Tumoricidal, Temozolomide- and Radiation-Sensitizing Effects of KCa3.1 K+ Channel Targeting In Vitro Are Dependent on Glioma Cell Line and Stem Cell Fraction
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Cell Culture
2.2. RNA Isolation and qPCR
2.3. Drug Treatment
2.4. Ionizing Radiation
2.5. Patch-Clamp on-Cell Recording
2.6. Clonogenic Survival
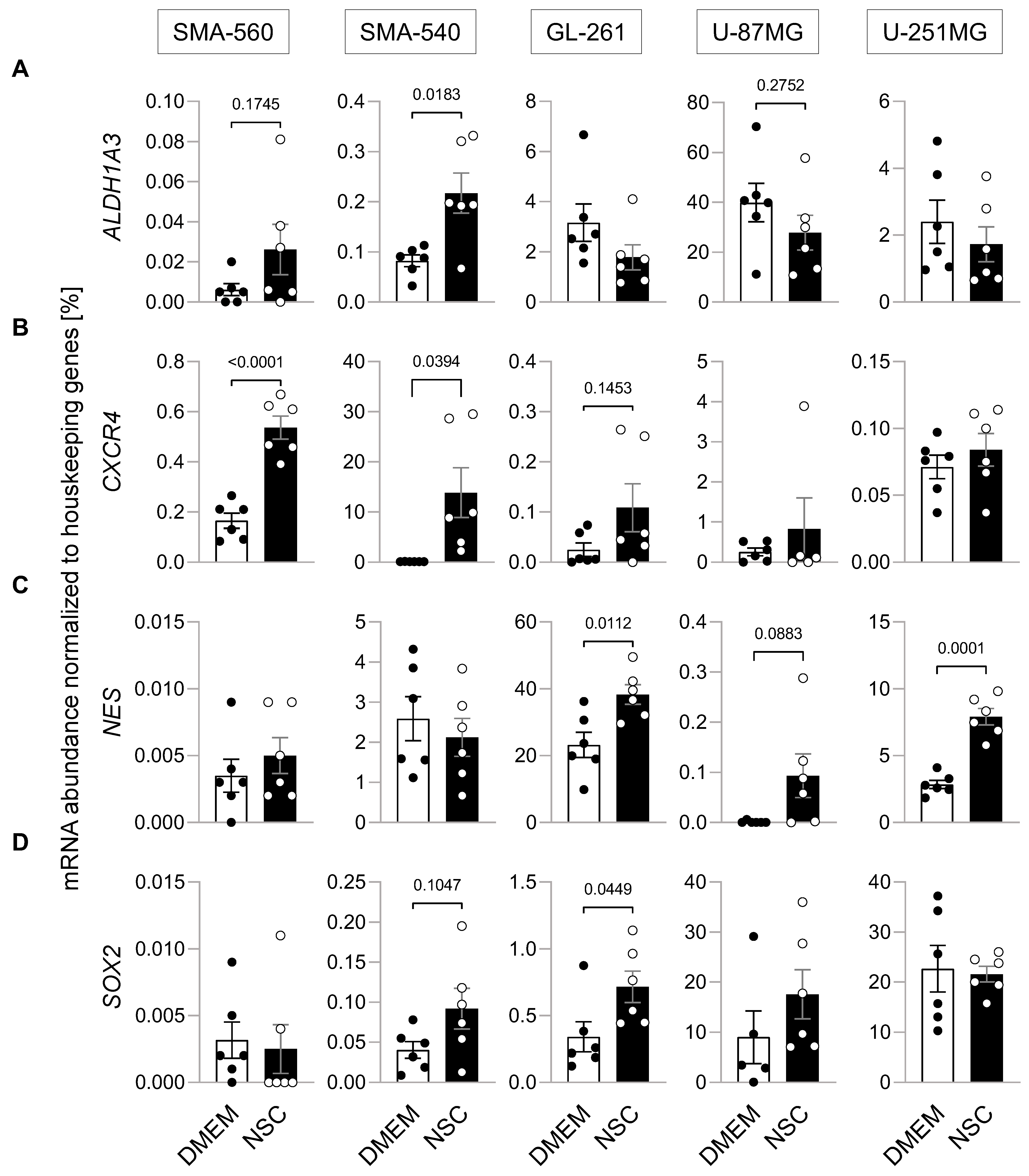
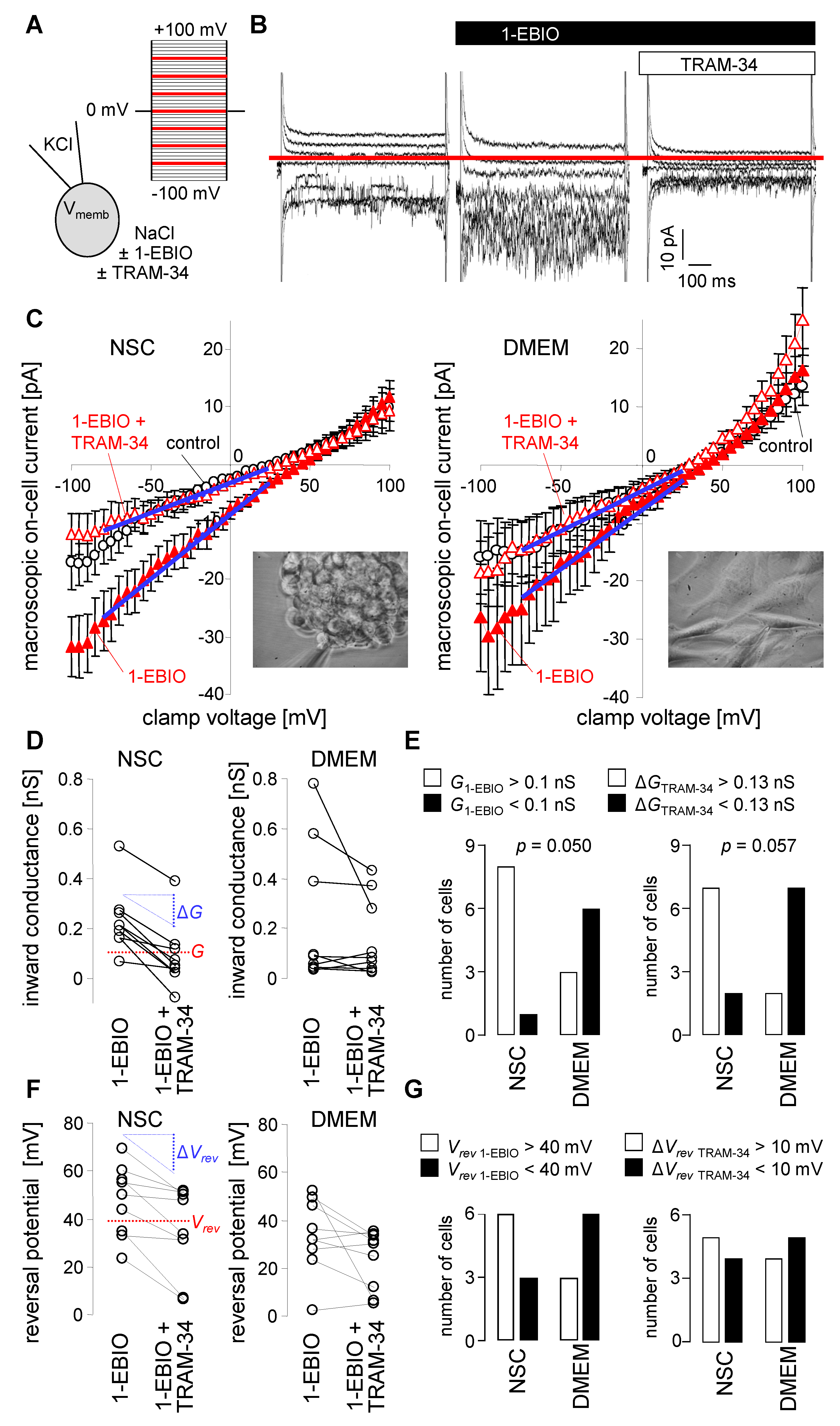
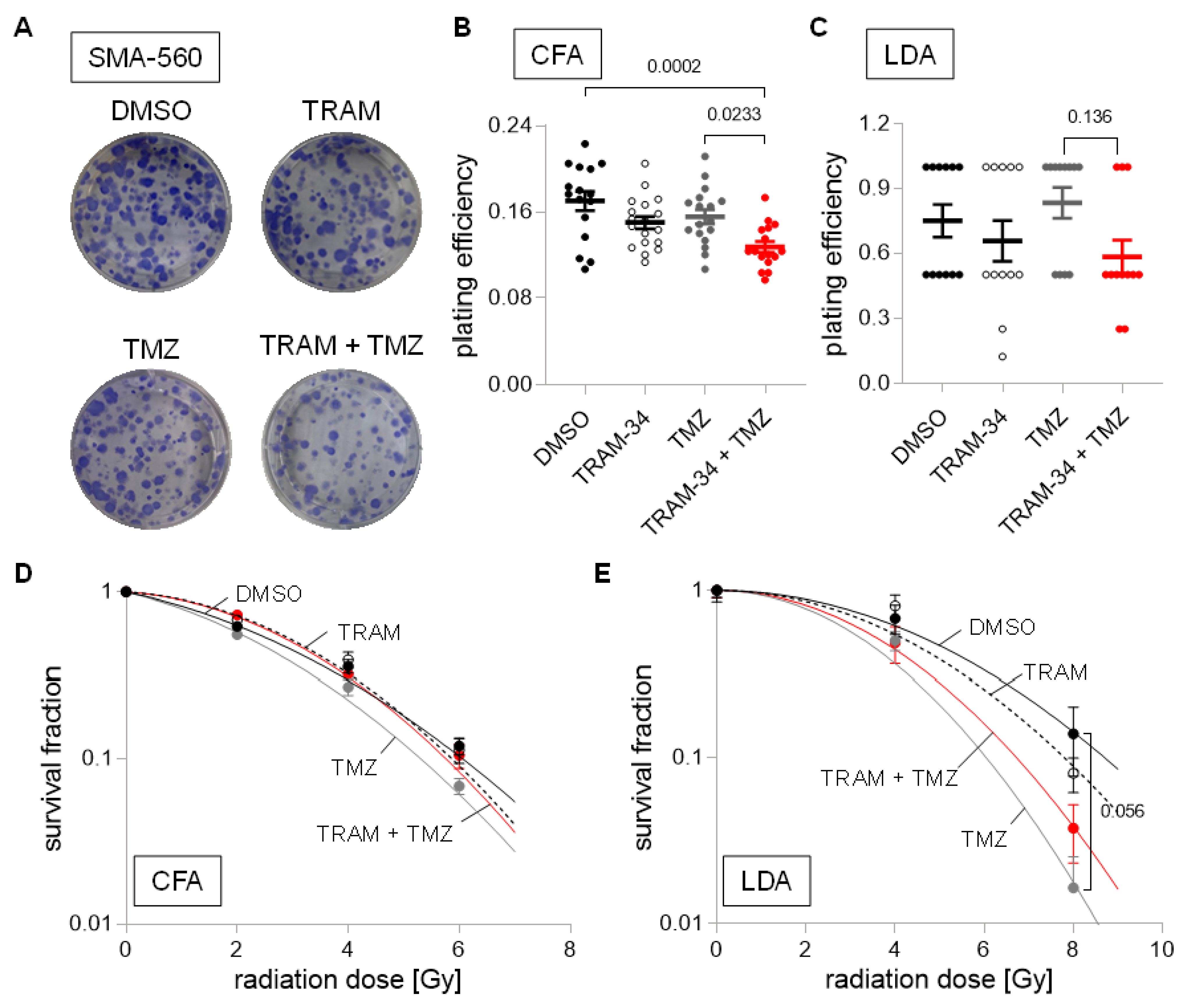
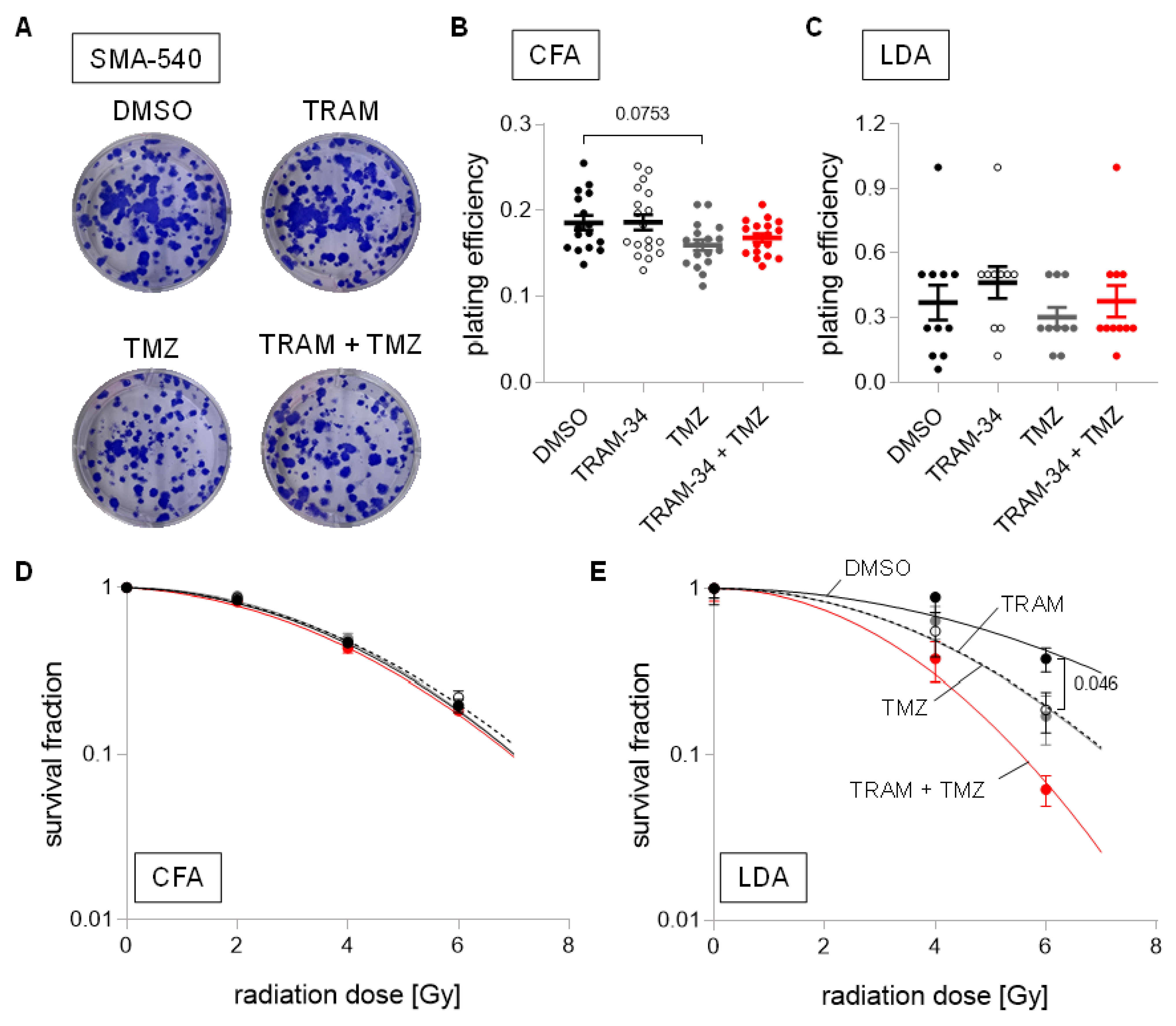
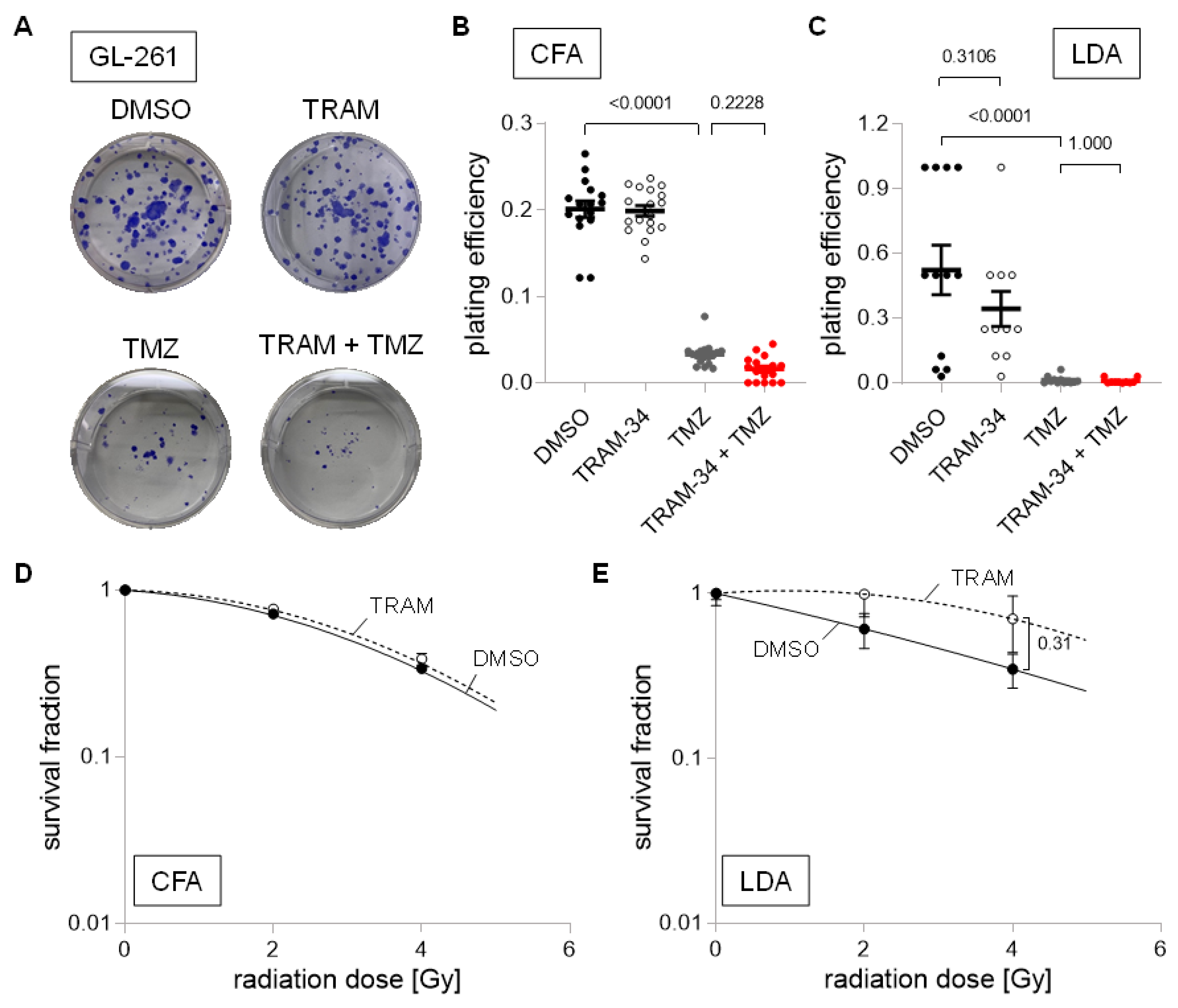
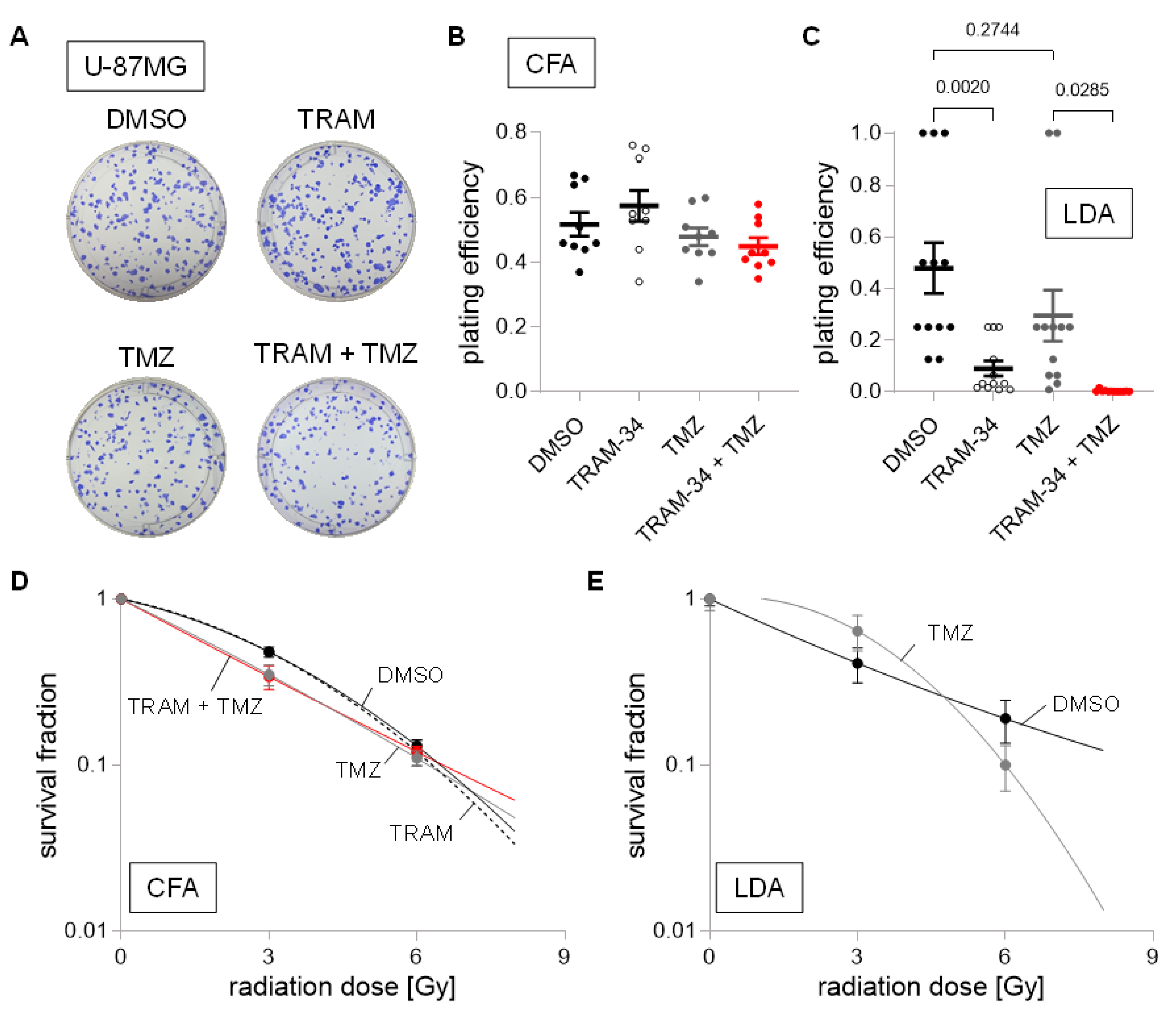
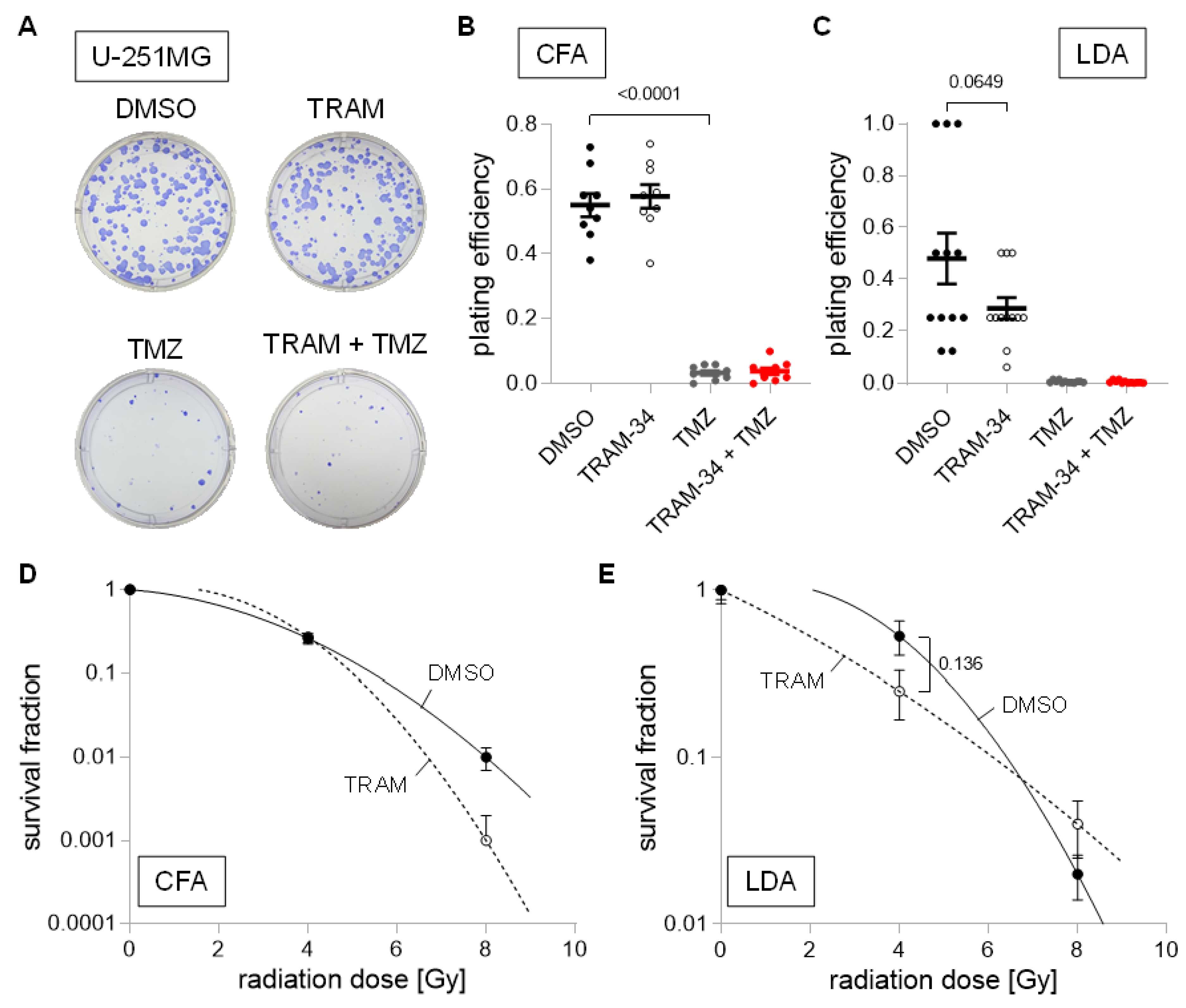
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| CFA | colony formation assay |
| DMEM | Dulbecco’s modified eagle medium |
| DMSO | dimethylsulfoxide |
| FBS | fetal Bovine Serum |
| LDA | limited dilution assay |
| NSC | neural stem cell inducing medium |
| qPCR | quantitative Polymerase Chain Reaction |
| RT | reverse transcriptase |
| TMZ | temozolomide |
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro.-Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs. Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef] [Green Version]
- Klumpp, L.; Sezgin, E.C.; Skardelly, M.; Eckert, F.; Huber, S.M. KCa3.1 Channels and Glioblastoma: In Vitro Studies. Curr. Neuropharmacol. 2018, 16, 627–635. [Google Scholar] [CrossRef]
- Catacuzzeno, L.; Sforna, L.; Esposito, V.; Limatola, C.; Franciolini, F. Ion Channels in Glioma Malignancy. In Transportome Malfunction in the Cancer Spectrum: Ion Transport in Tumor Biology; Stock, C., Pardo, L.A., Eds.; Springer International Publishing: Cham, Germany, 2021; pp. 223–267. [Google Scholar]
- Brown, B.M.; Pressley, B.; Wulff, H. KCa3.1 Channel Modulators as Potential Therapeutic Compounds for Glioblastoma. Curr. Neuropharmacol. 2018, 16, 618–626. [Google Scholar] [CrossRef]
- Ferreira, R.; Lively, S.; Schlichter, L.C. IL-4 type 1 receptor signaling up-regulates KCNN4 expression, and increases the KCa3.1 current and its contribution to migration of alternative-activated microglia. Front. Cell Neurosci. 2014, 8, 183. [Google Scholar] [CrossRef]
- Wei, T.; Yi, M.; Gu, W.; Hou, L.; Lu, Q.; Yu, Z.; Chen, H. The Potassium Channel KCa3.1 Represents a Valid Pharmacological Target for Astrogliosis-Induced Neuronal Impairment in a Mouse Model of Alzheimer’s Disease. Front. Pharm. 2016, 7, 528. [Google Scholar] [CrossRef] [Green Version]
- Faragó, N.; Kocsis, Á.K.; Braskó, C.; Lovas, S.; Rózsa, M.; Baka, J.; Kovács, B.; Mikite, K.; Szemenyei, V.; Molnár, G.; et al. Human neuronal changes in brain edema and increased intracranial pressure. Acta Neuropathol. Commun. 2016, 4, 78. [Google Scholar] [CrossRef] [Green Version]
- Ruggieri, P.; Mangino, G.; Fioretti, B.; Catacuzzeno, L.; Puca, R.; Ponti, D.; Miscusi, M.; Franciolini, F.; Ragona, G.; Calogero, A. The inhibition of KCa3.1 channels activity reduces cell motility in glioblastoma derived cancer stem cells. PLoS ONE 2012, 7, e47825. [Google Scholar] [CrossRef]
- Jäger, H.; Dreker, T.; Buck, A.; Giehl, K.; Gress, T.; Grissmer, S. Blockage of intermediate-conductance Ca2+-activated K+ channels inhibit human pancreatic cancer cell growth in vitro. Mol. Pharm. 2004, 65, 630–638. [Google Scholar] [CrossRef] [Green Version]
- Lallet-Daher, H.; Roudbaraki, M.; Bavencoffe, A.; Mariot, P.; Gackière, F.; Bidaux, G.; Urbain, R.; Gosset, P.; Delcourt, P.; Fleurisse, L.; et al. Intermediate-conductance Ca2+-activated K+ channels (IKCa1) regulate human prostate cancer cell proliferation through a close control of calcium entry. Oncogene 2009, 28, 1792–1806. [Google Scholar] [CrossRef]
- Cuddapah, V.A.; Turner, K.L.; Seifert, S.; Sontheimer, H. Bradykinin-induced chemotaxis of human gliomas requires the activation of KCa3.1 and ClC-3. J. Neurosci. 2013, 33, 1427–1440. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, G.; Catalano, M.; Sciaccaluga, M.; Chece, G.; Cipriani, R.; Rosito, M.; Grimaldi, A.; Lauro, C.; Cantore, G.; Santoro, A.; et al. KCa3.1 channels are involved in the infiltrative behavior of glioblastoma in vivo. Cell Death. Dis. 2013, 4, e773. [Google Scholar] [CrossRef] [Green Version]
- Turner, K.L.; Honasoge, A.; Robert, S.M.; McFerrin, M.M.; Sontheimer, H. A proinvasive role for the Ca(2+)-activated K(+) channel KCa3.1 in malignant glioma. Glia 2014, 62, 971–981. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, G.; Monaco, L.; Catacuzzeno, L.; Antonangeli, F.; Santoro, A.; Esposito, V.; Franciolini, F.; Wulff, H.; Limatola, C. Radiation Increases Functional KCa3.1 Expression and Invasiveness in Glioblastoma. Cancers 2019, 11, 279. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, G.; Grimaldi, A.; Chece, G.; Porzia, A.; Esposito, V.; Santoro, A.; Salvati, M.; Mainiero, F.; Ragozzino, D.; Di Angelantonio, S.; et al. KCa3.1 channel inhibition sensitizes malignant gliomas to temozolomide treatment. Oncotarget 2016, 7, 30781–30796. [Google Scholar] [CrossRef]
- Stegen, B.; Butz, L.; Klumpp, L.; Zips, D.; Dittmann, K.; Ruth, P.; Huber, S.M. Ca2+-Activated IK K+ Channel Blockade Radiosensitizes Glioblastoma Cells. Mol. Cancer Res. 2015, 13, 1283–1295. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.C.; Sheltzer, J.M. Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep. 2022, 38, 110569. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [Green Version]
- Serano, R.D.; Pegram, C.N.; Bigner, D.D. Tumorigenic cell culture lines from a spontaneous VM/Dk murine astrocytoma (SMA). Acta Neuropathol. 1980, 51, 53–64. [Google Scholar] [CrossRef]
- Pilkington, G.J.; Darling, J.L.; Lantos, P.L.; Thomas, D.G.T. Cell lines (VMDk) derived from a spontaneous murine astrocytoma: Morphological and immunocytochemical characterization. J. Neurol. Sci. 1983, 62, 115–139. [Google Scholar] [CrossRef]
- Ausman, J.I.; Shapiro, W.R.; Rall, D.P. Studies on the Chemotherapy of Experimental Brain Tumors: Development of an Experimental Model. Cancer Res. 1970, 30, 2394–2400. [Google Scholar]
- Pontén, J.; Macintyre, E.H. Long term culture of normal and neoplastic human glia. Acta Pathol. Microbiol. Scand. 1968, 74, 465–486. [Google Scholar] [CrossRef]
- Allen, M.; Bjerke, M.; Edlund, H.; Nelander, S.; Westermark, B. Origin of the U87MG glioma cell line: Good news and bad news. Sci. Transl. Med. 2016, 8, 354re3. [Google Scholar] [CrossRef]
- Torsvik, A.; Stieber, D.; Enger, P.; Golebiewska, A.; Molven, A.; Svendsen, A.; Westermark, B.; Niclou, S.P.; Olsen, T.K.; Enger, M.C.; et al. U-251 revisited: Genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med. 2014, 3, 812–824. [Google Scholar] [CrossRef]
- Lazic, S.E.; Clarke-Williams, C.J.; Munafò, M.R. What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol. 2018, 16, e2005282. [Google Scholar] [CrossRef] [Green Version]
- McMahon, S.J. The linear quadratic model: Usage, interpretation and challenges. Phys. Med. Amp. Biol. 2018, 64, 01TR01. [Google Scholar] [CrossRef]
- Huang, L.; Xu, S.; Hu, D.; Lu, W.; Xie, X.; Cheng, X. IQGAP1 is Involved in Enhanced Aggressive Behavior of Epithelial Ovarian Cancer Stem Cell-Like Cells During Differentiation. Int. J. Gynecol. Cancer 2015, 25, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Quiñones-Hinojosa, A.; Sanai, N.; Gonzalez-Perez, O.; Garcia-Verdugo, J.M. The human brain subventricular zone: Stem cells in this niche and its organization. Neurosurg. Clin. N Am. 2007, 18, 15–20, vii. [Google Scholar] [CrossRef]
- Soehngen, E.; Schaefer, A.; Koeritzer, J.; Huelsmeyer, V.; Zimmer, C.; Ringel, F.; Gempt, J.; Schlegel, J. Hypoxia upregulates aldehyde dehydrogenase isoform 1 (ALDH1) expression and induces functional stem cell characteristics in human glioblastoma cells. Brain. Tumor. Pathol. 2014, 31, 247–256. [Google Scholar] [CrossRef]
- Gatti, M.; Pattarozzi, A.; Bajetto, A.; Würth, R.; Daga, A.; Fiaschi, P.; Zona, G.; Florio, T.; Barbieri, F. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology 2013, 314, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jin, X.; Jung, J.-E.; Beck, S.; Kim, H. Cell surface Nestin is a biomarker for glioma stem cells. Biochem. Biophys. Res. Commun. 2013, 433, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Behnan, J.; Stangeland, B.; Hosainey, S.A.M.; Joel, M.; Olsen, T.K.; Micci, F.; Glover, J.C.; Isakson, P.; Brinchmann, J.E. Differential propagation of stroma and cancer stem cells dictates tumorigenesis and multipotency. Oncogene 2017, 36, 570–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.J.; Raman, G.; Bodendiek, S.; O’Donnell, M.E.; Wulff, H. The KCa3.1 blocker TRAM-34 reduces infarction and neurological deficit in a rat model of ischemia/reperfusion stroke. J. Cereb. Blood. Flow. Metab. 2011, 31, 2363–2374. [Google Scholar] [CrossRef] [Green Version]
- Wulff, H.; Miller, M.J.; Hansel, W.; Grissmer, S.; Cahalan, M.D.; Chandy, K.G. Design of a potent and selective inhibitor of the intermediate-conductance Ca2+-activated K+ channel, IKCa1: A potential immunosuppressant. Proc. Natl. Acad. Sci. USA 2000, 97, 8151–8156. [Google Scholar] [CrossRef] [Green Version]
- Sesen, J.; Dahan, P.; Scotland, S.J.; Saland, E.; Dang, V.T.; Lemarié, A.; Tyler, B.M.; Brem, H.; Toulas, C.; Moyal, E.C.-J.; et al. Metformin inhibits growth of human glioblastoma cells and enhances therapeutic response. PLoS ONE 2015, 10, e0123721. [Google Scholar] [CrossRef] [Green Version]
- McFaline-Figueroa, J.L.; Braun, C.J.; Stanciu, M.; Nagel, Z.D.; Mazzucato, P.; Sangaraju, D.; Cerniauskas, E.; Barford, K.; Vargas, A.; Chen, Y.; et al. Minor Changes in Expression of the Mismatch Repair Protein MSH2 Exert a Major Impact on Glioblastoma Response to Temozolomide. Cancer Res. 2015, 75, 3127–3138. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Gueble, S.E.; Sundaram, R.K.; Huseman, E.D.; Bindra, R.S.; Herzon, S.B. Mechanism-based design of agents that selectively target drug-resistant glioma. Science 2022, 377, 502–511. [Google Scholar] [CrossRef]
- Yi, G.Z.; Huang, G.; Guo, M.; Zhang, X.; Wang, H.; Deng, S.; Li, Y.; Xiang, W.; Chen, Z.; Pan, J.; et al. Acquired temozolomide resistance in MGMT-deficient glioblastoma cells is associated with regulation of DNA repair by DHC2. Brain 2019, 142, 2352–2366. [Google Scholar] [CrossRef] [Green Version]
- Stegen, B.; Klumpp, L.; Misovic, M.; Edalat, L.; Eckert, M.; Klumpp, D.; Ruth, P.; Huber, S.M. K+ channel signaling in irradiated tumor cells. Eur. Biophys. J. 2016, 45, 585–598. [Google Scholar] [CrossRef]
- Mohr, C.J.; Gross, D.; Sezgin, E.C.; Steudel, F.A.; Ruth, P.; Huber, S.M.; Lukowski, R. K(Ca)3.1 Channels Confer Radioresistance to Breast Cancer Cells. Cancers 2019, 11, 1285. [Google Scholar] [CrossRef]
- Ganser, K.; Eckert, F.; Riedel, A.; Stransky, N.; Paulsen, F.; Noell, S.; Krueger, M.; Schittenhelm, J.; Beck-Wödl, S.; Zips, D.; et al. Patient-individual phenotypes of glioblastoma stem cells are conserved in culture and associate with radioresistance, brain infiltration and patient prognosis. Int. J. Cancer 2022, 150, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Mao, P.; Joshi, K.; Li, J.; Kim, S.-H.; Li, P.; Santana-Santos, L.; Luthra, S.; Chandran, U.R.; Benos, P.V.; Smith, L.; et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. USA 2013, 110, 8644–8649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weichselbaum, R.R.; Liang, H.; Deng, L.; Fu, Y.-X. Radiotherapy and immunotherapy: A beneficial liaison? Nat. Rev. Clin. Oncol. 2017, 14, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; D’Alessandro, G.; Golia, M.T.; Grössinger, E.M.; Di Angelantonio, S.; Ragozzino, D.; Santoro, A.; Esposito, V.; Wulff, H.; Catalano, M.; et al. KCa3.1 inhibition switches the phenotype of glioma-infiltrating microglia/macrophages. Cell Death Dis. 2016, 7, e2174. [Google Scholar] [CrossRef] [Green Version]
- Becker, A.P.; Sells, B.E.; Haque, S.J.; Chakravarti, A. Tumor Heterogeneity in Glioblastomas: From Light Microscopy to Molecular Pathology. Cancers. 2021, 13, 761. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [Green Version]
- Brada, M.; Judson, I.; Beale, P.; Moore, S.; Reidenberg, P.; Statkevich, P.; Dugan, M.; Batra, V.; Cutler, D. Phase I dose-escalation and pharmacokinetic study of temozolomide (SCH 52365) for refractory or relapsing malignancies. Br. J. Cancer 1999, 81, 1022–1030. [Google Scholar] [CrossRef]
- Rosso, L.; Brock, C.S.; Gallo, J.M.; Saleem, A.; Price, P.M.; Turkheimer, F.E.; Aboagye, E.O. A New Model for Prediction of Drug Distribution in Tumor and Normal Tissues: Pharmacokinetics of Temozolomide in Glioma Patients. Cancer Res. 2008, 69, 120–127. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stransky, N.; Ganser, K.; Naumann, U.; Huber, S.M.; Ruth, P. Tumoricidal, Temozolomide- and Radiation-Sensitizing Effects of KCa3.1 K+ Channel Targeting In Vitro Are Dependent on Glioma Cell Line and Stem Cell Fraction. Cancers 2022, 14, 6199. https://doi.org/10.3390/cancers14246199
Stransky N, Ganser K, Naumann U, Huber SM, Ruth P. Tumoricidal, Temozolomide- and Radiation-Sensitizing Effects of KCa3.1 K+ Channel Targeting In Vitro Are Dependent on Glioma Cell Line and Stem Cell Fraction. Cancers. 2022; 14(24):6199. https://doi.org/10.3390/cancers14246199
Chicago/Turabian StyleStransky, Nicolai, Katrin Ganser, Ulrike Naumann, Stephan M. Huber, and Peter Ruth. 2022. "Tumoricidal, Temozolomide- and Radiation-Sensitizing Effects of KCa3.1 K+ Channel Targeting In Vitro Are Dependent on Glioma Cell Line and Stem Cell Fraction" Cancers 14, no. 24: 6199. https://doi.org/10.3390/cancers14246199
APA StyleStransky, N., Ganser, K., Naumann, U., Huber, S. M., & Ruth, P. (2022). Tumoricidal, Temozolomide- and Radiation-Sensitizing Effects of KCa3.1 K+ Channel Targeting In Vitro Are Dependent on Glioma Cell Line and Stem Cell Fraction. Cancers, 14(24), 6199. https://doi.org/10.3390/cancers14246199







