5. STEAP4 Correlates with IL22 in Patient Samples of HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinter, M.; Trauner, M.; Peck-Radosavljevic, M.; Sieghart, W. Cancer and liver cirrhosis: Implications on prognosis and management. ESMO Open 2016, 1, e000042. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Park, J.W.; Chen, M.; Colombo, M.; Roberts, L.R.; Schwartz, M.; Chen, P.J.; Kudo, M.; Johnson, P.; Wagner, S.; Orsini, L.S.; et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015, 35, 2155–2166. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.; Yang, X.R.; Chung, W.Y.; Dennison, A.R.; Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target. Ther. 2020, 5, 146. [Google Scholar] [CrossRef]
- Huppert, L.A.; Gordan, J.D.; Kelley, R.K. Checkpoint Inhibitors for the Treatment of Advanced Hepatocellular Carcinoma. Clin. Liver Dis. 2020, 15, 53–58. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Lucke, J.; Shiri, A.M.; Zhang, T.; Kempski, J.; Giannou, A.D.; Huber, S. Rationalizing heptadecaphobia: TH 17 cells and associated cytokines in cancer and metastasis. FEBS J. 2021, 288, 6942–6971. [Google Scholar] [CrossRef]
- Huber, S.; Gagliani, N.; Zenewicz, L.A.; Huber, F.J.; Bosurgi, L.; Hu, B.; Hedl, M.; Zhang, W.; O’Connor, W., Jr.; Murphy, A.J.; et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 2012, 491, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Gronke, K.; Hernandez, P.P.; Zimmermann, J.; Klose, C.S.N.; Kofoed-Branzk, M.; Guendel, F.; Witkowski, M.; Tizian, C.; Amann, L.; Schumacher, F.; et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 2019, 566, 249–253. [Google Scholar] [CrossRef]
- Sabihi, M.; Bottcher, M.; Pelczar, P.; Huber, S. Microbiota-Dependent Effects of IL-22. Cells 2020, 9, 2205. [Google Scholar] [CrossRef]
- Paget, C.; Ivanov, S.; Fontaine, J.; Renneson, J.; Blanc, F.; Pichavant, M.; Dumoutier, L.; Ryffel, B.; Renauld, J.C.; Gosset, P.; et al. Interleukin-22 is produced by invariant natural killer T lymphocytes during influenza A virus infection: Potential role in protection against lung epithelial damages. J. Biol. Chem. 2012, 287, 8816–8829. [Google Scholar] [CrossRef] [Green Version]
- Steinbach, S.; Vordermeier, H.M.; Jones, G.J. CD4+ and gammadelta T Cells are the main Producers of IL-22 and IL-17A in Lymphocytes from Mycobacterium bovis-infected Cattle. Sci. Rep. 2016, 6, 29990. [Google Scholar] [CrossRef] [Green Version]
- Gnirck, A.C.; Wunderlich, M.; Becker, M.; Xiong, T.; Weinert, E.; Meyer-Schwesinger, C.; Dumoutier, L.; Renauld, J.C.; Huber, S.; Panzer, U.; et al. Endogenous IL-22 is dispensable for experimental glomerulonephritis. Am. J. Physiol. Ren. Physiol. 2019, 316, F712–F722. [Google Scholar] [CrossRef]
- Perez, L.G.; Kempski, J.; McGee, H.M.; Pelzcar, P.; Agalioti, T.; Giannou, A.; Konczalla, L.; Brockmann, L.; Wahib, R.; Xu, H.; et al. TGF-beta signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat. Commun. 2020, 11, 2608. [Google Scholar] [CrossRef]
- Busman-Sahay, K.O.; Walrath, T.; Huber, S.; O’Connor, W., Jr. Cytokine crowdsourcing: Multicellular production of TH17-associated cytokines. J. Leukoc. Biol. 2015, 97, 499–510. [Google Scholar] [CrossRef] [Green Version]
- Duhen, T.; Geiger, R.; Jarrossay, D.; Lanzavecchia, A.; Sallusto, F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 2009, 10, 857–863. [Google Scholar] [CrossRef]
- Victor, A.R.; Nalin, A.P.; Dong, W.; McClory, S.; Wei, M.; Mao, C.; Kladney, R.D.; Youssef, Y.; Chan, W.K.; Briercheck, E.L.; et al. IL-18 Drives ILC3 Proliferation and Promotes IL-22 Production via NF-kappaB. J. Immunol. 2017, 199, 2333–2342. [Google Scholar] [CrossRef] [Green Version]
- Sabat, R. IL-10 family of cytokines. Cytokine Growth Factor Rev. 2010, 21, 315–324. [Google Scholar] [CrossRef]
- Xie, M.H.; Aggarwal, S.; Ho, W.H.; Foster, J.; Zhang, Z.; Stinson, J.; Wood, W.I.; Goddard, A.D.; Gurney, A.L. Interleukin (IL)-22, a novel human cytokine that signals through the interferon receptor-related proteins CRF2-4 and IL-22R. J. Biol. Chem. 2000, 275, 31335–31339. [Google Scholar] [CrossRef]
- Wolk, K.; Kunz, S.; Witte, E.; Friedrich, M.; Asadullah, K.; Sabat, R. IL-22 increases the innate immunity of tissues. Immunity 2004, 21, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Dumoutier, L.; Lejeune, D.; Colau, D.; Renauld, J.C. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J. Immunol. 2001, 166, 7090–7095. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Presnell, S.R.; Parrish-Novak, J.; Kindsvogel, W.; Jaspers, S.; Chen, Z.; Dillon, S.R.; Gao, Z.; Gilbert, T.; Madden, K.; et al. A soluble class II cytokine receptor, IL-22RA2, is a naturally occurring IL-22 antagonist. Proc. Natl. Acad. Sci. USA 2001, 98, 9511–9516. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.C.; Ho, T.W.; Liang, W.G.; Chen, G.Y.; Chang, M.S. Cloning and characterization of mouse IL-22 binding protein. Genes Immun. 2003, 4, 204–211. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, P.P.; Mahlakoiv, T.; Yang, I.; Schwierzeck, V.; Nguyen, N.; Guendel, F.; Gronke, K.; Ryffel, B.; Hoelscher, C.; Dumoutier, L.; et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015, 16, 698–707. [Google Scholar] [CrossRef] [Green Version]
- Kempski, J.; Giannou, A.D.; Riecken, K.; Zhao, L.; Steglich, B.; Lucke, J.; Garcia-Perez, L.; Karstens, K.F.; Wostemeier, A.; Nawrocki, M.; et al. IL22BP Mediates the Anti-Tumor Effects of Lymphotoxin Against Colorectal Tumors in Mice and Humans. Gastroenterology 2020, 159, 1417–1430. [Google Scholar] [CrossRef]
- Lucke, J.; Sabihi, M.; Zhang, T.; Bauditz, L.F.; Shiri, A.M.; Giannou, A.D.; Huber, S. The good and the bad about separation anxiety: Roles of IL-22 and IL-22BP in liver pathologies. Semin. Immunopathol. 2021, 43, 591–607. [Google Scholar] [CrossRef]
- Kudira, R.; Malinka, T.; Kohler, A.; Dosch, M.; de Aguero, M.G.; Melin, N.; Haegele, S.; Starlinger, P.; Maharjan, N.; Saxena, S.; et al. P2X1-regulated IL-22 secretion by innate lymphoid cells is required for efficient liver regeneration. Hepatology 2016, 63, 2004–2017. [Google Scholar] [CrossRef] [Green Version]
- Weber, G.F.; Gaertner, F.C.; Erl, W.; Janssen, K.P.; Blechert, B.; Holzmann, B.; Weighardt, H.; Essler, M. IL-22-mediated tumor growth reduction correlates with inhibition of ERK1/2 and AKT phosphorylation and induction of cell cycle arrest in the G2-M phase. J. Immunol. 2006, 177, 8266–8272. [Google Scholar] [CrossRef] [Green Version]
- Kirchberger, S.; Royston, D.J.; Boulard, O.; Thornton, E.; Franchini, F.; Szabady, R.L.; Harrison, O.; Powrie, F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 2013, 210, 917–931. [Google Scholar] [CrossRef]
- Kryczek, I.; Lin, Y.; Nagarsheth, N.; Peng, D.; Zhao, L.; Zhao, E.; Vatan, L.; Szeliga, W.; Dou, Y.; Owens, S.; et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 2014, 40, 772–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.H.; Cao, Y.F.; Jiang, Z.Y.; Zhang, S.; Gao, F. Th22 cell accumulation is associated with colorectal cancer development. World J. Gastroenterol. 2015, 21, 4216–4224. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gong, G.; Sheh, A.; Muthupalani, S.; Bryant, E.M.; Puglisi, D.A.; Holcombe, H.; Conaway, E.A.; Parry, N.A.P.; Bakthavatchalu, V.; et al. Interleukin-22 drives nitric oxide-dependent DNA damage and dysplasia in a murine model of colitis-associated cancer. Mucosal Immunol. 2017, 10, 1504–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudloff, I.; Jarde, T.; Bachmann, M.; Elgass, K.D.; Kerr, G.; Engel, R.; Richards, E.; Oliva, K.; Wilkins, S.; McMurrick, P.J.; et al. Molecular signature of interleukin-22 in colon carcinoma cells and organoid models. Transl. Res. 2020, 216, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, G.; Kim, J.Y.; Yun, H.J.; Lim, S.C.; Choi, H.S. Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis 2014, 35, 1352–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigt, C.; May, P.; Gottschlich, A.; Markota, A.; Wenk, D.; Gerlach, I.; Voigt, S.; Stathopoulos, G.T.; Arendt, K.A.M.; Heise, C.; et al. Cancer cells induce interleukin-22 production from memory CD4(+) T cells via interleukin-1 to promote tumor growth. Proc. Natl. Acad. Sci. USA 2017, 114, 12994–12999. [Google Scholar] [CrossRef] [Green Version]
- Katara, G.K.; Kulshrestha, A.; Schneiderman, S.; Riehl, V.; Ibrahim, S.; Beaman, K.D. Interleukin-22 promotes development of malignant lesions in a mouse model of spontaneous breast cancer. Mol. Oncol. 2020, 14, 211–224. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.; Gao, J.; Shao, S.; Cui, Y.; Yin, S.; Pan, B. IL-22 promotes tumor growth of breast cancer cells in mice. Aging 2020, 12, 13354–13364. [Google Scholar] [CrossRef]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019, 176, 998–1013. [Google Scholar] [CrossRef] [Green Version]
- Khosravi, N.; Caetano, M.S.; Cumpian, A.M.; Unver, N.; De la Garza Ramos, C.; Noble, O.; Daliri, S.; Hernandez, B.J.; Gutierrez, B.A.; Evans, S.E.; et al. IL22 Promotes Kras-Mutant Lung Cancer by Induction of a Protumor Immune Response and Protection of Stemness Properties. Cancer Immunol. Res. 2018, 6, 788–797. [Google Scholar] [CrossRef]
- Park, O.; Wang, H.; Weng, H.; Feigenbaum, L.; Li, H.; Yin, S.; Ki, S.H.; Yoo, S.H.; Dooley, S.; Wang, F.S.; et al. In vivo consequences of liver-specific interleukin-22 expression in mice: Implications for human liver disease progression. Hepatology 2011, 54, 252–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, R.; Tan, Z.; Deng, L.; Chen, Y.; Xia, Y.; Gao, Y.; Wang, X.; Sun, B. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology 2011, 54, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, Y.; Wang, F.; Zhu, Z.; Gao, Y.; Zhang, Q.; Du, Z. Interleukin 22 is related to development and poor prognosis of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2020, 44, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, O.; Kronenberger, B.; Scheiermann, P.; Koberle, V.; Muhl, H.; Piiper, A. Interleukin-22 serum levels are a negative prognostic indicator in patients with hepatocellular carcinoma. Hepatology 2014, 59, 1207. [Google Scholar] [CrossRef]
- Kuang, D.M.; Xiao, X.; Zhao, Q.; Chen, M.M.; Li, X.F.; Liu, R.X.; Wei, Y.; Ouyang, F.Z.; Chen, D.P.; Wu, Y.; et al. B7-H1-expressing antigen-presenting cells mediate polarization of protumorigenic Th22 subsets. J. Clin. Investig. 2014, 124, 4657–4667. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Tuteja, G.; Schug, J.; Kaestner, K.H. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 2012, 148, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Wangensteen, K.J.; Teta-Bissett, M.; Wang, Y.J.; Mosleh-Shirazi, E.; Buza, E.L.; Greenbaum, L.E.; Kaestner, K.H. Genetic lineage tracing analysis of the cell of origin of hepatotoxin-induced liver tumors in mice. Hepatology 2016, 64, 1163–1177. [Google Scholar] [CrossRef] [Green Version]
- Kishida, N.; Matsuda, S.; Itano, O.; Shinoda, M.; Kitago, M.; Yagi, H.; Abe, Y.; Hibi, T.; Masugi, Y.; Aiura, K.; et al. Development of a novel mouse model of hepatocellular carcinoma with nonalcoholic steatohepatitis using a high-fat, choline-deficient diet and intraperitoneal injection of diethylnitrosamine. BMC Gastroenterol. 2016, 16, 61. [Google Scholar] [CrossRef] [Green Version]
- Charni-Natan, M.; Goldstein, I. Protocol for Primary Mouse Hepatocyte Isolation. STAR Protoc. 2020, 1, 100086. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, S.; Ma, S.; Huang, X.; Lu, D.; Zhou, Y.; Jiang, H. Th22 cells are associated with hepatocellular carcinoma development and progression. Chin. J. Cancer Res. 2014, 26, 135–141. [Google Scholar] [CrossRef]
- Lejeune, D.; Dumoutier, L.; Constantinescu, S.; Kruijer, W.; Schuringa, J.J.; Renauld, J.C. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. Pathways that are shared with and distinct from IL-10. J. Biol. Chem. 2002, 277, 33676–33682. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Zhao, J.; Bulek, K.; Tang, F.; Chen, X.; Cai, G.; Jia, S.; Fox, P.L.; Huang, E.; Pizarro, T.T.; et al. Inflammation mobilizes copper metabolism to promote colon tumorigenesis via an IL-17-STEAP4-XIAP axis. Nat. Commun. 2020, 11, 900. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Bredell, B.X.; Anderson, E.R.; Martin, A.; Mays, C.; Nagao-Kitamoto, H.; Huang, S.; Gyorffy, B.; Greenson, J.K.; Hardiman, K.; et al. Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9608–E9617. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Wang, Y.; Xu, X.; Sun, H.; Tang, W. STEAP4 promoter methylation correlates with tumorigenesis of hepatocellular carcinoma. Pathol. Res. Pract. 2022, 233, 153870. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannou, A.D.; Lücke, J.; Kleinschmidt, D.; Shiri, A.M.; Steglich, B.; Nawrocki, M.; Zhang, T.; Zazara, D.E.; Kempski, J.; Zhao, L.; et al. A Critical Role of the IL-22–IL-22 Binding Protein Axis in Hepatocellular Carcinoma. Cancers 2022, 14, 6019. https://doi.org/10.3390/cancers14246019
Giannou AD, Lücke J, Kleinschmidt D, Shiri AM, Steglich B, Nawrocki M, Zhang T, Zazara DE, Kempski J, Zhao L, et al. A Critical Role of the IL-22–IL-22 Binding Protein Axis in Hepatocellular Carcinoma. Cancers. 2022; 14(24):6019. https://doi.org/10.3390/cancers14246019
Chicago/Turabian StyleGiannou, Anastasios D., Jöran Lücke, Dörte Kleinschmidt, Ahmad Mustafa Shiri, Babett Steglich, Mikolaj Nawrocki, Tao Zhang, Dimitra E. Zazara, Jan Kempski, Lilan Zhao, and et al. 2022. "A Critical Role of the IL-22–IL-22 Binding Protein Axis in Hepatocellular Carcinoma" Cancers 14, no. 24: 6019. https://doi.org/10.3390/cancers14246019
APA StyleGiannou, A. D., Lücke, J., Kleinschmidt, D., Shiri, A. M., Steglich, B., Nawrocki, M., Zhang, T., Zazara, D. E., Kempski, J., Zhao, L., Giannou, O., Agalioti, T., Brockmann, L., Bertram, F., Sabihi, M., Böttcher, M., Ewald, F., Schulze, K., von Felden, J., ... Huber, S. (2022). A Critical Role of the IL-22–IL-22 Binding Protein Axis in Hepatocellular Carcinoma. Cancers, 14(24), 6019. https://doi.org/10.3390/cancers14246019







